Abstract
BH3 mimetics are small molecules designed or discovered to mimic the binding of BH3-only proteins to the hydrophobic groove of antiapoptotic BCL2 proteins. The selectivity of these molecules for BCL2, BCL-XL, or MCL1 has been established in vitro; whether they inhibit these proteins in cells has not been rigorously investigated. In this study, we used a panel of leukemia cell lines to assess the ability of seven putative BH3 mimetics to inhibit antiapoptotic proteins in a cell-based system. We show that ABT-737 is the only BH3 mimetic that inhibits BCL2 as assessed by displacement of BAD and BIM from BCL2. The other six BH3 mimetics activate the endoplasmic reticulum stress response inducing ATF4, ATF3, and NOXA, which can then bind to and inhibit MCL1. In most cancer cells, inhibition of one antiapoptotic protein does not acutely induce apoptosis. However, by combining two BH3 mimetics, one that inhibits BCL2 and one that induces NOXA, apoptosis is induced within 6 h in a BAX/BAK-dependent manner. Because MCL1 is a major mechanism of resistance to ABT-737, these results suggest a novel strategy to overcome this resistance. Our findings highlight a novel signaling pathway through which many BH3 mimetics inhibit MCL1 and suggest the potential use of these agents as adjuvants in combination with various chemotherapy strategies.
Keywords: Anticancer Drug, Apoptosis, ER Stress, Leukemia, Microtubules, BCL2, BH3 Mimetics, MCL1, Vinblastine
Introduction
Proteins of the BCL2 family have been implicated in both tumor development and the resistance of tumors to anticancer therapies (1). Members of this family are functionally defined as either antiapoptotic or proapoptotic, based on their ability to either inhibit or induce the release of cytochrome c from the mitochondria. The BCL2 family of proteins share sequence homology in up to four conserved regions termed the BCL2 homology (BH)2 domains (BH1–BH4) (2). Most antiapoptotic proteins, including BCL2, BCL-XL, and MCL1, contain all four BH domains and sequester proapoptotic proteins via protein-protein interaction. Their BH1–BH3 domains form a hydrophobic groove to which the BH3 domain of a proapoptotic protein binds. Once sequestered, proapoptotic proteins are prevented from exerting their apoptotic function within the cell. Proapoptotic proteins can be further divided into two groups: 1) BAX and BAK multi-domain proteins and 2) BH3-only proteins. BH3-only proteins respond to environmental cues and activate apoptosis through BAX and BAK. Activated BAX and BAK oligomerize and permeabilize the mitochondrial outer membrane, an event critical for the release of cytochrome c into the cytosol and subsequent caspase activation. Once activated, caspases can cleave hundreds of cellular proteins, including poly(ADP-ribose) polymerase (PARP), resulting in the hallmark morphological characteristics of apoptosis such as chromatin condensation, cellular membrane blebbing, and cell shrinkage. Because of the critical role BAX and BAK play in permeabilizing the mitochondrial outer membrane, the mitochondrial apoptotic pathway is traditionally considered to be BAX/BAK-dependent.
The evasion of apoptosis has been recognized as one of the hallmarks of cancer (3), making pharmacological inhibition of antiapoptotic proteins a potential strategy to induce apoptosis in cancer cells. We recently demonstrated that the anticancer drug vinblastine can induce rapid, cell cycle phase-independent apoptosis in the ML-1 human myeloblastic leukemia cell line when MCL1 expression is suppressed (4). Because the majority of leukemia and lymphoma cell lines did not appear to rely on MCL1, we hypothesized that BCL2 and/or BCL-XL might protect them from this acute vinblastine-mediated apoptosis. Accordingly, in the current study, we assessed the impact of small molecule inhibitors of the BCL2 family on the response of leukemia cells to vinblastine. Initial results seriously questioned the reported targets for some of these inhibitors, thereby leading us to investigate their targets in intact cells.
Several small molecules, including ABT-737 and GX15-070, have been designed to mimic the binding of BH3-only proteins to the hydrophobic groove of antiapoptotic proteins (5, 6). The binding specificity of ABT-737 has been extensively studied via competitive fluorescence polarization assays and the use of recombinant BCL2 family proteins. ABT-737 preferentially binds BCL2 and BCL-XL but not MCL1 (7–9) and as a consequence displaces any proapoptotic proteins that are bound (10). It is generally believed that specific inhibitors of antiapoptotic BCL2 proteins should induce apoptosis in a BAX/BAK-dependent manner. Although ABT-737 does kill cells in a BAX/BAK-dependent manner, many other purported BH3 mimetics have been shown to kill cells in a BAX/BAK-independent manner, leading to the conclusion that they function through alternative undefined targets (8, 11).
The selectivity of these BH3 mimetics for BCL2, BCL-XL, or MCL1 has been established in vitro; whether they inhibit these proteins in cells has not been rigorously investigated. In this study, we assessed the ability of seven putative BH3 mimetics to affect the interaction of BCL2 family members in a cell-based system. We show that ABT-737 is the only BH3 mimetic that inhibits BCL2 in cells, whereas all of the other BH3 mimetics elevate NOXA to antagonize MCL1. As a consequence, dual inhibition of BCL2 and MCL1 induces BAX/BAK-dependent apoptosis as rapidly as 6 h, suggesting the potential use of these combinations to overcome cellular resistance to ABT-737, which has been attributed to high levels of MCL1 (8).
EXPERIMENTAL PROCEDURES
Cell Culture
Sources of most of the cell lines have been described recently (4). JurkatΔBAK cells were kindly provided by Dr. Hanna Rabinowich (Pittsburgh, PA). Wild-type K562 and K562 cells expressing S peptide-tagged BCL2 were kindly provided by Dr. Scott Kaufmann (Rochester, MN). All of the cell lines were maintained in RPMI 1640 medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Isolation of Chronic Lymphocytic Leukemia Cells
Chronic lymphocytic leukemia (CLL) cells were obtained from consented patients at the Norris Cotton Cancer Center (Lebanon, NH), purified by centrifugation in Ficoll-Paque PLUS, and immediately incubated with drugs as described previously (4).
Quantification of Apoptosis
The cells were incubated with 2 μg/ml Hoechst 33342 for 20 min at 37 °C and visualized with a fluorescent microscope. The cells with condensed and fragmented chromatin were considered apoptotic and were quantified by scoring at least two hundred cells per sample. The data are expressed as the percentages of cells with chromatin condensation.
Digitonin Permeabilization
Separation of membrane fraction and supernatant was performed based on modifications of previously described methods (12, 13). Briefly, the cells were incubated with digitonin buffer (8.75 μg/106 cells digitonin, 75 mm NaCl, 1 mm NaH2PO4, 8 mm Na2HPO4, 250 mm sucrose, and protease/phosphatase inhibitor mixture) for 1 min followed by centrifugation for 1 min. The supernatant was removed and supplemented with an equal volume of 2× urea buffer. The pellet was resuspended in an equal volume of digitonin buffer and 2× urea buffer. The samples were subsequently boiled for 5 min prior to Western blot analysis.
Western Blot Analysis
The proteins from cell lysates were separated by SDS-PAGE and subjected to Western blot analysis as described previously (4). Antibodies are detailed in the supplemental information.
