Abstract
Enzymes that scan single-stranded (ss) DNA have been studied far less extensively than those that scan double-stranded (ds) DNA. Activation-induced deoxycytidine deaminase (AID) deaminates C to U on single-stranded DNA to initiate immunological diversity. Except for processive deaminations favoring WRC hot motifs (W = (A/T) and R = (G/C)), the rules governing AID scanning remain vague. Here, we examine the patterns of deaminations on naked single-stranded DNA and during transcription of dsDNA by embedding cassettes containing combinations of motifs within a lacZ mutational reporter gene. Deaminations arise randomly, spatially distributed as isolated events and in clusters. The deamination frequency depends on the motif and its surrounding sequence. We propose a random walk model that fits the data well, having a deamination probability of 1–7% per motif encounter. We suggest that inefficient, haphazard deamination produces antibody diversity associated with AID.
Keywords: Antibodies, DNA Damage, DNA Enzymes, DNA-Protein Interaction, DNA Repair, Immunology, Mutagenesis Mechanisms, Nucleic Acid Enzymology, Protein-DNA Interaction, Activation-induced Deoxycytidine Deaminase
Introduction
An understanding of AID2 scanning and C deamination is central to deciphering the many mysteries surrounding its essential role in initiating antibody diversity. AID deaminates C to U in B cells in variable (V) and switch (S) regions of immunoglobulin genes that are undergoing active transcription. Deaminations in the V region are responsible for initiating somatic hypermutation, and those in the S region initiate class switch recombination (1–3). These regulatory processes are multifaceted with poorly defined basic properties. How is AID targeted to the V and S regions, for example? What are its principal partnering proteins and regulatory elements? Why does AID fail to target concurrently transcribed non-V and non-S regions? Or if AID were to attack other transcribed regions, do these undergo efficient base excision and mismatch repair (1–3)? AID is also involved in restriction of retroviral transposition (4) and in editing of hepatitis B viral DNA (5). Recently, AID has taken on an entirely new “life,” appearing to play a central role in active demethylation processes in stem cells, involving the deamination of 5-methylcytosine (6–9). These recent developments and open questions about AID are discussed in several reviews (10–13).
We have previously shown that AID acts on ssDNA processively by catalyzing numerous C to U deaminations in trinucleotide target motifs on the same substrate molecule prior to acting on another DNA substrate (14, 15). WRC hot motifs (W = (A/T) and R = (A/G)) are favored targets for deamination over SYC cold motifs (S = (G/C) and Y = (T/C)) (14–17). In light of the uncertainties of how AID functions in complex biological systems, as an initial step it is both essential and timely to study the behavior of AID in the simplest cases, when it acts on naked ssDNA and on actively transcribed dsDNA. It is timely because AID is the subject of numerous recent studies, and enzymes that scan ssDNA are poorly understood compared with those that scan dsDNA. It is essential because a fundamental knowledge of how AID scans DNA in the absence of putative binding proteins, e.g. RNA exosome proteins or Spt5 (18, 19), is a prerequisite to understanding the behavior of AID in more complex biological systems.
In this study, we examine the mechanism used by AID to scan ssDNA and dsDNA undergoing transcription. The processive behavior of dsDNA-scanning enzymes has been studied with single molecule resolution (20–26) and in bulk studies that typically measure reactions occurring at target sites or motifs located at varying distances on the same DNA substrate (27–29). Here, we have taken an approach that detects processive AID-catalyzed C deaminations occurring at large numbers of targets, including combinations of hot, cold, and neutral motifs on the same ssDNA substrate. We enumerate the distribution of deaminations along individual ssDNA molecules, with each substrate acted on by a single AID molecule. The deaminated motifs occur as isolated singletons and as clusters of various lengths and spaces between clusters. We have determined how neighboring motifs alter C deamination patterns and probabilities and have formulated a random walk model to analyze the statistical properties of AID-catalyzed deamination. The model is indispensable in dissecting scanning and specifically how individual mutations along with mutational clusters are likely to arise while AID moves along the DNA and to determine deamination efficiencies and motion distances based on a model fit to the data.
Uracil DNA glycosylase and formamidopyrimidine DNA glycosylases that scan dsDNA to find and excise rare aberrant bases are catalytically efficient (23, 29). In contrast, when scanning ssDNA, AID encounters many C deamination motifs, while leaving many of these untouched. We suggest that a catalytically inefficient enzyme is well suited for generating antibody diversity.
