Abstract
Naïve CD4+ T helper (Th) cells differentiate into distinct subsets of effector cells (Th1, Th2, Th17, and induced regulatory T cells (iTreg)) expressing different sets of cytokines upon encounter with presented foreign antigens. It has been well established that Th1/Th2 balance is critical for the nature of the following immune responses. Previous reports have demonstrated important roles of c-Jun N-terminal kinase (JNK) in Th1/Th2 balance, whereas the regulatory mechanisms of JNK activity in Th cells have not been elucidated. Here, we show that dual specificity phosphatase 16 (DUSP16, also referred to as MKP-M or MKP-7), which preferentially inactivates JNK, is selectively expressed in Th2 cells. In the in vitro differentiation assay of naïve CD4+ cells, DUSP16 expression is up-regulated during Th2 differentiation and down-regulated during Th1 differentiation. Chromatin immunoprecipitation revealed the increased acetylation of histone H3/H4 at the dusp16 gene promoter in CD4+ T cells under the Th2 condition. Adenoviral transduction of naïve CD4+ T cells with DUSP16 resulted in increased mRNA expression of IL-4 and GATA-3 in Th2 and decreased expression of IFNγ and T-bet in Th1 differentiation. In contrast, transduction of a dominant negative form of DUSP16 had the reverse effects. Furthermore, upon immunization, T cell-specific dusp16 transgenic mice produced antigen-specific IgG2a at lower amounts, whereas DN dusp16 transgenic mice produced higher amounts of antigen-specific IgG2a accompanied by decreased amounts of antigen-specific IgG1 and IgE than those of control mice. Together, these data suggest the functional role of DUSP16 in Th1/Th2 balance.
Keywords: Cytokine, Differentiation, Immunology, Jun N-terminal kinase (JNK), Lymphocyte, Phosphatase, Signal Transduction
Introduction
The activation and expansion of CD4+ T helper (Th)2 cells are crucial in establishing effective immune responses toward invading pathogens and antigens. Upon first encounter with foreign antigens presented by antigen-presenting cells, naïve Th cells can differentiate into at least four distinct subsets (Th1, Th2, Th17, and induced regulatory T cells (iTreg)) of effector cells (1, 2). Th1 cells produce mainly interferon-γ (IFNγ), induce cell-mediated immunity, and provide protection against intracellular pathogens. Th2 cells produce interleukin-4 (IL-4), IL-5, IL-10, and IL-13, mediate humoral immunity, and provide protection against extracellular pathogens, including helminthes. The recently identified Th17 cells produce IL-17 and participate in the protection against extracellular bacterial and fungi through neutrophil recruitment and the induction of antimicrobial peptides (3). Further, some of the autoimmune responses formally attributed to Th1 cells, such as experimental autoimmune encephalomyelitis, have been demonstrated to be mediated, at least partly, by Th17 cells. On the other hand, iTreg play important roles in the maintenance of immune tolerance and the negative regulation of immune responses by producing TGF-β, IL-10, and IL-35 (4).
It has been well established that the balance of Th1 and Th2 subsets is critical to cellular and humoral immunity. Conversely, the imbalance of Th1/Th2 subsets often causes pathological conditions. More specifically, an excess of Th1 cells is associated with organ-specific autoimmunity, whereas an excess of Th2 cells is associated with hypersensitivity as well as allergy (5).
The regulatory mechanisms of Th1/Th2 cell polarization have been studied extensively. One of the most important factors that influence the differentiation of Th cells is cytokines. Most notably, IL-12 directs differentiation of Th1, whereas IL-4 drives differentiation of Th2 cells (6, 7). Inside the Th cells, transcription factors play critical roles instructing either Th1 or Th2 differentiation. Some transcription factors are expressed selectively by Th1 or Th2 and are identified as Th1- or Th2-specific transcription factors. When expressed, these transcription factors regulate the expression of the Th1- or Th2-specific cytokine genes. For example, a T-box transcription factor, T-bet, is selectively expressed in Th1 cells and is responsible for the transcription of the gene encoding IFNγ (8). In contrast, the GATA-binding protein 3 (GATA-3) is selectively expressed in Th2 cells and is responsible for the transcription of the gene encoding IL-4 (5, 9, 10).
The intracellular signaling pathways via mitogen-activated kinases (MAPKs) are implicated in various cellular events, including the differentiation and the activation of Th cells (11). The c-Jun N-terminal kinases (JNK) and p38 kinases are considered especially important for the responses of Th1 effector cells (12, 13). The p38 MAPK activity is specifically induced in Th1, but not in Th2 cells. Inhibition of p38 by pyridinyl imidazole compound, a specific inhibitor of p38 MAPK, or by the expression of a dominant negative version of p38 blocked IFNγ production and impaired Th1 responses (12). Similarly, JNK is selectively activated in Th1, but not in Th2 cells. Jnk2-deficient CD4+ T cells showed defective IFNγ production and impairment in Th1 differentiation (13). On the other hand, jnk1-deficient CD4+ T cells exhibited increased IL-4 production and preferential differentiation of Th cells into Th2 type (14).
MAPK activation is a reversible phosphorylation process, and a subclass of dual specificity phosphatases (DSPs) is responsible for the inactivation of MAPKs through dephosphorylation of both threonine and tyrosine residues that are essential for the enzymatic activity (15). Among DSP family members, some show selective substrate specificity, whereas others efficiently inactivate all classes of MAPKs: extracellular signal-regulated kinase (ERK), JNK, and p38 kinase.
We have previously reported a MAPK phosphatase (MKP) termed DUSP16 (also referred to as MKP-M/MKP-7) (16–18). DUSP16 has been shown as a MKP that preferentially inactivates JNK (16, 17). Here, we show that the expression of DUSP16 is selectively up-regulated during Th2 differentiation. Furthermore, we have analyzed T cell-specific dusp16 transgenic (Tg) mice to provide evidence that DUSP16 functionally contributes to the regulation of Th1/Th2 balance in vivo.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Recombinant mouse IL-2, IL-4, and IL-12 were purchased from Petrotech Corporation (Seattle, WA). Anti-mouse IFNγ antibody was from Endogen (Woburn, MA). Anti-mouse CD3 antibody was from Cedarlane Laboratories Ltd. (Ontario, Canada). Anti-mouse IL-12 antibody was from Genzyme Diagnostics (Cambridge, MA). Anti-mouse IL-4 antibody was from PharMingen. Anti-acetylhistone H3 and anti-acetylhistone H4 antibodies were from Upstate Biotechnology Inc. (Lake Placid, NY). Anti-mouse IgG1 and IgG2a antibodies were from Invitrogen. Anti-mouse IgE antibody was from Bethyl Laboratories Inc. (Montgomery, TX). Anti-mouse DUSP16 antibody was prepared as described previously (17). Ovalbumin (OVA), complete Freund's adjuvant, and LPS from Escherichia coli serotype B5: 055 were obtained from Sigma.