Immunoprecipitation
Lysates were prepared based on modifications of previously described methods (14–16). Briefly, the cells were lysed in 1% CHAPS lysis buffer (1% CHAPS, 150 mm NaCl, 10 mm HEPES, pH 7.4, and protease/phosphatase inhibitor) for 30 min. Lysates were passed through a 26.5-gauge needle and centrifuged to remove insoluble matter. The supernatant was incubated with rabbit anti-MCL1 (Santa Cruz, Santa Cruz, CA) followed by incubation with protein A/G-agarose beads (Santa Cruz). Immunoprecipitated proteins were pelleted and eluted by boiling in urea lysis buffer prior to Western blot analysis.
S Peptide Tag Pulldown
The cells were processed based on modifications of a previously described method (17). Briefly, K562 cells expressing S peptide-tagged BCL2 were lysed in pulldown buffer (1% CHAPS, 20 mm HEPES, pH 7.4, 150 mm NaCl, 1% glycerol, and protease/phosphatase inhibitor mixture inhibitors) for 30 min and centrifuged to remove insoluble matter. The supernatant was incubated with S protein-agarose (Novagen, Madison, WI) for 0–16 h, followed by three washes in pulldown buffer. The proteins bound to the beads were pelleted and boiled in urea lysis buffer prior to Western blot analysis.
Protease Activity Assay
Proteasome activity was measured in freshly prepared cell lysates using the Proteasome-GloTM cell-based assay (Promega, Madison, WI) according to the manufacturer's protocol. Luminescence was measured by LMAX II 384 (Molecular Devices, Silicon Valley, CA).
Quantitative Reverse Transcription-PCR
Total mRNA was extracted using the RNeasy Plus mini kit (Qiagen) and reverse transcribed using iScriptTM cDNA synthesis kit (Bio-Rad). The DNA was analyzed via quantitative PCR using the iQTM SYBR® Green Supermix (Bio-Rad) according to the manufacturer's protocol. The expression ratios for MCL1 and NOXA relative to GAPDH were calculated according to the equation of Pfaffl (18) using untreated cells as reference.
Microarray
Total mRNA was extracted using the RNeasy Plus mini kit (Qiagen). Reverse transcription using an oligo(dT) primer bearing a T7 promoter and the high yield Array ScriptTM reverse transcriptase were used to make double-stranded cDNA as a template for in vitro transcription with T7 RNA polymerase and the biotin-NTP mix. Labeled cRNA were purified and used for hybridization to the bead array for 16 h at 55 °C. Following hybridization, the bead arrays were washed and stained with streptavidin-Cy3 (GE Healthcare). Bead arrays with probes for all known human genes (Illumina, San Diego, CA) were used for RNA profiling. Fluorescent images were obtained with Illumina 500GX scanner and processed with BeadScan software (Illumina).
RESULTS
ABT-737 and GX15-070 Sensitize Leukemia Cells to Vinblastine
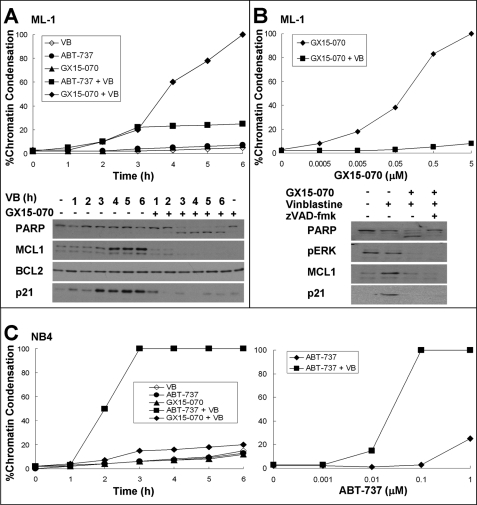
Vinblastine is a chemotherapeutic drug that dissociates microtubules and causes an accumulation of cells in mitosis prior to apoptosis. We have recently shown that several leukemia and lymphoma cell lines are acutely sensitive to vinblastine and die within 6 h from all phases of the cell cycle (4). In addition, a few cell lines, including the ML-1 leukemia, can be induced to undergo this rapid vinblastine-mediated apoptosis when the antiapoptotic MCL1 protein is pharmacologically or genetically repressed. We hypothesized that many other cell lines relied on BCL2 and/or BCL-XL for protection from acute vinblastine-mediated apoptosis. Accordingly, we tested the ability of the BH3 mimetics ABT-737 and GX15-070 to sensitize cells to vinblastine. ABT-737 does not inhibit MCL1, and consequently it did not sensitize ML-1 cells to vinblastine (Fig. 1A). NB4 human acute promyelocytic leukemia cells, on the other hand, were acutely sensitized to vinblastine with 100% of the cells dying within 3 h when combined with 0.1 μm ABT-737 (Fig. 1C). Because these cells express predominately BCL2 (4), this result can be attributed to the inhibition of BCL2. Many other leukemia and lymphoma cell lines, including U937, THP-1, and Jurkat, were not sensitized to vinblastine by ABT-737 (data not shown), but this could be due to concurrent expression of MCL1, which is a known mechanism of resistance to ABT-737 (5, 8, 9).
FIGURE 1.
Sensitization of leukemia cells to vinblastine by ABT-737 or GX15-070. A, ML-1 cells were incubated with 2.2 μm vinblastine (VB) with or without 0.1 μm ABT-737 or 5 μm GX15-070 for 0–6 h. The cells were stained with Hoechst 33342 and scored for apoptosis (top panel). Lysates from cells incubated with or without GX-15–070 were also analyzed for protein expression as indicated (bottom panel). B, ML-1 cells were incubated with 0–5 μm GX-15–070 for 6 h with or without 2.2 μm vinblastine and analyzed for apoptosis (top panel). Lysates from cells incubated with or without 2.2 μm vinblastine or 5 μm GX15-070 were analyzed for protein expression (bottom panel). The caspase inhibitor z-VAD-fmk was also added where indicated. C, NB4 cells were incubated with 2.2 μm vinblastine with or without 0.1 μm ABT-737 or 5 μm GX15-070 for 0–6 h. The cells were stained with Hoechst 33342 and scored for apoptosis (left panel). NB4 cells were incubated with 0–1 μm ABT-737 for 6 h with or without 2.2 μm vinblastine and analyzed for apoptosis (right panel).
GX15-070 has been reported to inhibit BCL2, BCL-XL, and MCL1 (6). We thus anticipated that such a pan-BCL2 inhibitor would overcome the protective effect of antiapoptotic BCL2 proteins when combined with vinblastine. Indeed, GX15-070 sensitized ML-1 cells to vinblastine (Fig. 1, A and B). However, it did not sensitize NB4 cells (Fig. 1C) or U937, THP-1, Jurkat, and many of the other leukemia cell lines tested (data not shown). These findings are inconsistent with the reported function of GX15-070 as a pan-BCL2 inhibitor.
Vinblastine induces a rapid induction of MCL1 in ML-1 cells, which consequently protects the cells from acute apoptosis. This induction is dependent on the MEK/ERK pathway such that the combination of vinblastine with the MEK inhibitor PD98059 prevents MCL1 induction resulting in acute apoptosis (4). GX15-070 prevented the vinblastine-mediated up-regulation of MCL1 and had no impact on BCL2 level (Fig. 1A). To determine whether this loss of MCL1 induction was a cause or consequence of dying cells, ML-1 cells were incubated with vinblastine and GX15-070, whereas caspase activity was inhibited by z-VAD-fmk (Fig. 1B). Even in the absence of caspase activity, vinblastine-mediated induction of MCL1 was suppressed. Another protein induced by vinblastine in ML-1 cells is the cyclin-dependent kinase inhibitor p21waf1, the induction of which was also inhibited by GX15-070. Because the MEK/ERK pathway is required for the induction of both MCL1 and p21waf1, we determined whether GX15-070 also impacted ERK signaling. GX15-070 inhibited the phosphorylation of ERK, which could explain its impact on MCL1 and p21waf1. Hence, it appears that GX15-070 has a previously unidentified off-target effect that prevents the expression of MCL1.