EXPERIMENTAL PROCEDURES
Substrates and Enzymes
Oligonucleotides containing various AID C deamination trinucleotide motifs and their complementary strands were synthesized on an Applied Biosystems 3400 DNA/RNA synthesizer, purified, and 5′-end phosphorylated using T4 polynucleotide kinase (Affymetrix Corp.). Double-stranded DNA cassettes, containing a 5′-TTAA overhang, were formed by annealing each oligonucleotide with its complementary strand at a 1:1 molar ratio. M13mp2 phage DNA constructs, with either two or three cassettes, were constructed after several ligation steps. First, the dsDNA cassette with 30 alternating AAC and CAG motifs was ligated into a unique EcoRI site of M13mp2 phage DNA in a lacZα mutational reporter gene creating a construct with a single inserted cassette (AAC AGC)15. In the next ligation, dsDNA cassettes containing either 30 alternating AAC and GTC motifs, 30 alternating AAC and GAC motifs, or AAC and AGC motifs with an irregular composition were inserted into a newly formed EcoRI site on the right-hand side of the (AAC AGC)15 cassettes. The construct containing two cassettes with single AGC motifs (AGC32-AGC24) was constructed by a similar procedure. The recombinant M13 phage DNAs were used to prepare gap substrates located within an ssDNA region containing the inserted cassettes flanked by the lacZα sequence (270 nt on the 5′-side and 95 nt on the 3′-side) as described previously (14). The construct (AAC AGC)15-(AAC GTC)15 containing a 24-nt dsDNA region (see Fig. 3d) was formed by annealing a complementary 24-nt oligonucleotide with the gapped DNA at 3:1 molar ratio. The constructs for transcription-dependent deamination were constructed by replacing 18 nt (positions −89 to −72) upstream of lacZα with the T7 promoter sequence using a Phusion site-directed mutagenesis kit (New England Biolabs).
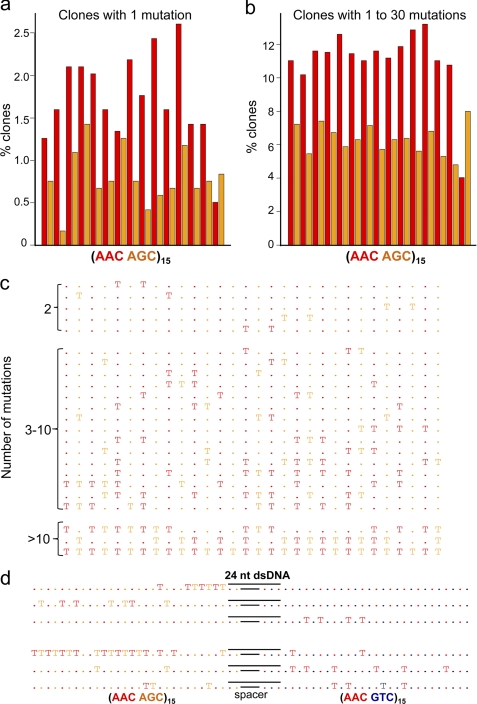
FIGURE 3.
Mutation spectra for AAC hot-AGC hot′ motifs. The spectra were obtained by sequencing individual clones from the constructs (Fig. 1, b–d) and combining the data from all of the left-hand cassettes composed of alternating AAC and AGC motifs. a, mutation spectrum for clones with a single deamination. b, mutation spectrum for clones with 1–30 deaminations. c, representative clones having 2, 3–10, and >10 mutations. The bars shown in a and b give the percent of clones with mutations at each AAC motif (red) or AGC motif (orange). c, T shown in red or orange denotes deaminated AAC and AGC motifs, respectively; the dots in red or orange denote nondeaminated AAC and AGC motifs, respectively. d, representative mutated clones on the construct composed of AAC hot-AGC hot′ and AAC hot-GTC cold motifs containing a 24-nt dsDNA region. Clones with mutations on both sides of the dsDNA region imply jumping.
Wild type GST-tagged AID protein was expressed in baculovirus Sf9-infected insect cells and purified as described previously (15). Purified GST-tagged AID was dialyzed in a buffer containing 20 mm Tris-HCl (pH 7.5), 25 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, and 10% glycerol and stored at −70 °C.
Deamination Reactions
Time course reactions for AID-catalyzed deaminations of C to U were performed as follows. GST-tagged AID (300 ng) was preactivated by incubation with 300 ng of RNase A (Sigma) (30) containing 10 mm Tris (pH 8.0), 1 mm dithiothreitol, and 1 mm EDTA in a total volume of 90 μl for 2 min at 37 °C. The deamination reactions were initiated by transferring the entire enzyme mixture into a pre-warmed tube containing 3 μg of a gapped DNA substrate in 90 μl of a buffer solution containing 10 mm Tris (pH 8.0), 1 mm dithiothreitol, and 1 mm EDTA. In deamination experiments performed in the presence of salt, 150 mm NaCl was included in the reaction. Following incubations at 37 °C for 15, 30, and 45 s and 1, 2, and 5 min, 30-μl aliquots were withdrawn and quenched by a double extraction with phenol/chloroform/isoamyl alcohol (25:24:1). The DNA products at each time point were transfected into uracil glycosylase-deficient (ung−) E. coli cells and plated on α-complementation host cells in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) and isopropyl β-d-1-thiogalactopyranoside as described previously (14, 15). T7 RNA polymerase transcription-dependent AID deamination was measured using a covalently closed circular dsDNA M13mp substrate containing the inserted CAG32-CAG24 cassettes with an upstream T7 promoter. In a typical reaction (30 μl volume), dsDNA substrate (50 ng) was incubated with T7 RNA polymerase (1 μl), AID (300 ng), and RNase A (300 ng) in a reaction buffer containing HEPES (50 mm, pH 7.5), dithiothreitol (1 mm), MgCl2 (10 mm), and rNTPs (250 μm) at 37 °C for 30 min. Reactions were terminated by extracting twice with phenol/chloroform/isoamyl alcohol (25:24:1). The DNA reaction products were transfected into ung− E. coli competent cells and plated on α-complementation host cells. The occurrence of C deaminations within the inserted cassettes create stop codon(s) within the lacZα reading frame giving rise to clear mutant M13 plaques. Mutant M13 phage DNA was isolated, and the inserted cassettes and the lacZα portion on the 3′-side of the cassettes were sequenced. C→U deaminations were detected as C→T transition mutations.