Animals
Female C57BL/6 and BALB/c mice were obtained from Japan SLC (Shizuoka, Japan). All mice were housed under specific pathogen-free conditions, fed with autoclaved food and water, and handled in laminar airflow hoods. All experiments on the animals were done in accordance with the Animal Care and Use of the Graduate School of Medical and Dental Sciences, Kagoshima University.
Th Cell Clones and in Vitro T Cell Differentiation
Mouse Th1 clones 28-4 and Th2 clone MS-SB were gifts from Dr. M. Kubo (Tokyo University of Sciences, Chiba, Japan). The two clones have been established from (B6C3N) F1 and C3N/HeN mice, respectively, as described elsewhere (19). 28-4 is an H-2k-restricted, keyhole limpet hemocyanin-specific Th1 clone, and MS-SB is an I-Ak-restricted autoreactive Th2 clone. Cell clones were grown continuously in RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma) and 10% concanavalin A-stimulated mouse spleen cell supernatant for Th1 clone 28-4 or 0.4 ng/ml recombinant mouse IL-4 for Th2 clone MS-SB.
Differentiation of Mouse Primary CD4+ T Cells in Vitro
CD4+ T cells were purified from enriched splenic T cells of the mice using the magnetic cell sorting system (Miltenyi Biotec Inc., Sunnyvale, CA) according to the manufacturer's instruction. The obtained CD4+ T cells that were >95% positive for CD4 by flow cytometry were used for the differentiation experiment, which was performed as described previously (20, 21). Briefly, CD4+ T cells were cultured for 7 days with 2 μg/ml plate-bound anti-CD3 mAb. For Th1 differentiation, the culture was supplemented with 10 ng/ml anti-IL-4 and 10 ng/ml IL-12. For Th2 differentiation, the culture medium was supplemented with 10 μg/ml anti-IFNγ, 10 μg/ml anti-IL-12, and 10 ng/ml IL-4. Both cultured conditions were supplied with 10 μg/ml IL-2 on day 2 and day 4 to maintain cell survival. Cells were harvested, washed, and cultured in new medium without any antibody or cytokine for 16 h before the assays.
Northern Blot Analysis
Total cellular RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instruction. 15-μg aliquots of the total RNA were fractionated, transferred, and hybridized as described previously (17). For DUSP16, PAC-1, MKP-1, M3/6, and β-actin gene expressions, probes were prepared as described previously (17). For T-bet, IFNγ, GATA-3, and IL-4 mRNA gene expressions, probes were prepared as described previously (22, 23) and labeled with 32P.
Immunoblotting
Preparation of total cellular lysate and Western blotting were performed as previously described (17).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described previously (24, 25). The primers used for ChIP assay were: DUSP16 sense, GAAAAGCCCCGGATTTGGGA and antisense, CTCTCTGCTAGTCAGCTGCT; PAC-1 sense, TCTAGACTCCAGGCCGACAC and antisense, GGTTCTGGGCTCTTCGTCGA; IL-4 sense, TTGGTCTGATTTCACAGG and antisense, AACAATGCAATGCTGGC; and IFNγ sense, GCTCTGTGGATGAGAAT and antisense, AAGATGGTGACAGATAGG (25).
Generation and Transduction of Adenoviral Constructs
The wild-type (WT) and the C244S mutated (dominant negative) DUSP16 cDNAs (specifically mutated in the phosphatase catalytic domain) were generated as described previously (17). These cDNAs were cloned into pEGFP-C1 vector (Clontech) to add the N-terminal EGFP tag. The EGFP-tagged DUSP16 cDNAs were then cloned into Shuttle vector (Stratagene), and co-transformed into BJ5183 with pAdEasy-1 vector (Stratagene) to prepare the recombinant Ad-EGFP-DUSP16 constructs. The generated adenoviral vectors were transfected into an adenovirus packaging cell line, AD293, using LipofectamineTM (Invitrogen) according to the manufacturer's instruction, and adenovirus was prepared. Purified CD4+ T cells from C57BL/6 mice stimulated with anti-CD3 mAb were infected using equal titers of adenovirus for 24 h. Cells were washed, supplied with new culture media, and differentiated under the Th1 or Th2 condition as described above. The infection ratio was measured by flow cytometry, and the expression of transgene was evaluated by Western blotting using anti-GFP (Invitrogen) or anti-DUSP16 antibodies.
Reverse Transcription-PCR (RT-PCR)
Total cellular RNA was isolated using TRIzol reagent. Reverse transcription was carried out essentially as described previously (26). The PCR of synthesized cDNA was done as reported previously (27, 28). The synthesized PCR products were separated by electrophoresis on a 1.2% agarose gel and visualized by ethidium bromide staining. The primers were: IFNγ sense, CATTGAAAGCCTAGAAAGTCTG and antisense, CTCATGAATGCATCCTTTTTCG (27, 28); T-bet sense, TGCCTGCAGTGCTTCTAACA and antisense, TGCCCCGCTTCCTCTCCAACCAA (29); IL-4 sense, CATCGGCATTTTGAACGAGGTCA and antisense, CTTATCGATGAATCCAGGCATCG (27, 28); GATA-3 sense, GAAGGCATCCAGACCCGAAAC and antisense, ACCCATGGCGGTGACCATGC (10); β-actin sense, TGGAATCCTGTGGCATCCATGAAAC and antisense, TAAAACGCAGCTCAGTAACAGTCCG (26).
Generation of T Cell-specific Tg Mice
To generate Tg mice, the C-terminally truncated DUSP16WT and DUSP16C244S (17) were N-terminally tagged with FLAG epitope and cloned into the BamHI site of the p1017 vector (provided by Dr. Chuzo Kishimoto, Osaka University) which contains the lck proximal promoter. We have demonstrated previously that these FLAG-tagged forms of DUSP16 work similarly to the untagged forms of DUSP16 in various cell types (17, 30). We also confirmed the functions of these FLAG-tagged forms of DUSP16 in mouse T cells by transient expression experiments (data not shown). The transgene DNA fragments were used in pronuclear injection of C57BL/6 embryos to produce T cell-specific dusp16WT and dusp16C244S Tg mice. Tg founder mice were identified by Western blotting of the thymus and spleen cell lysates with anti-DUSP16 and anti-FLAG antibodies. Mice with wild-type or C244S mutated dusp16 transgene were bred for more than seven generations on C57BL/6 or BALB/C background. Genotyping was performed by PCR from tail DNA.
Flow Cytometry
Splenocytes were prepared from spleens of the mice, and cells were stained with a combination of phycoerythrin-conjugated anti-CD3 and FITC-conjugated anti-CD4 mAbs by standard procedures. Flow cytometry was performed using EPICS XLTM (Beckman Coulter). Data were analyzed by System IITM software (Beckman Coulter).