These results seriously question whether GX15-070 is a direct inhibitor of any of the BCL2 proteins. We therefore sought a more definitive assay to determine whether putative BH3 mimetics could inhibit BCL2 and/or MCL1 in cells. Concurrently, we compared the efficacy of seven small molecules reported as BH3 mimetics (supplemental Fig. S1).
BH3 Mimetics Induce Cytotoxicity in a Concentration- and Time-dependent Manner
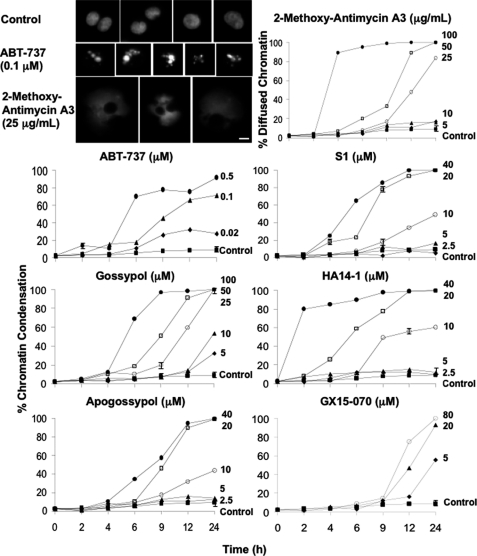
NB4 cells were incubated for 0–24 h with a range of concentrations of seven putative BH3 mimetics, and the percentage of cells dying was determined (Fig. 2). When stained with Hoechst 33342, the chromatin of undamaged NB4 cells appeared evenly dispersed within the nucleus, whereas incubation with ABT-737 induced multiple bright spheres indicative of apoptotic chromatin condensation. This morphology was also induced by the other BH3 mimetics, with the exception of 2-methoxy-antimycin A3, which exhibited swollen, perhaps ruptured, nuclei. We thus scored 2-methoxy-antimycin A3 for nonapoptotic cell death and the rest of the BH3 mimetics for apoptosis.
FIGURE 2.
Cytotoxicity of BH3 mimetics. NB4 cells were incubated with each of the BH3 mimetics at the indicated concentrations for 0–24 h and subsequently stained with Hoechst 33342. The morphology of cells incubated with ABT-737 and 2-methoxy-antimycin A3 was photographed on a fluorescent microscope. The scale bar represents 10 μm. The cells were scored for chromatin condensation, except in the case of 2-methoxy-antimycin A3, which was scored for diffused chromatin morphology. The results reflect the averages of three independent experiments at most time points. S.E. are presented where n = 3, and they exceed the size of the symbol.
All of the tested BH3 mimetics induced a concentration- and time-dependent cell death. Cell death generally occurred within 6 h at high concentrations of the BH3 mimetics or by 24 h at lower concentrations. The two extremes within this data were HA14-1, where the highest concentration (40 μm) induced extensive apoptosis within 2 h, and GX15-070, where the highest concentration (80 μm) did not exhibit any apoptosis until 12 h.
ABT-737 Is the Only BH3 Mimetic That Inhibits BCL2
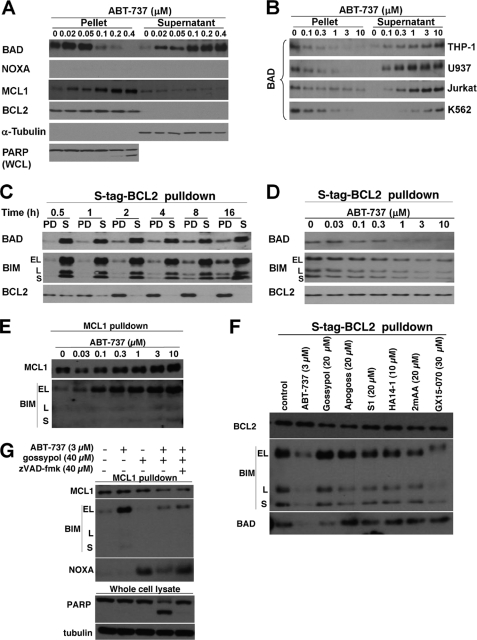
Most antiapoptotic proteins, including BCL2, are membrane-bound, whereas BH3-only proteins are often cytosolic (19). We therefore assessed the localization of BCL2 family members during the induction of apoptosis. NB4 cells were incubated with ABT-737 for 6 h, permeabilized with digitonin, fractionated into pellet and supernatant, and probed for the localization of BH3-only proteins (Fig. 3). Prior to drug incubation, BCL2 was found in the pellet fraction, consistent with it being a membrane-bound protein. BAD, BIM, and PUMA were also found in the pellet (Fig. 3A and data not shown). Upon incubation with ABT-737, BAD was displaced from the pellet to the supernatant in a concentration-dependent manner. BCL2, BIM, and PUMA remained in the pellet fraction regardless of treatment (Fig. 3 and data not shown). The changes observed in NOXA and MCL1 are discussed below.
FIGURE 3.
Effect of ABT-737 on the localization of BCL2 family members. A, NB4 cells were incubated with 0–0.4 μm ABT-737 for 6 h, fractioned into pellet and supernatant, and probed for the indicated proteins. PARP was analyzed in whole cell lysates (WCL). B, THP-1, U937, Jurkat, and K562 cells were incubated with 0–10 μm ABT-737 for 6 h, fractionated, and analyzed for the localization of BAD. C, untreated K562 cells expressing S peptide-tagged BCL2 were lysed and incubated with S protein-agarose for 0–16 h, separated into pulldown fraction (PD) and leftover supernatant (S), and probed for the indicated proteins. D, K562 cells expressing S peptide-tagged BCL2 were incubated with 0–10 μm ABT-737 prior to pulldown with S protein-agarose for 16 h. The pulldown fractions were probed for the indicated proteins. E, cells treated as in D were immunoprecipitated with anti-MCL1 and probed for associated BIM. F, K562 cells were incubated with the indicated BH3 mimetics for 6 h and then incubated with S protein-agarose to pulldown BCL2 bound proteins. The lysates were then probed for associated BIM and BAD. G, K562 cells were incubated with ABT-737 alone, gossypol alone, or both in combination for 6 h, then immunoprecipitated for MCL1, and probed for associated BIM and NOXA. To prevent caspase-mediated proteolysis induced by the drug combination, the cells were also incubated in z-VAD-fmk (far-right lane) to demonstrate that the decrease in BIM was not due to caspase activity.
BAD is usually considered to be phosphorylated and sequestered by protein 14-3-3 in the cytosol of viable cells (20, 21); hence it was surprising to find it in the membrane fraction. Accordingly, we repeated these experiments in four other cell lines: THP-1, U937, Jurkat, and K562 (Fig. 3B). As shown in NB4 cells, BAD was also primarily present in the pellet of untreated cells. Upon the addition of ABT-737, BAD was displaced from the pellet to the supernatant in a concentration-dependent manner. In most of the cell lines tested, 0.1–0.3 μm ABT-737 displaced most of the BAD, whereas K562 cells required 1–3 μm ABT-737 (Fig. 3, A and B). Compared with the other leukemia cell lines, K562 cells have a higher basal level of BCL-XL (data not shown). Because BAD has been suggested to have a higher affinity for BCL-XL than BCL2 (22), the higher concentration of ABT-737 required to displace most of BAD in K562 cells may be due to the higher level of BCL-XL expressed in these cells.