Statistical Analysis and Modeling
We have written a computer program to simulate the various models. We tested these models and their parameter values via the parametric bootstrap. For each set of parameter values for each model, and for each number of mutations per clone between 2 and 10, we separately simulated 10,000 independent clones. We then calculated the likelihood of a clone from the data set by comparing the statistics for this clone to the statistics for those simulations with the same number of mutations per clone. For the distance between mutations statistics, we assumed the distances were independent. To calculate the likelihood of a clone, we measured the distances between mutations in this clone, found the fraction of times each distance was found in the simulations (for one particular model and set of parameter values these fractions are shown in Table 2), and multiplied these fractions together. The calculation was different for the mutation cluster size statistic (we did not use the fractions in Table 1), because clusters were not independent, e.g. if there are two mutations per clone and one mutation is a singleton then the other mutation has to be a singleton too. For this statistic, we computed the unordered set of cluster sizes found in the clone and counted the fraction of simulations that have this same set. To calculate the likelihood of the entire data set, we multiplied the likelihoods for each of the clones. To test whether these data were consistent with both the model and the parameter values, we independently simulated the same number of clones with the same number of mutations per clone as in the actual data set. We then calculated the likelihood of this simulated data set. We repeated this process many times to generate a distribution of simulated likelihoods and compared the likelihood of the actual data to this distribution. We repeated the entire process for different parameter values and for different models.
TABLE 2.
Distance between mutated motifs in clones with 2–10 mutations in the combined (AAC AGC)15 cassettes
| Mutations/clone | Distribution of distance between mutated motifs (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5–10 | 11–15 | 16–20 | 21–29 | ||
| 2 | Data (209)a | 2.4 | 11.0 | 5.7 | 9.1 | 4.8 | 27.8 | 19.1 | 12.4 | 7.7 |
| 2 | Simulatedb | 8.5 | 9.0 | 6.7 | 7.9 | 6.1 | 26.3 | 16.2 | 11.6 | 7.7 |
| 3 | Data (113) | 8.8 | 8.8 | 5.3 | 9.3 | 8.4 | 31.4 | 20.4 | 5.8 | 1.8 |
| 3 | Simulated | 11.5 | 11.4 | 9.1 | 9.0 | 6.9 | 27.9 | 14.5 | 7.0 | 2.7 |
| 4 | Data (64) | 14.6 | 14.6 | 6.8 | 8.9 | 6.3 | 31.8 | 16.1 | 1.0 | 0.0 |
| 4 | Simulated | 13.8 | 14.5 | 10.4 | 10.7 | 7.5 | 27.1 | 11.3 | 3.9 | 0.8 |
| 5 | Data (55) | 15.9 | 17.7 | 9.5 | 12.7 | 6.4 | 25.9 | 10.5 | 1.4 | 0.0 |
| 5 | Simulated | 17.3 | 17.0 | 11.4 | 11.6 | 7.8 | 24.8 | 8.0 | 2.0 | 0.2 |
| 6 | Data (37) | 15.1 | 20.5 | 8.6 | 11.4 | 16.2 | 23.8 | 3.8 | 0.5 | 0.0 |
| 6 | Simulated | 20.3 | 18.8 | 12.8 | 11.6 | 8.0 | 22.1 | 5.4 | 0.9 | 0.0 |
| 7 | Data (27) | 16.7 | 23.5 | 13.6 | 8.6 | 7.4 | 27.8 | 2.5 | 0.0 | 0.0 |
| 7 | Simulated | 23.4 | 20.8 | 13.3 | 12.0 | 7.8 | 18.9 | 3.5 | 0.4 | 0.0 |
| 8 | Data (19) | 21.8 | 21.8 | 11.3 | 12.0 | 12.0 | 20.3 | 0.8 | 0.0 | 0.0 |
| 8 | Simulated | 26.5 | 22.7 | 14.0 | 11.8 | 7.1 | 15.6 | 2.1 | 0.1 | 0.0 |
| 9 | Data (16) | 25.8 | 27.3 | 12.5 | 9.4 | 5.5 | 19.5 | 0.0 | 0.0 | 0.0 |
| 9 | Simulated | 29.5 | 24.2 | 14.4 | 11.4 | 6.7 | 12.7 | 1.2 | 0.0 | 0.0 |
| 10 | Data (12) | 30.6 | 25.9 | 12.0 | 13.9 | 5.6 | 12.0 | 0.0 | 0.0 | 0.0 |
| 10 | Simulated | 32.9 | 25.1 | 14.2 | 10.9 | 6.0 | 10.2 | 0.6 | 0.0 | 0.0 |
a Numbers in parentheses are the number of sequenced clones.
b Simulated refers to the modified random walk model (see text) with the following parameters: AID-catalyzed deamination efficiency in AAC motif = 3%; average slide distance obtained from geometric distribution = 30 nt (10 motifs).