In Vivo Immunization and Determination of Antigen-specific Immunoglobulin Isotypes
Six-week-old dusp16WT Tg, dusp16C244S mutated Tg mice and C57BL/6 or BALB/c control mice were injected subcutaneously with 50 μg of OVA emulsified 1:1 in complete Freund's adjuvant. Then, the mice were reimmunized intraperitoneally on days 14, 21, and 28. Serum was collected from the mice on days 0, 10, and 35, and the serum concentrations of OVA-specific IgG2a, IgG1, and IgE antibodies were determined as described previously (31).
The mice were killed on day 35; CD4+ T cells were isolated from splenic cells and cultured with 2 μg/ml plate-bound anti-CD3 mAb in the presence of 2 μg/ml anti-CD28 mAb for 3 days. Cells then were harvested, washed, and left unstimulated or stimulated with either 100 or 1000 μg of OVA in vitro for 24 h. The culture supernatants were collected, and IFNγ and IL-4 productions were measured with commercial enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer's instructions (R&D Systems). All samples were assayed in triplicate, and the data are presented as the mean ± S.D.
RESULTS
DUSP16 Is Expressed Differentially in Th1/Th2 Clones
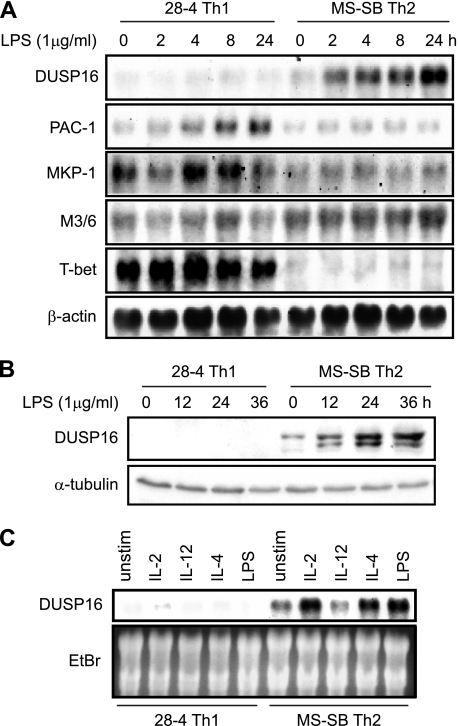
To elucidate the role of DUSP16 in Th1/Th2 cells, we analyzed gene expression of DUSP16 in a mouse Th1 clone (28-4) and a Th2 clone (MS-SB). The mouse Th clones were left untreated or treated with LPS, a potent stimulant of DUSP16 mRNA expression in macrophages, and total RNAs were isolated for Northern blot analysis. These Th clones respond to LPS presumably through Toll-like receptor 4 expressed on the cell surface (26). Using a probe coding the C-terminal domain of the DUSP16, which is not homologous to any other MKP family members, we found that DUSP16 was barely detectable in 28-4 Th1, but was constitutively expressed at a low level in MS-SB Th2 clone (Fig. 1A). Moreover, DUSP16 mRNA expression was substantially increased after LPS stimulation in the MS-SB Th2 clone, but not in the 28-4 Th1 clone (Fig. 1A). The same blot was stripped and was used for gene expression analysis of other MKP members (PAC-1, MKP-1 and M3/6). Unlike DUSP16, PAC-1 and MKP-1 mRNA levels were increased after LPS stimulation in 28-4 Th1 cells, but not in MS-SB Th2. M3/6 mRNA remained constant after LPS treatment in both 28-4 Th1 and MS-SB Th2 clones (Fig. 1A).
FIGURE 1.
DUSP16 is differentially expressed in Th clones. A, 28-4, a Th1 clone, and MS-SB, a Th2 clone, were starved overnight without growth factors and either left untreated or treated with 1 μg/ml LPS for indicated times. Total RNA was extracted for Northern blot analysis using a 32P-labeled cDNA DUSP16 probe. The blot was stripped and rehybridized with a mouse PAC-1, MKP-1, M3/6, T-bet, or β-actin cDNA probe. B, 28-4 and MS-SM Th clones were starved and left untreated or treated as described in A. Total cellular lysates were prepared, and protein levels of DUSP16 were analyzed by Western blotting using anti-DUSP16 polyclonal antibody. As a control, 10% of each lysate was used to detect α-tubulin using anti-α-tubulin mAb. C, 28-4 and MS-SB Th clones were starved as described in A, and cells were left untreated (unstim) or treated for 4 h with IL-2 (10 ng/ml), IL-12 (10 ng/ml), IL-4 (10 ng/ml), or LPS (1 μg/ml). Total RNA was extracted, and gene expression of DUSP16 was analyzed. The ethidium bromide-stained gel is shown.
To analyze the levels of the DUSP16 protein after LSP treatment of these clones, we used a polyclonal antibody specific to DUSP16 (17). MS-SB cells constitutively expressed DUSP16 protein at a moderate level (Fig. 1B). After LPS stimulation, the DUSP16 protein level increased further. In contrast, DUSP16 protein was not detected in 28-4 Th1, and its level was not increased by LPS. These results were consistent with the Northern blotting results and demonstrated that DUSP16 is expressed preferentially in the Th2 clone, unlike the other MKPs examined.
To determine the effects of various cytokines on the DUSP16 mRNA expression in Th clones, 28-4 and MS-SB cells were treated for 4 h with IL-2, IL-12, or IL-4, and total RNAs were isolated. IL-2 and IL-4 but not IL-12 treatment of MS-SB significantly increased DUSP16 gene expression, whereas the DUSP16 mRNA level was not increased in the 28-4 Th1 clone after the cytokine stimulations (Fig. 1C).