Because BAD lacks a transmembrane domain, our data suggest that it may be bound to its antiapoptotic partners BCL2 or BCL-XL, which would explain why ABT-737 is able to induce its translocation to the cytosol. To further validate this, we investigated the effect of ABT-737 on the interaction between BAD and BCL2 by co-precipitation from K562 cells expressing S peptide-tagged BCL2. We initially determined that at least 4 h was required to pull down all BCL2 from cell lysates (Fig. 3C). Both BAD and BIM were co-precipitated with BCL2, although a majority of both proteins remained in solution. The addition of ABT-737 disrupted the binding of both BAD and BIM to S peptide-tagged BCL2 in a concentration-dependent manner (Fig. 3D). The concentration required to displace all BAD was 1 μm ABT-737, which correlated with the cell fractionation assay (Fig. 3B). Considering that ABT-737 displaced all of BAD from the membrane fraction to the cytosol of intact cells, we believe that all of BAD is indeed bound to BCL2, but the co-precipitation assay cannot quantitatively detect this interaction. Furthermore, we chose this co-precipitation method because initial experiments to immunoprecipitate endogenous BCL2 in NB4 cells failed to co-immunoprecipitate BAD or BIM. We surmise that this inefficiency relates to the need to extract BCL2 from the membrane, which in turn destabilizes its interaction with BAD and BIM.
NB4 cells normally express little MCL1, but incubation with ABT-737 led to a significant increase (Fig. 3). This increase is probably due to the release of BIM from BCL2, which then binds to and stabilizes MCL1. We pursued this possibility further in the K562 cells containing S peptide-tagged BCL2 because these cells have considerably more MCL1. The cells were incubated with 0–10 μm ABT-737 for 6 h, and then MCL1 was immunoprecipitated. Although MCL1 bound little BIM in the absence of ABT-737, there was significant co-immunoprecipitation of these two proteins when the cells were incubated with ABT-737 (Fig. 3E). The concentration of ABT-737 that dissociated BAD and BIM from BCL2 was consistent with the concentration that resulted in BIM binding to MCL1. This result demonstrates the importance of MCL1 in influencing the response of cells to ABT-737.
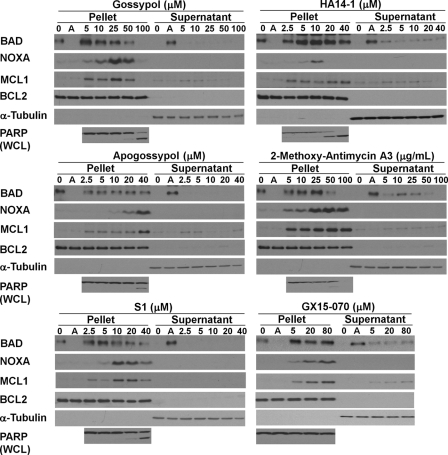
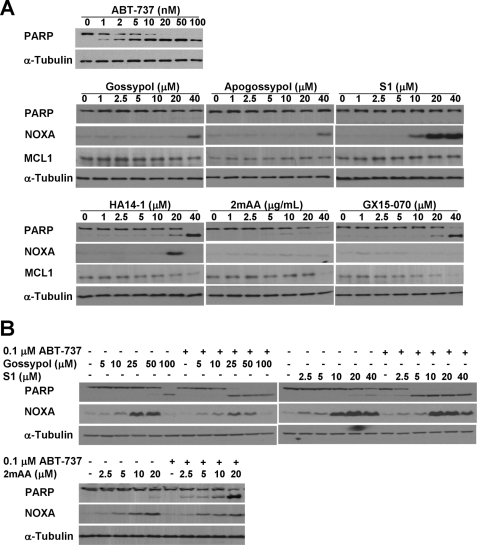
In sum, these results suggest that BAD is constitutively bound to BCL2 in many cell lines and that the disruption of this binding by ABT-737 provides a rapid and quantitative readout for the functional inhibition of BCL2 in a cell-based system. We then used this assay to determine the ability of other BH3 mimetics to inhibit BCL2 (Fig. 4). We found that none of these BH3 mimetics displaced BAD from the pellet in NB4 cells, suggesting that they do not inhibit BCL2. Furthermore, we found that these other BH3 mimetics did not dissociate BAD or BIM from S peptide-tagged BCL2 in K562 cells (Fig. 3F). In this figure, GX15-070 induced a band shift in BIM, possibly because of phosphorylation, and may reflect the impact of this compound on an alternate target.
FIGURE 4.
Effect of BH3 mimetics on expression and localization of BCL2 family members. NB4 cells were incubated with each of the indicated BH3 mimetics for 6 h, fractionated into pellet and supernatant, and probed for the indicated proteins. PARP was analyzed in whole cell lysates (WCL). Pellet and supernatant fractions from NB4 cells incubated with 0.1 μm ABT-737 for 6 h (lanes A) were added as controls to each panel. The Western blot of MCL1 is a shorter exposure than presented in Fig. 3; hence MCL1 is only marginally visible when cells were incubated with ABT-737.
PARP is a substrate of caspase 3 and is cleaved during apoptosis. The highest concentration of most of the BH3 mimetics induced PARP cleavage within 6 h (Fig. 4), which correlated with the cytotoxicity data (Fig. 2). High concentrations of gossypol that killed NB4 cells resulted in the loss of BAD, which was prevented by the addition of z-VAD-fmk (data not shown). However, BAD was recovered in the pellet, suggesting that this BH3 mimetic did not inhibit BCL2. In the case of 2-methoxy-antimycin A3, both BAD and PARP were lost, and neither were recovered upon incubation with z-VAD-fmk (data not shown) consistent with the nonapoptotic death that occurred.
Most BH3 Mimetics Induce NOXA
While investigating the translocation of BAD, we observed that NOXA was elevated in a concentration-dependent manner by all of the BH3 mimetics except ABT-737; MCL1 was concurrently induced (Figs. 3A and 4A). NOXA was also induced in THP-1, U937, Jurkat, and K562 cells within the same concentration range (see Fig. 8 below, and data not shown). This finding was puzzling because BH3 mimetics are thought to physically disrupt the interaction between pro- and antiapoptotic proteins and are not expected to alter protein expression.
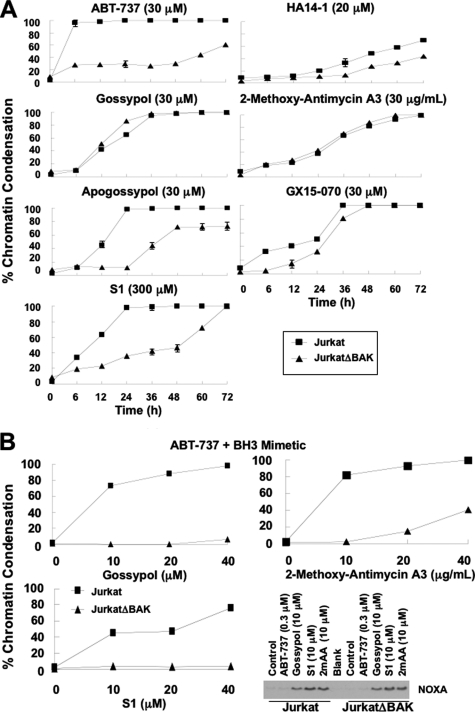
FIGURE 8.