TABLE 1.
Mutation cluster patterns in clones with 2–10 mutations in the combined (AAC AGC)15 cassettes
| Mutations/clone | Cluster | Mutation clusters (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 2 | Data (209)a | 92.8 | 7.2 | ||||||||
| 2 | Simulatedb | 90.4 | 9.6 | ||||||||
| 3 | Data (113) | 87.6 | 11.4 | 1.0 | |||||||
| 3 | Simulated | 83.7 | 14.6 | 1.7 | |||||||
| 4 | Data (64) | 77.0 | 19.0 | 3.0 | 1.0 | ||||||
| 4 | Simulated | 77.5 | 18.5 | 3.5 | 0.5 | ||||||
| 5 | Data (55) | 71.1 | 23.9 | 2.5 | 2.0 | 0.5 | |||||
| 5 | Simulated | 71.0 | 21.9 | 5.6 | 1.3 | 0.1 | |||||
| 6 | Data (37) | 67.3 | 24.4 | 7.1 | 1.3 | 0.0 | 0.0 | ||||
| 6 | Simulated | 65.8 | 23.4 | 7.9 | 2.2 | 0.5 | 0.1 | ||||
| 7 | Data (27) | 62.1 | 25.0 | 11.3 | 1.6 | 0.0 | 0.0 | 0.0 | |||
| 7 | Simulated | 60.4 | 25.1 | 9.7 | 3.4 | 1.1 | 0.3 | 0.0 | |||
| 8 | Data (19) | 57.4 | 29.8 | 7.4 | 4.3 | 1.1 | 0.0 | 0.0 | 0.0 | ||
| 8 | Simulated | 55.3 | 25.5 | 11.4 | 4.8 | 2.0 | 0.7 | 0.2 | 0.0 | ||
| 9 | Data (16) | 48.7 | 28.9 | 11.8 | 6.6 | 2.6 | 1.3 | 0.0 | 0.0 | 0.0 | |
| 9 | Simulated | 50.6 | 25.6 | 13.0 | 6.2 | 2.9 | 1.1 | 0.4 | 0.1 | 0.0 | |
| 10 | Data (12) | 49.2 | 25.4 | 11.9 | 3.4 | 6.8 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | Simulated | 45.5 | 25.7 | 14.4 | 7.3 | 4.0 | 1.8 | 0.8 | 0.4 | 0.1 | 0.0 |
a Numbers in parentheses are the number of sequenced clones.
b Simulated refers to the modified random walk model (see text) with the following parameters: AID-catalyzed deamination efficiency in AAC motif = 3%; average slide distance obtained from geometric distribution = 30 nt (10 motifs).
RESULTS
Slow Time Evolution of AID-catalyzed C Deamination
We used human AID, containing an N-terminal GST tag, purified from baculovirus-infected insect cells (14, 15). DNA clones containing a lacZ reporter gene acted on by a single AID molecule were found previously to contain widely varying numbers of mutations, ranging from 1 to 80 mutations per clone (14, 15). The mutations, which were initiated by AID-catalyzed deamination of C→U, were distributed either as single isolated mutations (singletons) or as mutational clusters of variable lengths (14, 15). The presence of clusters containing as many as five consecutive triplet target motifs and as many as four consecutive deaminated C residues (14, 15) indicated that clustering is likely to occur when AID slides along ssDNA. Our objective was to explore the properties of a stochastic scanning mechanism in which C deaminations either may or may not occur when a motif is encountered by AID and to determine motif deamination efficiencies per encounter. What was required was an ssDNA substrate containing a defined series of detectable target motifs and a mathematical model to analyze the stochastic nature of the scanning and deamination events.
We have synthesized a series of ssDNA substrates containing combinations of WRC hot motifs and non-WRC motifs inserted as in-frame cassettes embedded within a lacZ reporter gene (Fig. 1). The conversion of C→U in any of these motifs generated a stop codon in lacZα, causing a lacZ+ → lacZ− mutation, which results in a colorless M13 mutant phage plaque. We considered two hot motifs AAC and AGC, the latter one we call hot′, one cold motif GTC, and one neutral (i.e. neither WRC nor SYC) GAC motif. To ensure that virtually all mutated cassettes were acted on by no more than a single AID molecule, AID and ssDNA concentrations were chosen so that colorless (mutant)/blue (wild type) plaque ratios were always less than about 2%, as prescribed by Poisson statistics (14, 15).
FIGURE 1.
M13mp2 phage dsDNA constructs having an ssDNA gapped region containing trinucleotide motif cassettes inserted in-frame within a lacZα mutational reporter gene. a, 32 AGC hot motifs on the left-hand side and 24 AGC hot motifs on the right-hand side separated by a 9-nt linker region. b–d, each construct contains an identical cassette composed of alternating AAC hot and AGC hot′ motifs on the left-hand side and two cassettes on the right-hand side containing AAC hot motifs and GAC neutral motifs (b and c) or one cassette on the right-hand side containing AAC hot motifs and GTC cold motifs (d). AID deaminations within the inserted cassettes create stop codons in the lacZα reading frame of M13 phage progeny resulting in colorless mutant plaques. Unmutated gap molecules give rise to “wild type” dark blue plaques. Deaminations in the inserted cassettes are detected as C→T mutations by sequencing individual DNA clones isolated from the mutant colorless phage plaques.