Expression Kinetics of DUSP16 during the Th1/Th2 Differentiation of Naïve CD4+ T Cells
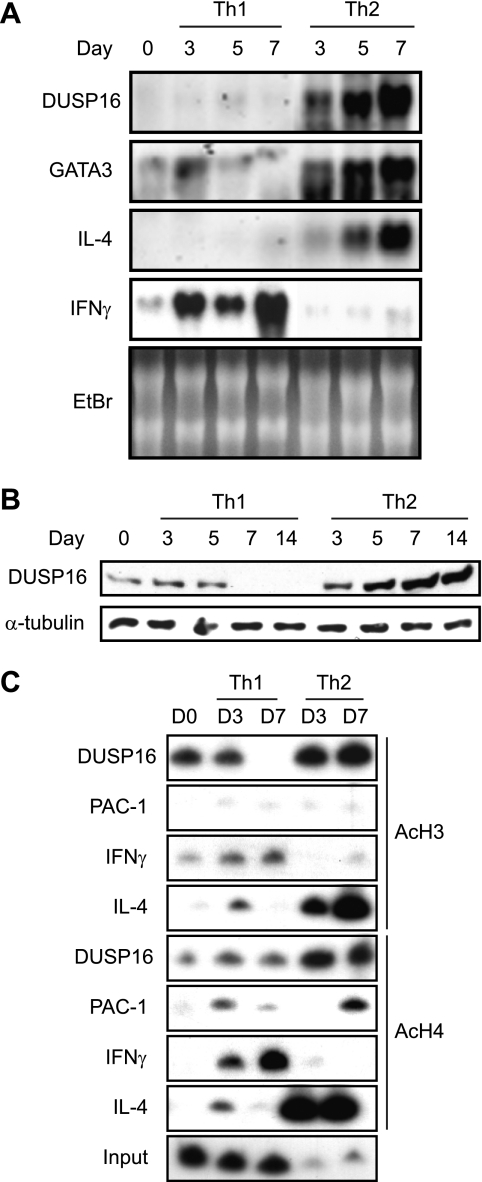
To determine the expression profiles of DUSP16 in the Th1/2 differentiation of the primary CD4+ T cells, naïve CD4+ T cells were purified from C57BL/6 mice and induced to differentiate in vitro. CD4+ T cells were stimulated with immobilized anti-CD3 and differentiated into either Th1 or Th2 by adding the combinations of cytokines and neutralizing anti-cytokine antibodies (20, 21). Total RNAs were isolated on days 0, 3, 5, and 7. DUSP16 mRNA was barely detectable in the naïve CD4+ T cells. The DUSP16 mRNA expression level was gradually increased in CD4+ T cells cultured under the Th2 condition (the combination of IL-4 and anti-IL-12 antibody), but was not induced under the Th1 condition (the combination of IL-12 and anti-IL-4 antibody) (Fig. 2A). The same RNA samples were also analyzed for the gene expression of the Th1/Th2 differentiation markers (GATA-3, IL-4, and IFNγ). As expected, mRNA expressions of GATA-3 and IL-4 were induced in CD4+ T cells cultured under the Th2 condition, whereas IFNγ transcription was induced under the Th1 condition (Fig. 2A), indicating that the Th1/Th2 differentiation was successfully induced.
FIGURE 2.
Selective expression of DUSP16 and histone acetylation at a regulatory region of DUSP16 in differentiated Th2. A, purified CD4+ T cells were differentiated to Th1 or Th2 as described under “Experimental Procedures.” Total RNA was extracted on the indicated days, and DUSP16 gene expression was analyzed by Northern blotting using DUSP16 cDNA probe. The same blot was stripped and rehybridized with a mouse GATA-3, IL-4, or IFNγ cDNA probe. The ethidium bromide-stained gel is also shown. B, CD4+ T cells were differentiated as described in A. Total cellular lysates were prepared on the indicated days, and protein levels of DUSP16 were analyzed by Western blotting. As a control, 10% of each lysate was used to detect α-tubulin. C, purified CD4+ T cells were differentiated to Th1 or Th2 for the indicated days. Chromatin was extracted and immunoprecipitated with either anti-acetylhistone H3 or anti-acetylhistone H4 antibody. PCR analyses of DNA products for DUSP16, PAC-1, or IL-4 from immunoprecipitation reactions were carried out as described under “Experimental Procedures.”
We then prepared total cellular lysates from these differentiating CD4+ T cells on days 0, 3, 5, 7, and 14, and the lysates were used to analyze the protein expression of DUSP16. Consistent with the Northern blots, DUSP16 protein was moderately expressed in naïve CD4 T cells and gradually increased when they differentiated toward Th2 cells (Fig. 2B). In contrast, DUSP16 protein disappeared by day 7 following Th1 differentiation (Fig. 2B).
Chromatin Remodeling of dusp16 Gene during the Th2 Type Differentiation
The Th1/Th2 cell differentiation is associated with chromatin remodeling and histone acetylation at Ifng and Il-4 promoters (25). We have reported previously that the transcription of the dusp16 gene in LPS-stimulated macrophages is accompanied by an increased histone acetylation and chromatin remodeling of the dusp16 gene promoter (24). Thus, we assessed the histone acetylation status at the regulatory region of dusp16 gene during Th1/Th2 differentiation. The isolated CD4+ T cells were differentiated under the Th1 or Th2 condition for 0, 3, and 7 days, and the chromatin was extracted for the immunoprecipitation with either anti-acetylhistone H3 or anti-acetylhistone H4 antibody. The presence of dusp16 gene DNA in the immunoprecipitated chromatin was analyzed by PCR using the radiolabeled primers specific to the dusp16 gene promoter region. The ChIP assay result revealed the increased acetylation of both histone H3 and H4 at the dusp16 gene promoter in the chromatin of CD4+ T cells cultured under the Th2 but not under the Th1 condition (Fig. 2C). We then examined the acetylation of histones H3 and H4 at the Ifng and Il-4 promoter as the differentiation controls. In agreement with a previous study (25), the acetylation of histones H3 and H4 on the Ifng gene was found in CD4+ T cells cultured under the Th1 condition, whereas the histones H3/H4 acetylation on the Il-4 gene was found in CD4+ T cells cultured under the Th2 condition (Fig. 2C).
We then addressed whether histone H3/H4 acetylation also occurs at the gene promoter of Pac-1, another MKP. Pac-1 is abundantly expressed in T cells and efficiently dephosphorylates ERK and p38, unlike DUSP16 (32). The same chromatin precipitates were used for the PCR using a pair of primers designed from the regulatory region of the Pac-1 gene. As a result, no remarkable change of histone H3/H4 acetylation was detected at the Pac-1 promoter in either Th1 or Th2 differentiation (Fig. 2C). These data suggest that DUSP16 gene expression is regulated differently in Th1 and Th2 cells.
Effects of Adenoviral Transduction of dusp16 on Th1/Th2-specific Gene Expression
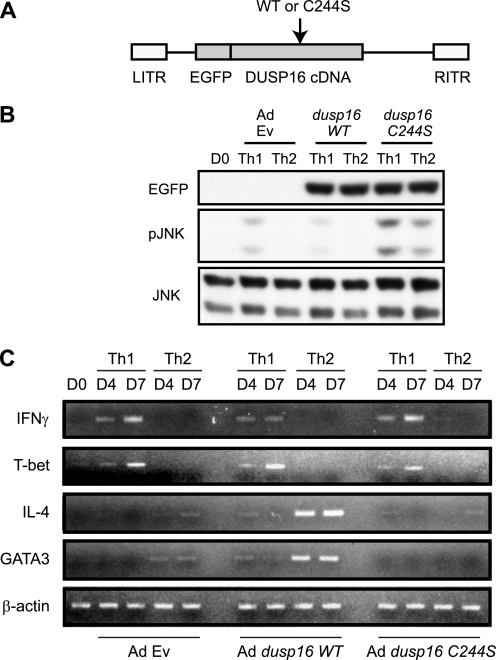
To demonstrate the functional involvement of DUSP16 in the differentiation processes of Th cells, adenoviral gene transduction of the WT and dominant negative (cysteine 244 to serine mutation, thus referred to as the C244S mutation) forms of DUSP16 was utilized. We generated adenoviral vectors containing WT or C244S-mutated DUSP16 cDNA (17) tagged with the N-terminal GFP (Fig. 3A). The adenoviral vector containing GFP cDNA alone was used as a control for the effects of adenoviral transduction itself on the cells. Anti-CD3 mAb-stimulated CD4+ T cells from C57BL/6 mice were infected with one of the adenoviruses for 24 h, followed by the induction of differentiation under either the Th1 or Th2 condition. The expression levels of exogenous DUSP16 protein were evaluated by anti-GFP antibody (Fig. 3B). The phosphorylation of JNK was undetectable in the unstimulated CD4+ T cells (Fig. 3B). In the control infection experiment, phosphorylation of JNK was moderately increased under the Th1 differentiation condition, but not under the Th2 condition. Transduction of dusp16WT decreased the phosphorylation level of JNK in the Th1 condition. In contrast, the JNK phosphorylation level was further increased in Th1 and was detectable in the Th2 condition when the cells were transduced with dusp16C244S. We then analyzed the effects of DUSP16 overexpression on the mRNA expression of the Th1/Th2-specific genes by semiquantitative RT-PCR. As the result, we found that the transduction of naïve CD4+ T cells with the dusp16C244S mildly decreased the mRNA expression of IL-4 and GATA-3 in the Th2 differentiation. In contrast, transduction of naïve CD4+ T cells with the DUSP16WT resulted in the dramatically increased mRNA expression of IL-4 and GATA-3 in the Th2 differentiation (Fig. 3C). On the other hand, the mRNA expression of IFNγ and T-bet under the Th1 condition was modestly decreased by the DUSP16WT, whereas it was increased by the DUSP16C244S (Fig. 3C).