BAX/BAK dependence of BH3 mimetic-induced apoptosis. A, wild-type Jurkat and JurkatΔBAK cells were incubated with each of the BH3 mimetics for 0–72 h, stained with Hoechst 33342, and scored for chromatin condensation. The results reflect the average of three independent experiments at most time points. The S.E. values are presented where n = 3, and they exceed the size of the symbol. B, wild-type Jurkat and JurkatΔBAK cells were incubated with 0.3 μm ABT-737 plus gossypol, S1, or 2-methoxy-antimycin A3 for 6 h, stained with Hoechst 33342, and scored for chromatin condensation. The cells were also incubated with ABT-737, gossypol, S1, or 2-methoxy-antimycin A3 for 6 h and lysed for Western blot analysis.
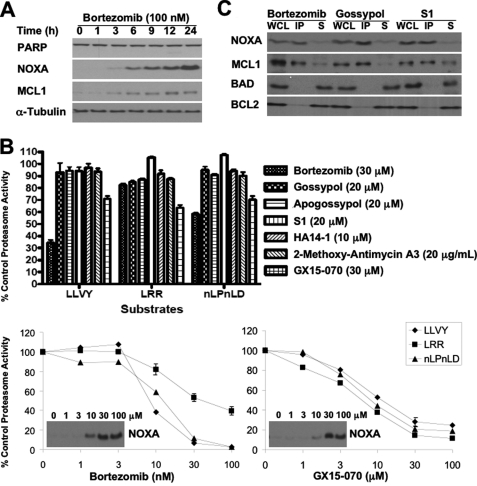
Untreated NB4 cells have very low levels of NOXA and MCL1. Because MCL1 is a short-lived protein, we first determined whether NOXA and MCL1 were constitutively degraded in NB4 cells (Fig. 5A). Upon incubation with the proteasome inhibitor bortezomib, NOXA and MCL1 concurrently accumulated within 6 h. This suggests that NB4 cells constitutively synthesize both NOXA and MCL1 proteins, which are then rapidly degraded via the proteasome (but see below).
FIGURE 5.
Effect of BH3 mimetics on proteasome activity. A, NB4 cells were incubated with bortezomib for 0–24 h, and lysates were probed for the indicated proteins. B, NB4 cells were incubated with bortezomib or each of the BH3 mimetics for 6 h and assayed for proteasome activity (top panel). The error bars represent S.E. of at least three independent experiments. NB4 cells were also incubated with a range of concentrations of bortezomib or GX15-070 and assayed for proteasome activity and induction of NOXA (bottom panel). C, NB4 cells were incubated with 100 nm bortezomib, 20 μm gossypol, or 20 μm S1 for 6 h. Whole cell lysates (WCL) were made from which MCL1 was immunoprecipitated. The immunoprecipitated fraction (IP) and the remaining nonimmunoprecipitated supernatant (S) were probed for the indicated proteins. The error bars represent S.E. of three independent experiments where the value exceeds the size of the symbol.
To determine whether any of the BH3 mimetics elevated NOXA and MCL1 as a consequence of proteasome inhibition, we incubated NB4 cells with a concentration of each BH3 mimetic that markedly elevated NOXA within 6 h and assayed for the three protease activities found within the proteasome (Fig. 5B). Bortezomib inhibited proteasome activity at all three protease sites in a concentration-dependent manner; the lowest concentration that induced NOXA inhibited the β5 subunit (substrate LLVY) by 60%, the β1 subunit (substrate nLPnLD) by 40%, and the β2 subunit (substrate LRR) by 15%. This inhibitory profile is consistent with previous reports on the selectivity of bortezomib (23, 24). None of the BH3 mimetics inhibited any of these proteasome activities, with one exception: GX15-070 induced ∼30% inhibition at 3 μm. Upon further analysis, GX15-070 inhibited proteasome activity in a concentration-dependent manner with no selectivity for any of the three protease activities. Accordingly, although NOXA and MCL1 elevation by GX15-070 may be a consequence of proteasome inhibition, this cannot explain the increase observed with the other BH3 mimetics.
NOXA binds MCL1 and targets the complex for proteasomal degradation (25). Because most of the BH3 mimetics cause the accumulation of both NOXA and MCL1, we hypothesized that these BH3 mimetics may inhibit MCL1 and thus disrupt the formation of the NOXA-MCL1 complex to prevent their subsequent degradation. Accordingly, we incubated NB4 cells with gossypol or S1, immunoprecipitated MCL1, and probed for its binding partner NOXA (Fig. 5C). We used NB4 cells treated with bortezomib as a positive control. Upon incubation with bortezomib, all the NOXA co-immunoprecipitated with MCL1, suggesting that there was no pool of free NOXA in NB4 cells. These data are consistent with NOXA binding MCL1 and targeting the NOXA-MCL1 for proteasomal degradation. Unexpectedly, NOXA and MCL1 were also co-immunoprecipitated upon incubation with gossypol or S1, showing that neither of these two BH3 mimetics dissociated NOXA from MCL1. As a negative control, we show that neither BAD nor BCL2 co-immunoprecipitated with MCL1.
Although we initially assumed that bortezomib elevates NOXA as a direct consequence of inhibiting NOXA degradation, bortezomib has been reported to induce NOXA mRNA, possibly through the stabilization of c-Myc (26) or as the result of endoplasmic reticulum (ER) stress, also known as the unfolded protein response or the integrated stress response (27). We therefore assessed the potential transcriptional regulation of NOXA and MCL1 by the BH3 mimetics via quantitative RT-PCR (Fig. 6). We were surprised to discover that all of the BH3 mimetics, except ABT-737, induced NOXA mRNA. We assessed c-Myc protein levels to determine whether it might be involved in the up-regulation of NOXA. Although bortezomib induced a slight increase in c-Myc protein, none of the other compounds increased c-Myc (supplemental Fig. S2); rather most decreased expression, which is consistent with the translational inhibition discussed below. This observation suggests that NOXA is regulated at the transcriptional level in response to BH3 mimetics via a pathway independent of c-Myc. In contrast, there was little induction of MCL1 mRNA (<2-fold) by any of the mimetics, except in the case of GX15-070, which induced more than a 5-fold increase.
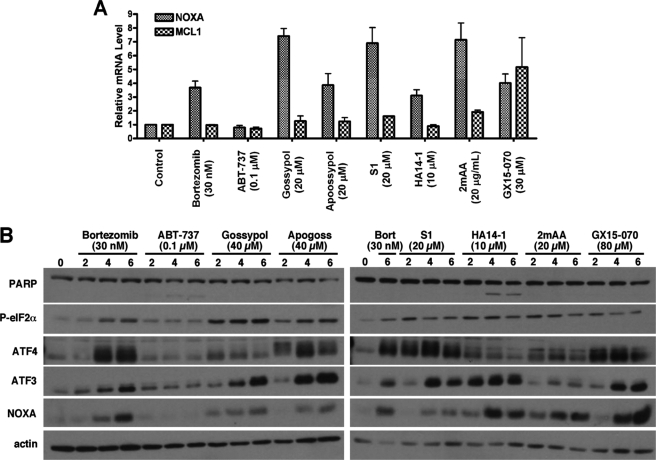
FIGURE 6.
Relative changes in mRNA and proteins levels in response to BH3 mimetics. A, NB4 cells were incubated with bortezomib or each of the BH3 mimetics for 6 h. Total mRNA was purified from the cells and analyzed for relative mRNA expression levels of NOXA and MCL1 via quantitative RT-PCR. The error bars represent S.E. of three independent experiments. B, cells were incubated with bortezomib (Bort) or each of the BH3 mimetics for 0–6 h. Cell lysates were probed for expression of ER stress response proteins. 2mAA, 2-methoxy-antimycin A3.