In the AGC32-AGC24 construct (Fig. 1a), the number of mutations/clone occurring as a function of length of exposure to AID shows that there are three or fewer mutations for short (15 s) incubations with AID, with about 75% of the clones containing one mutation (Fig. 2a). A 30-s exposure results in about 60–65% of the clones having 1 mutation, 15 to 20% with 2 mutations, about 10% with 3 mutations, and about 5% with ≥5 mutations (Fig. 2a). At longer incubations with AID, there is an increase in the clones with more mutations and a decrease in clones with fewer mutations, and at 5 min, about 65% of the clones contain ≥5 mutations (Fig. 2a). Similar results are obtained for another construct (AAC AGC)15-(AAC GAC)15-(AAC GAC)15 (Fig. 2a). There is a linear increase in mutations with time for both constructs, with an average rate of about 1 mutation per 46 s to 1 mutation per 63 s (Fig. 2b). The concomitant increase in multiple mutations/clone at the expense of fewer mutations/clone was further evidence that AID acts processively; otherwise, a more rapid dissociation of AID in bulk solution, enabling it to gain access to new ssDNA substrates, would result in a persisting higher percentage of clones with fewer mutations.
FIGURE 2.
AID-initiated mutations in AGC32-AGC24 and (AAC AGC)15-(AAC GAC)15-(AAC GAC)15 measured as a function of time. a, percentage of clones having 1–4 or ≥5 mutations for exposures to AID ranging from 15 s to 5 min. b, average numbers of mutations per clone accumulating with time. The average mutation rates are 1 per 46 s in AGC32-AGC24 and 1 per 63 s in (AAC AGC)15-(AAC GAC)15-(AAC GAC)15. The incubation conditions, including AID and ssDNA concentrations, are described under “Experimental Procedure.”
These slow average rates at which deaminations accumulate suggest that AID is spending most of the time either immobile or much more likely moving along the ssDNA backbone, although only occasionally in a catalytically competent conformation suitable for C deamination. There are often numerous mutations appearing in the same clone suggesting that AID is actively mobile on the DNA (Fig. 3c).
Analyzing the Mechanism of AID Scanning and C Deamination on ssDNA Containing Hot Motifs
To deduce how AID scans ssDNA, we have combined the data from the left-hand cassettes of three constructs composed of alternating AAC hot and AGC hot′ motifs (Fig. 1, b–d), containing a total of >1100 sequenced DNA clones. About half the clones have a single mutation and about half contain from 2 to 10 mutations, with a small number of additional clones containing >10 mutations. The mutation spectrum obtained by pooling data from clones containing exactly one mutation shows that the C deaminations occur in an approximately spatially uniform manner along the ssDNA, with every motif undergoing deamination (Fig. 3a). The AAC motif has a deamination frequency that is consistently ∼1.8-fold higher than the AGC motif. When clones with 2–10 mutations are combined with clones with 1 mutation, we find that these deaminations are also uniform, and the deamination frequency of the AAC motif is 1.8-fold higher than the AGC motif (Fig. 3b). Favored deamination of AAC over AGC occurs at 14/15 motif sites, the sole exception occurring at the junction between the alternating hot spot motifs and the 9-nt spacer sequence (Fig. 3, a and b).
The mutational patterns were determined by sequencing every DNA clone. Representative clones are shown in Fig. 3c, where the symbol T indicates that a mutation, i.e. deamination has taken place at that motif, and the symbol “.” denotes that no deamination has occurred. In Fig. 3c, red represents AAC hot motifs, and orange represents the AGC hot′ motifs. The data show that some mutations are isolated, and others are clustered (Fig. 3c). We define a cluster as a series of mutated motifs where the separation between the mutated motifs is one nonmutated motif or less. A singleton is defined as an isolated cluster of exactly one mutated motif. For example, in the 3–10 number of mutations grouping, the first three example clones each contain one singleton and one cluster of size 2 (Fig. 3c). Other clones in this 3–10 mutation group contain a cluster as large as size 6, along with smaller sized clusters and singletons. Our definition of cluster is intuitively reasonable. The definition depends on the arbitrary separation distance of no more than one nonmutated motif. However, our subsequent statistic of the distance between mutations is not arbitrary.
The distribution of mutation clusters is shown in Table 1. The data are separated into nine rows labeled “Data,” where each row corresponds to the total number of mutations per clone (2–10), with the number of clones shown in parentheses. Clones with more mutations are likely to have bigger clusters. For example, for clones with two mutations, about 93% of the mutations are singletons and 7% are clusters of size 2. As the number of mutations per clone increases, the fraction of singletons decreases; for clones with 10 mutations, about 49% of the clusters are singletons, 25% doublets, 12% triplets, and up to 3% as clusters having six deaminated AAC/AGC motifs. The rows labeled “Simulated” contain the fractions of clusters predicted by a stochastic model (modified random walk model) that will be described below.