FIGURE 3.
Adenoviral gene transduction of dusp16 into CD4+ T cells affects gene expression of cytokines. A, schematic representation of adenoviral constructs. B, adenoviral transduction of CD4+ T cells with control EGFP-Ad Ev, EGFP-Ad dusp16WT, and EGFP-Ad dusp16C244S. Purified CD4 T cells from C57BL/6 mice were infected with the indicated adenoviruses as described under “Experimental Procedures.” Infected cells were induced to differentiate under the Th1 or Th2 condition. Cell lysate was harvested on day 4 for Western blot analysis using anti-EGFP, phosphor-specific anti-JNK, or anti-JNK antibody. C, purified CD4 T cells from C57BL/6 mice infected with the indicated adenoviruses and induced to differentiate as in B. Cells were harvested on the indicated days, total RNAs were extracted, and IFNγ, T-bet, IL-4, GATA-3, and β-actin expressions were analyzed by semiquantitative RT-PCR.
T Cell Development in Transgenic Mice Expressing a Dominant Negative Form of DUSP16
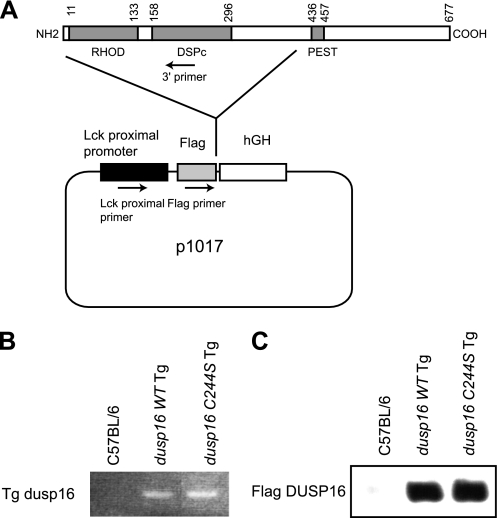
To determine the role of DUSP16 in Th cell differentiation in vivo, we generated T cell-specific Tg mice specifically expressing the WT or the dominant negative (C244S) form of DUSP16 in T cells. The expression constructs encoding the FLAG-tagged truncated DUSP16WT or DUSP16C244S mutant under the control of the lck promoter were microinjected into the C57BL/6 mouse embryos to generate the T cell-specific Tg mice (Fig. 4A). All Tg mice were born healthy and appeared phenotypically normal. Using anti-FLAG mAb, we found that the similar amounts of DUSP16 proteins were observed in T cells of dusp16WT and dusp16C244S Tg mice (Fig. 4C).
FIGURE 4.
Generation of the transgenic mice and genotyping. A, schematic representation of constructs for transgenic mice. B, PCR identifying the expected genotypes of Tg mice using DNA extracted from mouse tails. C, Western blot analysis of total cellular protein (50 μg each) prepared from splenic T cells from the mice using anti-FLAG mAb. The expression levels of DUSP16 in the Tg mice are shown.
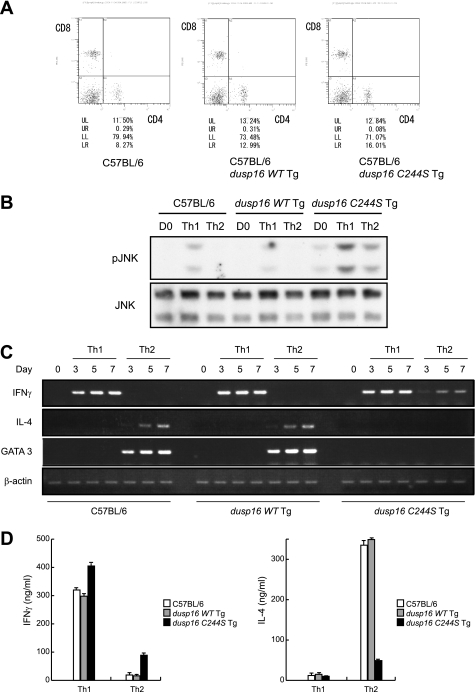
We then determined the development of CD4+ T cells in periphery of these Tg mice. Isolated spleen cells were stained with a combination of phycoerythrin-conjugated anti-CD8α and FITC-conjugated anti-CD4 mAbs and subjected to flow cytometry analysis. The peripheral B/T and CD4/8 ratio seems to be normal in dusp16WT and dusp16C244S Tg mice (Fig. 5A).
FIGURE 5.
Altered CD4+ T cell differentiation in T cell-specific dusp16 Tg mice. A, flow cytometric analyses of peripheral T cells for control C57BL/6, dusp16WT Tg, and dusp16C244S Tg mice. Splenic cells were prepared from 6-week-old mice and stained with phycoerythrin-conjugated anti-CD8α and FITC-conjugated anti-CD4 mAb. The percentages of quadrant fractions are indicated. These data represent one of several repeated experiments. B, purified CD4+ T cells from control C57BL/6 or T cell-specific dusp16 Tg mice differentiated under the Th1 or Th2 condition. Cell lysate was prepared on day 3 for Western blotting with the indicated antibodies. C, purified CD4+ T cells from control C57BL/6 or T cell-specific dusp16 Tg mice differentiated as in B. Total RNA was extracted on the indicated days, and gene expression of IFNγ, IL-4, GATA-3, and β-actin was analyzed by semiquantitative RT-PCR. D, day 7. The differentiated cells in B were collected and restimulated with 2 μg/ml plate-bound anti-CD3 mAb for 24 h. Supernatants were analyzed for IFNγ or IL-4 by ELISA. Data are expressed as mean ± S.D. (error bars) of three independent experiments.