In an attempt to understand the pathways that might be transcriptionally activating NOXA, we performed a gene expression analysis on NB4 cells that had been incubated with either 20 μm gossypol or 20 μm S1 for 6 h. At a cut-off of 2-fold induction, gossypol and S1 induced 42 and 199 genes, respectively, of which 31 were common (supplemental Table S1). The number of common genes induced by gossypol and S1 was increased from 31 to 72 genes if the cut-off was reset at a 1.5-fold induction. A larger number of genes were repressed by 2-fold: 186 by gossypol and 436 by S1, of which 146 were common. The commonly induced genes were very informative because they included DDIT3 (CHOP/GADD153), PPPIR15A (GADD34), ATF3, GADD45a, ASNS, and SLC3A2, which are genes commonly induced upon activation of the ER stress response (28).
By inhibiting the proteasome, bortezomib induces the accumulation of unfolded proteins and initiates ER stress, resulting in ATF3-mediated up-regulation of NOXA (27). Accordingly, our gene expression analysis suggests that gossypol and S1 are also inducing NOXA because of the activation of the ER stress response. To extend these observations further and determine whether other BH3 mimetics also activate the ER stress response, we assessed the expression of additional stress response proteins. All of the BH3 mimetics except ABT-737 induced phospho-eIF2α, ATF4, and ATF3, as did bortezomib (Fig. 6B). The extent of induction of each component of the pathway, particularly ATF4, varied with mimetic, suggesting that they may induce ER stress by different mechanisms or that there are additional signals that impact this pathway. We have also observed induction of this pathway in U937, THP-1, and K562 cells incubated with gossypol or S1 (data not shown). Although bortezomib induces ER stress by inhibiting the proteasome, we have demonstrated that BH3 mimetics, with the exception of GX15-070, do not inhibit the proteasome. These findings suggest that BH3 mimetics are inducing the ER stress-ATF4/ATF3-NOXA pathway via a potentially novel signaling mechanism(s) distinct from the proteasomal pathway.
Functional Analysis of the Selectivity of BH3 Mimetics
CLL cells are particularly sensitive to ABT-737 and undergo apoptosis within 4 h. This acute apoptosis has been attributed to the constitutive binding of BIM to BCL2 at the mitochondria, such that inhibition of BCL2 causes an immediate induction of apoptosis (10). To functionally determine whether other BH3 mimetics inhibit BCL2, we isolated CLL cells from patients, incubated the cells with each BH3 mimetic for 6 h, and scored for apoptosis (Fig. 7). As expected, CLL cells were extremely sensitive to ABT-737, with 100% cleavage of PARP at 20 nm (Fig. 7A). In contrast, CLL cells were insensitive to gossypol, apogossypol, and S1 at the micromolar range that elevated NOXA, although changes in MCL1 level were not apparent. This further suggests that gossypol, apogossypol, and S1 do not inhibit BCL2 within the concentration range that elevates NOXA.
FIGURE 7.
Functional evidence for the selectivity of BH3 mimetics. A, CLL cells isolated from patients were incubated with each of the BH3 mimetics for 6 h, and then the lysates were probed by Western blot analysis. B, NB4 cells were incubated with gossypol, S1, or 2-methoxy-antimycin A3 (2mAA) with or without ABT-737 for 6 h prior to Western blot analysis.
2-Methoxy-antimycin A3 induced the loss of PARP and MCL1 in CLL cells and the same swollen nuclear morphology as observed in NB4 cells. However, 2-methoxy-antimycin A3 did not elevate NOXA in CLL cells, suggesting that NOXA elevation is not involved in this morphological change. Both HA14-1 and GX15-070 induced a concentration-dependent apoptosis. HA14-1 slightly elevated NOXA at the concentration where apoptosis was first observed, whereas GX15-070 did not induce NOXA. Because CLL cells can tolerate NOXA induced by gossypol, apogossypol, and S1, the cytotoxicity observed with HA14-1 is likely due to it targeting another pathway. Similarly, GX15-070 could be killing CLL cells through the inhibition of other targets identified above, i.e. the MEK/ERK pathway or the proteasome.
Incubation of NB4 cells with 0.1 μm ABT-737 appeared to completely inhibit BCL2 as judged by BAD translocation, yet this was insufficient to kill the cells (Fig. 3). Because MCL1 is recognized as the major mechanism of resistance to ABT-737 (8), we questioned whether concurrent incubation with other BH3 mimetics would induce apoptosis. In combination with 0.1 μm ABT-737, gossypol, S1, and 2-methoxy-antimycin A3 induced apoptosis at concentrations that elevated NOXA (Fig. 7B). It is important to emphasize that although higher concentrations of 2-methoxy-antimycin A3 alone caused nonapoptotic cell death (Fig. 2), lower concentrations of 2-methoxy-antimycin A3 were not cytotoxic and induced NOXA within 6 h in NB4 cells (Figs. 2, 4, and 7B). Furthermore, this low concentration of 2-methoxy-antimycin A3, when combined with ABT-737, induced chromatin condensation within 6 h, indicative of apoptotic death.
We showed above that incubation of K562 cells with ABT-737 facilitated binding of BIM to MCL1 (Fig. 3E). We therefore surmised that up-regulation of NOXA might displace BIM, and this could explain why cells undergo apoptosis rapidly when both BCL2 and MCL1 are inhibited. Concurrent incubation with ABT-737 and gossypol resulted in the formation of a MCL1-NOXA complex and a dramatic decrease of the MCL1-BIM complex (Fig. 3G). As a consequence, BIM is now likely free to activate BAX and BAK.
Collectively, our findings suggest that most BH3 mimetics induce NOXA to antagonize MCL1 and do not inhibit BCL2. Furthermore, the dual inhibition of BCL2 and MCL1 overcomes cellular resistance to ABT-737 and induces apoptosis. These results further support the potential therapeutic value of combining a BCL2 inhibitor with a MCL1 inhibitor as a combination therapeutic regimen.
BH3 Mimetics Induce BAX/BAK-dependent Apoptosis when Combined with ABT-737
ABT-737 is considered a bona fide BH3 mimetic, and as a consequence, it induces BAX/BAK-dependent apoptosis. Most of the other BH3 mimetics investigated here have been proposed to have off-target effects, because they kill mouse embryonic fibroblasts that lack BAX and BAK (8, 11). Because our work has been done in human leukemia cell models, we thus compared the BAX/BAK dependence of apoptosis induced by the seven BH3 mimetics in the wild-type Jurkat human acute T cell leukemia cell line that constitutively lacks BAX versus the JurkatΔBAK cell line, a Jurkat subline that additionally has no detectable BAK (29) (supplemental Fig. S3). To confirm the lack of functional BAX/BAK in the JurkatΔBAK cells, we show that anisomycin killed wild-type Jurkat but not JurkatΔBAK cells (supplemental Fig. S3). Upon incubation with ABT-737, BAX/BAK-dependent apoptosis was induced, although this required 30 μm ABT-737 (Fig. 8A; lower concentrations were tested but did not induce apoptosis). In contrast, only 0.1–0.3 μm ABT-737 was needed to translocate most of BAD in Jurkat cells (Fig. 3B), suggesting that the functional inhibition of BCL2 alone is insufficient to induce apoptosis and that the apoptosis induced by high concentrations of ABT-737 is the consequence of effects on additional targets. Because 30 μm ABT-737 did not induce NOXA in Jurkat or JurkatΔBAK cells (data not shown), NOXA is not involved in apoptosis induced by ABT-737 alone.