The distribution of distances between deaminated motifs is contained in Table 2. In Table 2 we do not use the arbitrary threshold of one to define a cluster, rather we count the exact distance between mutations. For example, in Fig. 3c in the 3–10 number of mutations grouping, the first clone has three mutations and therefore two spaces between mutations, one of length seven and one of length zero. Clone 11, in the 3–10 number of mutations grouping, has nine mutations with eight spaces, one of length six, one of length seven, two of length two, and four of length zero. As in Table 1, the data are separated into rows based on the total number of mutation per clones. Clones with more mutations are more likely to have shorter spaces between mutations. For example, for clones with two mutations, about 2% of spaces are length zero, 11% are length one, 6% are length two, and 9% are length three, etc. (Table 2). As the number of mutations per clone increases, the spaces decrease in size. For clones with 10 mutations, about 31% of spaces are length zero, 26% are length one, 12% are length two, and 14% are length three, 6% are length 4, and 12% are length 5 (Table 2).
The stochastic behavior of AID is clearly seen from the numerous clones in which multiple deamination clusters, varying in length, are separated by nondeaminated motifs with variable spacing (Fig. 3c). Our objective was to deduce a scanning mechanism for AID using a stochastic model to fit the data in Tables 1 and 2.
A Stochastic Model Describing AID Clonal Deamination Patterns
We investigated several related models to analyze the data. In these models, AID slides along the ssDNA, and each time the enzyme encounters a C, it deaminates that site with a specified probability. This probability is a parameter of the model. We considered various values for this parameter, but in accordance with the data, we fixed the probability for the AAC hot motif to be 1.8-fold higher than the probability for the AGC hot′ motif. The models differ by the details of how the enzyme traverses the ssDNA. Under the “Experimental Procedures,” we discussed a parametric bootstrap procedure that estimates the model parameters and tests whether or not computer simulations of the model are consistent with the data in Tables 1 and 2.
First, we calculated an upper boundary for the deamination probability. For an individual clone, we do not know how many times (if ever) the enzyme has visited a given site on the DNA. But clearly, if a site has been mutated it was visited, and we assumed that AID slid to one of its neighboring motifs at least once. Because ∼10% of the mutated motifs have an immediate neighbor motif that is also mutated, 10% is an upper boundary for the deamination probability. If the deamination probability were greater, then we would observe more consecutive deaminations. If the enzyme visited the neighboring motif more than once, then the deamination probability would be less than 10%.
Based on Fig. 3a, the process begins when the enzyme lands in a random position on the ssDNA. In the first model, the enzyme follows a simple random walk, i.e. with probability one-half the enzyme slides one motif to the right, and with probability one-half the enzyme slides one motif to the left. This sliding iterates, with possible deaminations based on the parameter, until the simulation achieves the prescribed number of mutations. A higher value of the deamination probability parameter results in larger clusters and smaller spaces between mutations. Even when this parameter is as low as 1% for the AAC hot motif (and therefore 0.57% for the AGC hot′ motif), the clusters are larger, and the spaces are smaller than is observed in the data. Thus, we rejected this model (p value = 3 × 10−6).
Smaller sized clusters and larger spaces between mutations were obtained using a modified random walk model. In this model, with probability one-half the enzyme still slides to the right, and with probability one-half the enzyme slides to the left, but now the slide distance is not just one motif. In this model, the slide distance is a random number of motifs selected from a geometric distribution. The geometric distribution (31) is the discrete analog of the exponential distribution. So short slides are more likely than long slides. At the end of a slide, the process repeats itself, and the enzyme either slides to the right or to the left, and the slide distance is obtained as an independent draw from the geometric distribution. This model has two parameters as follows: the deamination probability and the average slide length. During a slide, when the enzyme encounters a C there might be a deamination based on the probability for that motif. Unlike the simple random walk model, this model is consistent with the data (p values uniform between 0 and 1) when deaminations occur infrequently, between 1 and 7% for AAC hot motifs.
For a deamination probability of 3% for the AAC hot motif (1.7% for the AGC hot′ motif) and an average sliding distance of 10 motifs (30 nt), the model shows excellent agreement with the data for the distribution of clusters (Table 1) and the distances between deaminated motifs (Table 2). By taking, for example, clones with four mutations (Table 1), 77% occur as singletons (77.5% predicted), 19% doublets (18.5% predicted), 3% triplets (3.5% predicted), and 1% quadruplets (0.5% predicted). The spacing between deaminated motifs for clones with four mutations (Table 2) are as follows: 14.6% have a space of 1 nondeaminated motif (13.8% predicted), 14.6% with a spacing of 2 (14.5% predicted), 6.8% with a spacing of 3 (10.4% predicted), 8.9% with a spacing of 4 (10.7% predicted), etc. An examination of the two tables for clones with 2–10 deaminations shows similarly good agreement for virtually all cluster frequencies and spacing distances. We have made a movie illustrating how C deaminations arise randomly as prescribed by the modified random walk model (supplemental movie 1).