Constitutive Expression of Dominant Negative DUSP16 Inhibited Th2 Responses in Vivo and in Vitro
Naïve CD4+ T cells were purified from spleens of C57BL/6, dusp16WT, or dusp16C244S Tg mice and were differentiated under the Th1 or Th2 condition. The phosphorylation of JNK was measured by Western blotting on days 0 and 3 (Fig. 5B). In the control mouse CD4+ T cell-phosphorylated JNK was undetectable in the unstimulated condition and moderately increased in Th1 but not in Th2 differentiation. The JNK phosphorylation level in Th1 differentiation was lower in CD4+ T cells from dusp16WT Tg mice. In contrast, phosphorylated JNK was detectable at a low level in unstimulated dusp16C244S Tg CD4+ T cells. The JNK phosphorylation level in dusp16C244S Tg CD4+ T cells was increased significantly in Th1 and mildly in Th2 differentiation. Total RNAs were extracted on days 0, 3, 5, or 7, and mRNA of IFNγ, IL-4, and GATA-3 was analyzed by semiquantitative RT-PCR. Under the Th1 condition, CD4+ T cells from C57BL/6, dusp16WT, and dusp16C244S Tg expressed similar amounts of IFNγ mRNA (Fig. 5C). In contrast, IL-4 and GATA-3 mRNA expression by CD4+ T cells of dusp16C244S Tg cultured under the Th2 condition was significantly decreased (Fig. 5C). Interestingly, CD4+ T cells from dusp16C244S Tg cultured under the Th2 condition expressed significant amounts of IFNγ mRNA (Fig. 5C). On the other hand, GATA-3 mRNA expression by CD4+ T cells of dusp16WT Tg cultured under the Th2 condition was moderately increased (Fig. 5C). We then restimulated these cells with immobilized anti-CD3 mAb and examined IFNγ and IL-4 production into the culture supernatants by ELISA. The Th2-differentiated cells from dusp16C244S Tg produced 85% less IL-4 than that from normal background controls (Fig. 5D). These results were consistent with the adenoviral transduction data and further demonstrated that the forced expression of WT DUSP16 could enhance the course of Th2 differentiation, although it alone did not induce the onset of Th2 differentiation. Conversely, the forced expression of the dominant negative DUSP16 impaired Th2 differentiation.
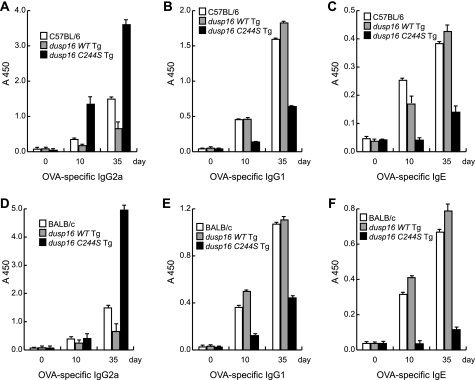
We further analyzed the abilities of dusp16WT and dusp16C244S Tg mice to generate the antigen-specific immune response in vivo. The dusp16WT or dusp16C244S Tg (C57BL/6 background) and C57BL/6 control mice were immunized with OVA emulsified in complete Freund's adjuvant. The production of OVA-specific IgG2a, IgG1, and IgE was analyzed at 0, 10, and 35 days after immunization. As shown in Fig. 6A, immunized dusp16WT Tg produced OVA-specific IgG2a isotype antibody at lower amounts than C57BL/6 control, whereas similar amounts of OVA-specific IgG1 and IgE antibodies were produced. In contrast, immunized dusp16C244S Tg mice produced dramatically higher amounts of OVA-specific IgG2a than those of control mice. This increased OVA-specific IgG2a production was accompanied by a decrease in production of OVA-specific IgG1 and IgE antibodies (Fig. 6, A–C). Moreover, we established T cell-specific dusp16WT or dusp16C244S Tg mice under the BALB/c background and immunized them together with normal BALB/c control mice. We observed similar patterns of OVA-specific IgG2a, IgG1, and IgE generated by the dusp16 Tg mice under the BALB/c background (Fig. 6, D–F).
FIGURE 6.
Effects of WT and dominant negative forms of DUSP16 on antigen-specific T helper cells response in vivo. OVA-specific immunoglobulin isotype production in the serum of immunized mice was analyzed by ELISA. A–C, C57BL/6 control and C57BL/6 background Tg mice were immunized with OVA absorbed in complete Freund's adjuvant, serum was collected before (day 0) and after immunization (days 10 and 35) and OVA-specific IgG2a (A), OVA-specific IgG1 (B) and OVA-specific IgE (C) were analyzed. D–F, BALB/c and BALB/c background Tg mice were also immunized as described above, and the production of OVA-specific IgG2a (D), OVA-specific IgG1 (E), and OVA-specific IgE (F) are shown. Data are expressed as mean ± S.D. (error bars) from three mice/group and are representative of three independent experiments.
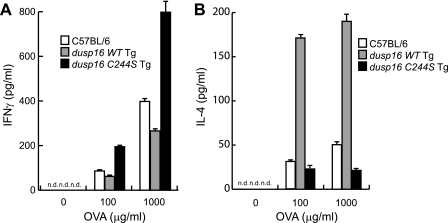
We then isolated CD4+ T cells from spleens of the immunized C57BL/6 background dusp16 Tg mice and cultured them for 3 days with plate-bound anti-CD3 and anti-CD 28 mAbs. The cells were restimulated with OVA in vitro, and the secreted IFNγ and IL-4 in the cultured supernatant were measured by ELISA. We found that CD4+ T cells from the immunized dusp16WT Tg produced slightly lower amount of IFNγ than the C57BL/6 control, whereas CD4+ T cells from the immunized dusp16C244S Tg produced a significantly higher amount of IFNγ than the C57BL/6 control (Fig. 7A). On the other hand, CD4+ T cells from dusp16WT Tg mice produced a marked higher amount of IL-4 than control mice, whereas CD4+ T cells from dusp16C244S Tg mice slightly decreased IL-4 production (Fig. 7B). Moreover, we observed similar results from BALB/c background dusp16 Tg mice (data not shown).
FIGURE 7.
Effects of WT and dominant negative forms of DUSP16 on antigen-specific IFNγ and IL-4 production by CD4+ T cells. CD4+ T cells were isolated from immunized mice and cultured in vitro for 3 days. Then, cells were collected and stimulated with indicated concentrations of antigen (OVA). Production of IFNγ (A) and IL-4 (B) in the supernatants of antigen-stimulated cells was determined by ELISA. Data are mean ± S.D. (error bars) from three mice/group and are representative of three independent experiments. n.d., not detected.