Apogossypol and S1 also induced extensive BAX/BAK-dependent apoptosis by 24 h, although this dependence became less apparent at later time points (Fig. 8A). The concentrations of apogossypol and S1 were 3- and 30- fold higher, respectively, than that required to induce NOXA within 6 h (Fig. 8B and data not shown). BAX/BAK-dependent apoptosis induced by HA14-1 and GX15-070 was very limited, whereas apoptosis induced by gossypol and 2-methoxy-antimycin A3 was independent of BAX/BAK. These findings suggest that neither the inhibition of BCL2 nor the induction of NOXA alone is sufficient to kill Jurkat cells within 6 h. Apoptosis induced by higher concentrations is likely the result of the BH3 mimetics impacting alternate pathways or targets.
Because our experiments with NB4 cells suggest that dual inhibition of BCL2 and MCL1 induces apoptosis (Fig. 7B), we next determined whether ABT-737 in combination with gossypol, S1, or 2-methoxy-antimycin A3 would induce apoptosis in a BAX/BAK-dependent manner (Fig. 8B). When combined with a concentration of ABT-737 that displaced BAD (0.3 μm; Fig. 3B), Jurkat cells were sensitized to the BH3 mimetics within the concentration range that induced NOXA. JurkatΔBAK cells, on the other hand, were almost completely resistant to the combinations, showing that these combinations induced BAX/BAK-dependent apoptosis. Importantly, whereas gossypol and 2-methoxy-antimycin A3 alone induced cell death independent of BAX/BAK at later time points (Fig. 8A), when combined with ABT-737, both BH3 mimetics induced apoptosis within 6 h in a BAX/BAK-dependent manner (Fig. 8B).
DISCUSSION
ABT-737 is a BH3 mimetic that specifically inhibits BCL2 and BCL-XL but not MCL1 and induces BAX/BAK-dependent apoptosis (5, 8). The ability of other BH3 mimetics to inhibit antiapoptotic proteins is currently questioned because they kill cells in a BAX/BAK-independent manner (8, 11). When used as a single agent, some of the BH3 mimetics induce BAX/BAK-dependent apoptosis within 24 h, although this dependence diminishes at later time points. However, when we combined these BH3 mimetics with ABT-737, there was a rapid induction of BAX/BAK-dependent apoptosis. These results suggest that the pathway of cell death induced by BH3 mimetics as single agents may be unrelated to their ability to induce NOXA and inhibit MCL1, a pathway that alone does not induce apoptosis.
Although binding affinities between BH3-only proteins and their antiapoptotic partners have been previously characterized (30), there is evidence that suggests affinities established in vitro may not reflect the actual affinities found in cells. For example, the NOXA BH3 peptide was reported to bind to the recombinant antiapoptotic protein MCL1 at least 4,000-fold more potently than BCL2 or BCL-XL. However, two recent independent studies reported that the binding of NOXA to BCL-XL but not MCL1 dictates cellular sensitivity to apoptosis (31, 32). Different binding affinities could result from alternate conformations when the proteins are inserted in the membrane as has been reported for BCL2 (33, 34). These results emphasize the need for biological validation of biochemically defined protein-protein interactions.
In this study, we assessed the specificity of seven putative BH3 mimetics using a cell-based assay and showed that the displacement of BAD is a cellular event indicative of the functional inhibition of BCL2. ABT-737 is the only BH3 mimetic that displaces BAD from BCL2. In viable cells, BAD is often reported to be phosphorylated and sequestered by protein 14-3-3 in the cytosol (20). However, most studies reporting the requirement for this phosphorylation were done in systems that over-express BAD. Our work with multiple cell lines showed that endogenous BAD appears to be sequestered by BCL2, suggesting that it does not need to be inactivated via phosphorylation, nor does it need to be sequestered away from the mitochondria for cells to remain viable.
Compared with ABT-737, none of the other BH3 mimetics induce BAD translocation, and we conclude that they do not inhibit BCL2 in cells. These BH3 mimetics, however, elevate NOXA, the consequence of which is expected to be the inhibition of MCL1 (25). NB4 cells have low basal levels of NOXA and MCL1, which concurrently accumulate upon proteasomal inhibition. Intuitively, this would suggest that NOXA and MCL1 are constitutively synthesized and degraded, which is consistent with newly synthesized NOXA immediately binding to the available MCL1 and targeting the complex for degradation (25). We thus hypothesized that MCL1 inhibitors would disrupt the formation of the NOXA-MCL1 complex and prevent its degradation. Unexpectedly, we found that gossypol and S1 do not disrupt the binding between NOXA and MCL1; rather they induce NOXA mRNA. The induction of NOXA mRNA was seen with all of the BH3 mimetics tested except ABT-737, and gene expression analysis further identified a signature of induced genes indicative of activation of the ER stress response. This same stress pathway has been implicated in bortezomib-induced NOXA mRNA. Specifically, activation of eIF2α leads to the induction of ATF4 and ATF3, both members of the CREB family. The ATF4-ATF3 complex has been shown to bind to a CRE reporter element in the promoter of the noxa gene, and suppression of either ATF4 or ATF3 reduced expression of NOXA (27). Furthermore, suppression of NOXA protected cells from bortezomib-induced apoptosis. Because we observed activation of the same pathway by the putative BH3 mimetics, it is logical to assume that NOXA is playing an equally important role in the apoptosis induced by these compounds. This conclusion is supported by the co-immunoprecipitation experiments in which ABT-737 was shown to dissociate BIM from BCL2, which then bound to MCL1. Induction of NOXA dissociated BIM from MCL1. BIM would then be free to activate BAX/BAK and induce apoptosis.
Although these BH3 mimetics induce NOXA, ATF4, and ATF3, this is achieved independent of the proteasome pathway because none of the BH3 mimetics, with the exception of GX15-070, inhibited the proteasome. The fact that these six different compounds were all thought to be BH3 mimetics is intriguing and raises the question as to whether they might impact some other function of BCL2 proteins that in turn could induce ER stress. For example, BAX and BAK can modulate the ER stress response by directly binding to IRE1, an endonuclease that splices the XBP1 mRNA (35). In addition, BCL2, BCL-XL, and MCL1 have been reported to regulate calcium flux through binding inositol 1,4,5-trisphosphate receptors in the ER (36). Whether these or other proteins are targeted by these BH3 mimetics remains to be determined.
We are cautious at this time to refer to this as the ER stress response (or integrated stress response) (37) rather than “ER stress” because we have not yet established whether the pathway is activated by unfolded proteins in the ER as occurs with bortezomib. We observe activation of eIF2α, but there are four different kinases that are known to accomplish this phosphorylation, each of which can respond to different stresses (38). Accordingly, there are multiple inputs to this pathway that will need to be investigated for each of these putative BH3 mimetics as a step in determining their specific molecular targets.
The success of ABT-737 in preclinical studies and the orally active counterpart ABT-263 in clinical trials is a proof-of-concept for developing BH3 mimetics as tools for studying apoptosis and as potential therapeutic agents. Our study provides evidence that cell-based assays are needed to validate in vitro-defined interactions between the BCL2 family members and to functionally assess the on-target inhibition by the BH3 mimetics. Our current functional analyses suggest that, of the seven putative BH3 mimetics tested, only ABT-737 inhibits BCL2. The other BH3 mimetics do not inhibit BCL2 but rather antagonize MCL1 through the induction of NOXA. When BH3 mimetics are used in appropriate combinations, concurrent inhibition of BCL2 and MCL1 induces BAX/BAK-dependent apoptosis within 6 h.