AID does not bind or deaminate dsDNA (30, 32). To determine whether AID can execute longer range movement on ssDNA, by jumping or possibly through intersegmental transfer (21, 33, 34), a partial dsDNA region was formed on the (AAC AGC)15-(AAC GTC)15 construct (Fig. 1d) by annealing a complementary 24-mer oligonucleotide to the region, covering the 9-nt spacer along with two deamination motifs (AAC AGC) on the left and three motifs (AAC GTC AAC) on the right (Fig. 3d). The annealing is virtually complete because there are no mutations in the blocked motifs (0 in 49 clones), although in contrast 30% of the clones (77 in 257 clones) have mutations in these same motifs on the ssDNA substrate. The presence of mutations in both hot-hot′ and hot-cold cassettes, situated to either side of the dsDNA, implies that some jumping is occurring. Several representative clones with mutations in one or both cassettes are shown in Fig. 3d. We reevaluated the random walk models by adding jumping, so that after a slide the enzyme can jump to a random position on the ssDNA and then continue to slide, deaminate, and possibly jump again. We simulated the model with low jumping probabilities. Because the deamination efficiency is low, implying there can be large gaps between mutations even without jumping, the jumps do not significantly alter the distribution of mutation clusters or spaces between mutations. Thus, this variant of the model remains consistent with the data.
The lengths of jumps for dsDNA-scanning enzymes tend to be restricted in the presence of high salt (28, 29, 35). To explore the issue of jumping further experimentally, we collected a separate mutational data set in the presence of 150 mm NaCl for the (AAC AGC)15-(AAC GAC)15-(AAC GAC)15 construct (Fig. 1b).The distribution of clusters and spaces between mutated motifs in the (AAC AGC)15 cassette is about the same in the absence or presence of salt (supplemental Tables S1 and S2). We conclude that whereas AID both slides and jumps, sliding appears to be the dominant process that determines the clustering patterns and spaces separating mutated motifs.
Deamination Efficiency Depends on DNA Sequence Context
Next, we considered both cassettes of the construct in Fig. 1d. The left-hand cassette has alternating AAC hot and AGC hot′ motifs. The right-hand cassette has alternating AAC hot and GTC cold motifs. Fig. 4a shows the mutation spectrum. The deamination frequencies of the AAC hot motifs are 1.8-fold greater than those of the AGC hot′ motifs (Fig. 4a), as observed for the combined data (Fig. 3b). The deamination frequency of the AAC hot motif is similar in the two cassettes and is about 4-fold greater than for the GTC cold motif (Fig. 4a). The cold motifs have no discernible influence on deaminations of AAC motifs. Mutations occur as isolated singletons and in clusters in clones with alternating hot and cold motifs. The modified random walk model is again consistent with the data when the deamination efficiency is low (supplemental Tables S3 and S4).
FIGURE 4.
Mutation spectra for alternating hot-hot′ and hot-cold, alternating hot-hot′ and hot-neutral, and irregular hot-hot′ and hot-neutral constructs. The composition of each cassette is shown below the bar graphs. The bars give the percent of clones with mutations at each motif position, AAC (red), AGC (orange), GTC (blue), and GAC (green). A mutation in the spacer region in an ATC motif is shown in black.
Fig. 4b shows the mutation spectrum for a construct with the following three cassettes: the left-hand cassette has alternating AAC hot and AGC hot′ motifs and the middle and right-hand cassettes have identical alternating AAC hot and GAC neutral motifs (Fig. 1b). The mutation frequencies for the AAC hot motifs in the left-hand cassette are consistently 1.8-fold greater than those in the AGC hot′ motifs, and 10-fold greater than those in the GAC neutral motifs (Fig. 4b). The novel result of this experiment is that the mutation frequency for the same AAC hot motif is ∼2-fold greater in the left-hand cassette than in the middle and right-hand cassettes (Fig. 4b). Unlike GTC cold motifs, neighboring GAC neutral motifs appear to depress the deamination in AAC hot motifs. This same GAC-mediated depression in AAC mutations was observed in a two-cassette construct containing only left hot-hot′ and middle hot-neutral cassettes (data not shown).
To investigate this surprising result further, we made an irregular cassette having four sets of three consecutive AAC hot motifs interspersed by one GAC neutral motif followed by regular alternating AAC hot and GAC neutral motifs (Fig. 1c); therefore, the AAC was surrounded on both sides either by GAC or by another AAC. Fig. 4c shows the mutation spectrum for this construct. As in Fig. 4b, the AAC hot motifs in the left-hand cassette are consistently 2-fold higher than the same AAC hot motifs in the middle and right-hand cassettes. Therefore, the reduction in deamination efficiency does not require that the two motifs be alternating, nor does it require that AAC be immediately adjacent to GAC; the three consecutive AAC motifs show the same 2-fold reduced deamination when sandwiched between two GAC motifs (Fig. 4c).
We tried with limited success to model the multiple cassette constructs, including hot-hot′ and hot-cold and hot-hot′ and hot-neutral cassettes, by having a separate deamination probability for each 6-base motif rather than for each 3-base motif. Thus, there would be similar probabilities for the AGCAAC hot′/hot motif and GTCAAC cold/hot motif (where C is the target site), which are each 2-fold higher than for the GACAAC neutral/hot motif and AACAAC hot/hot motif. The simulated mutation spectrum fits the data well (supplemental Fig. S1). The efficiency of the APOBEC3G C-deaminase has been shown to depend on a 6-base motif, where the YCC hot motif is strongly modulated by 3 nt to the 5′-side of the hot motif (36). Thus far, however, AID has only been shown to depend on 3 bases (14, 15). Whether or not AID is found to have a more extensive motif recognition region, it is clear that sequence context can alter hot motif deamination efficiency to a significant extent (Fig. 4, b and c).