DISCUSSION
By using both in vivo and in vitro experimental models, the present study has demonstrated an important role for DUSP16, a MKP that preferentially dephosphorylates JNK in the differentiation of Th cells into Th1/Th2 subsets. DUSP16 was first cloned from a macrophage cDNA library and was demonstrated to play an essential role in the down-regulation of JNK activity in LPS-stimulated macrophages (17). Our present data indicate that DUSP16 is also moderately expressed in naïve CD4+ T cells. The expression level of DUSP16 in CD4+ T cells significantly increases when cells differentiate into Th2 lineage, whereas it diminishes when cells differentiate into Th1 cells (Fig. 2, A and B). In the analysis of T cell clones, DUSP16 is induced by LPS, IL-2, and IL-4 in a Th2 clone, but not in a Th1 clone (Fig. 1). In contrast, two other MKPs, PAC-1 and MKP-1, which efficiently dephosphorylate all three types of MAPKs, are preferentially expressed in the Th1 clone. M3/6, an MKP structurally similar to DUSP16, is expressed at higher levels in the Th2 clone. The Th2 selectivity of M3/6, however, is less significant than that of DUSP16. Thus, the Th2-selective expression pattern of DUSP16 seems rather unique among MKP family members.
The difference of expression levels of DUSP16 in Th1/Th2 differentiation is correlated with the different acetylation levels of histone H3 and H4 in the transcriptional regulatory region of the dusp16 gene between Th1 and Th2 subsets (Fig. 2C). We have previously reported that the increased accessibility of the dusp16 gene promoter by histone acetylation is associated with increased DUSP16 expression in LPS-stimulated macrophages, in which two transcriptional regulatory elements, E-Box and CRE (cAMP-responsive element), play important roles (24). Further investigation is needed to demonstrate whether the same regulatory elements are also functional in the regulation of DUSP16 expression in Th cells. Notably, an essential role of CRE has been reported in the conserved noncoding sequence 1, which regulates the expression of IL-4, IL-5, and IL-13 in Th2-type cells, by recruiting CRE-binding protein (CREB)-CREB-binding protein (CBP) complex (33).
Because DUSP16 preferentially inactivates JNKs (17), the Th2-selective expression of DUSP16 may be considered as a regulatory mechanism of JNK activity in Th1/Th2 subpopulations. Previous reports have demonstrated that antigen stimulation causes rapid activation of JNKs in Th1 cells, whereas it does not up-regulate JNK activity in Th2 cells (13, 34). However, the mechanisms of this difference are not well revealed. Because a similar level of JNK protein is present in Th1 and Th2 cells (13), the different JNK activity between Th1 and Th2 is caused by the activation levels of JNKs. We hypothesize, based on our current results, that the difference of DUSP16 expression levels may cause the different degrees of JNK activity between Th1 and Th2 cells.
Our in vitro model of adenoviral transduction illustrates functional roles of DUSP16 in Th1- and Th2-type cells (Fig. 3C). In CD4+ T cells differentiating under the Th1 condition, exogenous overexpression of DUSP16 WT modestly decreased mRNAs of IFNγ and T-bet and slightly increased mRNA levels of IL-4 and GATA-3 compared with the control transduction. Overexpression of DUSP16C244S under the Th1 condition modestly increased the mRNA levels of IL-4 and GATA-3. Although these changes were consistently observed in repeated experiments, they were typically minimal. On the other hand, much more significant results were observed when the transduction was performed under the condition of Th2 differentiation. In CD4+ T cells differentiating under the Th2 condition, exogenous overexpression of DUSP16 WT significantly increased the mRNA levels of IL-4 and GATA-3 compared with the control transduction (Fig. 3C). In contrast, overexpression of DUSP16C244S effectively diminished GATA-3 mRNA expression. Notably, DUSP16C244S overexpression did not induce Th1 marker mRNAs under the Th2 differentiation condition. Thus, according to these adenoviral transduction results, it appears that DUSP16 strongly enhances Th2-type differentiation, whereas it only minimally affects the course of Th1-type differentiation in vitro.
The same type of in vitro differentiation experiment using CD4+ T cells from dusp16 Tg C57BL/6 mice produced similar but somewhat different results (Fig. 5C). Consistent with the adenoviral transduction experiment, CD4+ T cells from either dusp16WT or dusp16C244S Tg mice are not very different in Th1/Th2 marker expression from those of the control mice under the condition of Th1 differentiation. On the other hand, under the condition of Th2 differentiation, CD4+ T cells from dusp16WT Tg mice showed enhanced GATA-3 mRNA response, whereas CD4+ T cells from dusp16C244S Tg mice showed a significantly diminished response of IL-4 and GATA-3 compared with those from the control mice. These results are consistent with the results of the adenoviral transduction experiments. Unlike the adenoviral transduction experiment, however, IFNγ mRNA and protein were induced in the CD4+ T cells from dusp16C244S Tg even under the condition of Th2-type differentiation (Fig. 5, C and D). Although we currently do not know what exactly caused the difference between the adenoviral transduction and the Tg experiments, it seems possible that naïve CD4+ cells have somewhat different phenotypes without antigen stimulation (presumably more Th1-biased) in the dusp16C244S Tg mice.
The distribution patterns of the serum antigen-specific Igs of the dusp16 Tg C57BL/6 mice after OVA sensitization (Fig. 6A-C) were consistent with the characteristics of the CD4+ T cells discussed above, indicating that DUSP16 expressed in CD4+ T cells is functionally important in the Th1/Th2 balance of the antigen-specific immune responses in vivo. The phenotype was more pronounced for the dusp16C244S Tg C57BL/6 mice, in which the serum concentration of antigen-specific IgG2a was markedly increased, whereas antigen-specific IgG1and IgE were significantly decreased. Very similar results were observed in Tg mice of the BALB/c background (Fig. 6, D–F), indicating it is not a strain-specific finding. Furthermore, cytokine production data from antigen-stimulated CD4+ T cells (Fig. 7) also confirmed the Th1 dominance of dusp16C244S Tg mice and the Th2 dominance of the dusp16WT Tg mice.
According to previous reports, disruption of either the JNK1 or JNK2 gene does not cause abnormalities in the ratio of CD4+ to CD8+ T cell numbers, proliferative responses of CD4+ T cells, or IL-2 production (13, 14, 35). However, both JNK1−/− and JNK2−/− mice show a biased Th1/Th2 balance toward Th2 dominance. Interestingly, these two mice show rather different cytokine profiles. More specifically, IL-4 and IL-5 production by Th2 cells was dramatically increased, whereas IFNγ production by Th1 cells was normal in JNK1−/− mice (14). On the other hand, IFNγ production by Th1 cells was significantly reduced, but IL-4 production by Th2 cells was normal in JNK2−/− mice (13). The exact cause of these differences has not been resolved yet. Because DUSP16 dephosphorylates and inactivates both JNK1 and JNK2 efficiently (17), it seems reasonable that T cell-specific dusp16 Tg mice showed abnormal profiles of cytokine production in both Th1 and Th2 cells (Figs. 3C and 5, C and D). It should be noted, however, that Th2-type differentiation is affected more significantly than Th1 in DUSP16 Tg mice. It may be possible that JNK1 activity is generally dominant over JNK2 activity in T helper cells. Elucidation of this finding obviously needs further investigation.