This study began with the observation that ABT-737 dramatically sensitizes NB4 cells to vinblastine, with 100% of the cells undergoing apoptosis within 3 h. This provides an exciting therapeutic potential for certain tumors, but we have observed this phenotype in only a few other cell lines (e.g. three myeloma lines and one mantle cell lymphoma line; data not shown). Importantly, this rapid apoptosis occurs in cells from all phases of the cell cycle and contradicts the paradigm that vinca alkaloids are mitosis-specific drugs. Indeed, we recently reported that a few cell lines are acutely sensitive to vinblastine alone, without the need to otherwise inhibit antiapoptotic proteins (4). While seeking an explanation for these observations, we found that vinblastine also induces NOXA mRNA (data not shown), suggesting that there are multiple pathways to induce NOXA. In addition, bortezomib induces NOXA and acute apoptosis when combined with ABT-737 in NB4 cells (data not shown).
Overall, these results present a number of drug combinations that can rapidly induce apoptosis, all of which appear to center around NOXA induction. It is likely that different strategies would be effective for the treatment of different tumors based on their variable dependence on antiapoptotic proteins. Whether any of these strategies will be selective for the tumor remains to be determined.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA50224 (to A. E.). This work was also supported by Cancer Center Support Grant CA23108 to the Norris Cotton Cancer Center and National Institutes of Health Training Grant T32 09658 (to B. L. S., R. S. S., and D. J. P. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S3.
- BH
- BCL2 homology
- CLL
- chronic lymphocytic leukemia
- ER
- endoplasmic reticulum
- PARP
- poly(ADP-ribose) polymerase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Adams J. M., Cory S. (2007) Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walensky L. D. (2006) Cell Death Differ. 13, 1339–1350 [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 4. Salerni B. L., Bates D. J., Albershardt T. C., Lowrey C. H., Eastman A. (2010) Mol. Cancer Ther. 9, 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 6. Nguyen M., Marcellus R. C., Roulston A., Watson M., Serfass L., Murthy Madiraju S. R., Goulet D., Viallet J., Bélec L., Billot X., Acoca S., Purisima E., Wiegmans A., Cluse L., Johnstone R. W., Beauparlant P., Shore G. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Konopleva M., Contractor R., Tsao T., Samudio I., Ruvolo P. P., Kitada S., Deng X., Zhai D., Shi Y. X., Sneed T., Verhaegen M., Soengas M., Ruvolo V. R., McQueen T., Schober W. D., Watt J. C., Jiffar T., Ling X., Marini F. C., Harris D., Dietrich M., Estrov Z., McCubrey J., May W. S., Reed J. C., Andreeff M. (2006) Cancer Cell 10, 375–388 [DOI] [PubMed] [Google Scholar]
- 8. van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., Huang D. C. (2006) Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhai D., Jin C., Satterthwait A. C., Reed J. C. (2006) Cell Death Differ. 13, 1419–1421 [DOI] [PubMed] [Google Scholar]
- 10. Del Gaizo Moore V., Brown J. R., Certo M., Love T. M., Novina C. D., Letai A. (2007) J. Clin. Invest. 117, 112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vogler M., Weber K., Dinsdale D., Schmitz I., Schulze-Osthoff K., Dyer M. J., Cohen G. M. (2009) Cell Death Differ. 16, 1030–1039 [DOI] [PubMed] [Google Scholar]
- 12. Ganju N., Eastman A. (2003) Cell Death Differ. 10, 652–661 [DOI] [PubMed] [Google Scholar]
- 13. Single B., Leist M., Nicotera P. (1998) Cell Death Differ. 5, 1001–1003 [DOI] [PubMed] [Google Scholar]
- 14. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 15. Zhou P., Qian L., Kozopas K. M., Craig R. W. (1997) Blood 89, 630–643 [PubMed] [Google Scholar]
- 16. Oltvai Z. N., Milliman C. L., Korsmeyer S. J. (1993) Cell 74, 609–619 [DOI] [PubMed] [Google Scholar]
- 17. Meng X. W., Lee S. H., Dai H., Loegering D., Yu C., Flatten K., Schneider P., Dai N. T., Kumar S. K., Smith B. D., Karp J. E., Adjei A. A., Kaufmann S. H. (2007) J. Biol. Chem. 282, 29831–29846 [DOI] [PubMed] [Google Scholar]
- 18. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. (1997) Cell 91, 231–241 [DOI] [PubMed] [Google Scholar]
- 21. Downward J. (1999) Nat Cell Biol. 1, E33–E35 [DOI] [PubMed] [Google Scholar]
- 22. Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 23. Oerlemans R., Franke N. E., Assaraf Y. G., Cloos J., van Zantwijk I., Berkers C. R., Scheffer G. L., Debipersad K., Vojtekova K., Lemos C., van der Heijden J. W., Ylstra B., Peters G. J., Kaspers G. L., Dijkmans B. A., Scheper R. J., Jansen G. (2008) Blood 112, 2489–2499 [DOI] [PubMed] [Google Scholar]
- 24. Altun M., Galardy P. J., Shringarpure R., Hideshima T., LeBlanc R., Anderson K. C., Ploegh H. L., Kessler B. M. (2005) Cancer Res. 65, 7896–7901 [DOI] [PubMed] [Google Scholar]
- 25. Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikiforov M. A., Riblett M., Tang W. H., Gratchouck V., Zhuang D., Fernandez Y., Verhaegen M., Varambally S., Chinnaiyan A. M., Jakubowiak A. J., Soengas M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19488–19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q., Mora-Jensen H., Weniger M. A., Perez-Galan P., Wolford C., Hai T., Ron D., Chen W., Trenkle W., Wiestner A., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schröder M., Kaufman R. J. (2005) Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 29. Wang G. Q., Gastman B. R., Wieckowski E., Goldstein L. A., Gambotto A., Kim T. H., Fang B., Rabinovitz A., Yin X. M., Rabinowich H. (2001) J. Biol. Chem. 276, 34307–34317 [DOI] [PubMed] [Google Scholar]
- 30. Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 31. Hagenbuchner J., Ausserlechner M. J., Porto V., David R., Meister B., Bodner M., Villunger A., Geiger K., Obexer P. (2010) J. Biol. Chem. 285, 6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez H., Zhang L., George N. M., Liu X., Pang X., Evans J. J., Targy N. M., Luo X. (2010) J. Biol. Chem. 285, 15016–15026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim P. K., Annis M. G., Dlugosz P. J., Leber B., Andrews D. W. (2004) Mol. Cell 14, 523–529 [DOI] [PubMed] [Google Scholar]
- 34. Dlugosz P. J., Billen L. P., Annis M. G., Zhu W., Zhang Z., Lin J., Leber B., Andrews D. W. (2006) EMBO J. 25, 2287–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hetz C., Bernasconi P., Fisher J., Lee A. H., Bassik M. C., Antonsson B., Brandt G. S., Iwakoshi N. N., Schinzel A., Glimcher L. H., Korsmeyer S. J. (2006) Science 312, 572–576 [DOI] [PubMed] [Google Scholar]
- 36. Chen R., Valencia I., Zhong F., McColl K. S., Roderick H. L., Bootman M. D., Berridge M. J., Conway S. J., Holmes A. B., Mignery G. A., Velez P., Distelhorst C. W. (2004) J. Cell Biol. 166, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 38. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.