This work was supported in part by grants from Ministry of Education, Science, and Culture of the Japanese Government.
- Th
- T helper
- DSP
- dual specificity phosphatase
- EGFP
- enhanced green fluorescent protein
- GATA-3
- GATA-binding protein 3
- iTreg
- induced regulatory T cells
- MKP
- MAPK phosphatase
- OVA
- ovalbumin
- Tg
- transgenic.
REFERENCES
- 1. Mosmann T. R., Cherwinski H., Bond M. W., Giedlin M. A., Coffman R. L. (1986) J. Immunol. 136, 2348–2357 [PubMed] [Google Scholar]
- 2. Paul W. E., Seder R. A. (1994) Cell 76, 241–251 [DOI] [PubMed] [Google Scholar]
- 3. Ouyang W., Kolls J. K., Zheng Y. (2008) Immunity 28, 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu J., Paul W. E. (2008) Blood 112, 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murphy K. M., Reiner S. L. (2002) Nat. Rev. Immunol. 2, 933–944 [DOI] [PubMed] [Google Scholar]
- 6. Seder R. A., Gazzinelli R., Sher A., Paul W. E. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10188–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seder R. A., Paul W. E. (1994) Annu. Rev. Immunol. 12, 635–673 [DOI] [PubMed] [Google Scholar]
- 8. Szabo S. J., Kim S. T., Costa G. L., Zhang X., Fathman C. G., Glimcher L. H. (2000) Cell 100, 655–669 [DOI] [PubMed] [Google Scholar]
- 9. Ouyang W., Löhning M., Gao Z., Assenmacher M., Ranganath S., Radbruch A., Murphy K. M. (2000) Immunity 12, 27–37 [DOI] [PubMed] [Google Scholar]
- 10. Zheng W., Flavell R. A. (1997) Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 11. Glimcher L. H., Murphy K. M. (2000) Genes Dev. 14, 1693–1711 [PubMed] [Google Scholar]
- 12. Rincón M., Enslen H., Raingeaud J., Recht M., Zapton T., Su M. S., Penix L. A., Davis R. J., Flavell R. A. (1998) EMBO J. 17, 2817–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang D. D., Conze D., Whitmarsh A. J., Barrett T., Davis R. J., Rincón M., Flavell R. A. (1998) Immunity 9, 575–585 [DOI] [PubMed] [Google Scholar]
- 14. Dong C., Yang D. D., Wysk M., Whitmarsh A. J., Davis R. J., Flavell R. A. (1998) Science 282, 2092–2095 [DOI] [PubMed] [Google Scholar]
- 15. Camps M., Nichols A., Arkinstall S. (2000) FASEB J. 14, 6–16 [PubMed] [Google Scholar]
- 16. Masuda K., Shima H., Watanabe M., Kikuchi K. (2001) J. Biol. Chem. 276, 39002–39011 [DOI] [PubMed] [Google Scholar]
- 17. Matsuguchi T., Musikacharoen T., Johnson T. R., Kraft A. S., Yoshikai Y. (2001) Mol. Cell. Biol. 21, 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanoue T., Yamamoto T., Maeda R., Nishida E. (2001) J. Biol. Chem. 276, 26629–26639 [DOI] [PubMed] [Google Scholar]
- 19. Nakayama T., Kubo R. T., Kubo M., Fujisawa I., Kishimoto H., Asano Y., Tada T., Asao Y. (1988) Eur. J. Immunol. 18, 761–765 [DOI] [PubMed] [Google Scholar]
- 20. Venkataraman C., Schaefer G., Schindler U. (2000) J. Immunol. 165, 632–636 [DOI] [PubMed] [Google Scholar]
- 21. Kaplan M. H., Schindler U., Smiley S. T., Grusby M. J. (1996) Immunity 4, 313–319 [DOI] [PubMed] [Google Scholar]
- 22. Afkarian M., Sedy J. R., Yang J., Jacobson N. G., Cereb N., Yang S. Y., Murphy T. L., Murphy K. M. (2002) Nat. Immunol. 3, 549–557 [DOI] [PubMed] [Google Scholar]
- 23. Szabo S. J., Dighe A. S., Gubler U., Murphy K. M. (1997) J. Exp. Med. 185, 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Musikacharoen T., Yoshikai Y., Matsuguchi T. (2003) J. Biol. Chem. 278, 9167–9175 [DOI] [PubMed] [Google Scholar]
- 25. Avni O., Lee D., Macian F., Szabo S. J., Glimcher L. H., Rao A. (2002) Nat. Immunol. 3, 643–651 [DOI] [PubMed] [Google Scholar]
- 26. Matsuguchi T., Takagi K., Musikacharoen T., Yoshikai Y. (2000) Blood 95, 1378–1385 [PubMed] [Google Scholar]
- 27. Reiner S. L., Zheng S., Corry D. B., Locksley R. M. (1993) J. Immunol. Methods 165, 37–46 [DOI] [PubMed] [Google Scholar]
- 28. Reiner S. L., Zheng S., Corry D. B., Locksley R. M. (1994) J. Immunol. Methods 173, 133 [DOI] [PubMed] [Google Scholar]
- 29. Mullen A. C., High F. A., Hutchins A. S., Lee H. W., Villarino A. V., Livingston D. M., Kung A. L., Cereb N., Yao T. P., Yang S. Y., Reiner S. L. (2001) Science 292, 1907–1910 [DOI] [PubMed] [Google Scholar]
- 30. Matsuguchi T., Chiba N., Bandow K., Kakimoto K., Masuda A., Ohnishi T. (2009) J. Bone Miner. Res. 24, 398–410 [DOI] [PubMed] [Google Scholar]
- 31. Ishimitsu R., Nishimura H., Yajima T., Watase T., Kawauchi H., Yoshikai Y. (2001) J. Immunol. 166, 1991–2001 [DOI] [PubMed] [Google Scholar]
- 32. Chu Y., Solski P. A., Khosravi-Far R., Der C. J., Kelly K. (1996) J. Biol. Chem. 271, 6497–6501 [DOI] [PubMed] [Google Scholar]
- 33. Strempel J. M., Vercelli D. (2007) J. Biol. Chem. 282, 3738–3746 [DOI] [PubMed] [Google Scholar]
- 34. Rincón M., Dérijard B., Chow C. W., Davis R. J., Flavell R. A. (1997) Genes Funct. 1, 51–68 [DOI] [PubMed] [Google Scholar]
- 35. Sabapathy K., Hu Y., Kallunki T., Schreiber M., David J. P., Jochum W., Wagner E. F., Karin M. (1999) Curr. Biol. 9, 116–125 [DOI] [PubMed] [Google Scholar]