Distribution of Deaminations during Transcription
We investigated the distribution of deaminations occurring during transcription by T7 RNA polymerase (14, 15, 37, 38). We used a dsDNA version of the construct in Fig. 1a, with repeating AGC hot′ motifs. Fig. 5 shows example clones, all of which are deaminated on the nontranscribed strand. Unlike the previous experiments where the enzyme slides back and forth in a diffusive manner, in this experiment the enzyme slid across the DNA in one direction within the transcription bubble. There are more clusters for this experiment than in the previous experiments. A possible reason for the increased clustering might be that constricted movement of AID within the transcription bubble could localize the action of AID thereby forcing it to deaminate adjacent motifs. Transcriptional stalling (39) could have a similar effect; when the bubble stalls, the enzyme is confined to a region about 10 nucleotides long (40, 41), increasing the chance of clustered deaminations (42, 43).
FIGURE 5.
AID-induced mutations occurring on the nontranscribed strand of actively transcribed dsDNA. Representative mutated clones with ≥2 mutations. Clones with mutations near the 5′- and the 3′-end of the inserted cassettes (e.g. second clone from the top) indicate that AID can scan the entire region processively. The occurrence of a large excess of consecutive deaminations suggests that AID may be constrained in its ability to slide within the transcription bubble and that bubble stalling may also be occurring.
Because the transcription bubble slides across the DNA in a 5′ → 3′ direction, unlike the diffusive back and forth sliding of the enzyme in the earlier experiments, this experiment presents a way to estimate the deamination efficiency that does not depend on any model. There were 119 clones and each clone had 56 motifs. There were a total of 177 mutations, so an estimate of the deamination efficiency was 2.7% (=177/(119 × 56)) in the AGC hot′ motif and 4.9% (2.7 × 1.8) in the AAC hot motif. A deamination efficiency for AAC of 4.9% was in the range estimated from the modified random walk model (1–7%).
Deamination Efficiencies at Non-WRC Motifs Differ for Singly and Multiply Deaminated Clones
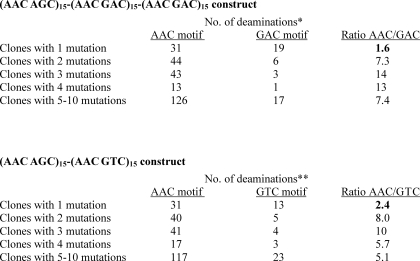
We now examine the relative deamination efficiencies for clones having just a single mutation compared with clones having multiple mutations (Table 3). It is striking that clones with only one mutation are much more likely to undergo deamination in non-WRC (GAC and GTC) motifs compared with clones containing multiple mutations. First, consider the AAC hot and GAC neutral cassette. For clones with one mutation, there are 1.6-fold more mutations at WRC than non-WRC sites, but for clones with more than one mutation this ratio ranges between 7 and 14. This difference is highly statistically significant (p value = 10−6, 2 × 2 χ2 test comparing clones with one mutation to clones with multiple mutations). A similar result holds for the AAC hot and GTC cold cassette (p value = 10−2). In contrast, the ratio of deaminations in WRC hot motifs AAC to AGC remains unchanged (∼1.8) for clones with either one or multiple mutations (compare Fig. 2, a and b). A possible explanation for the observed difference is that AID has a different motif-dependent deamination efficiency when it initially lands on the DNA than when it is sliding along the DNA.
TABLE 3.
AID-catalyzed deamination efficiencies for clones with one mutation compared with clones with multiple mutations
* Deaminations in AAC and GAC motifs were counted for the middle and the right cassettes (Fig. 1b).
** In Fig. 1d, a single deamination at the AAC motif in the right cassette cannot be detected, because it does not generate a stop codon; therefore, single deaminations in the GTC motif (which do generate stop codons) were compared with deamination in the AAC motif in the left cassette (which do generate stop codons).
DISCUSSION
Search and destroy enzymes that scan dsDNA processively to eliminate base mispairs, during mismatch repair, or damaged DNA bases, during base excision repair, have been studied extensively (20, 23, 27–29, 35, 44, 45). During base excision repair, for example, DNA glycosylases act processively on dsDNA to excise foreign or damaged DNA bases, U in the case of uracil DNA glycosylase (29, 35), and oxidized G in the case of Ogg (23, 26). Uracil or 8-oxoG may be present only at a level of about 1 in 105 to 106 bases (46). Therefore, to be effective in removing these rare but potently mutagenic bases, DNA glycosylases need to scan large regions of dsDNA rapidly and processively and then to act selectively and with high catalytic efficiency once an aberrant base is located (23, 29).
AID scans ssDNA processively and concomitantly deaminates C→U (14, 15). However, in contrast to the rare “needle-in-a-haystack” target sites for glycosylases, there are substantial numbers of trinucleotide target motifs containing cytosine. In our experiments, because <2% of the DNA molecules are mutated, even at the longest incubation time (5 min), virtually all of the individual clones were acted on by at most one AID molecule (14, 15). We cannot rule out that the presence of a GST tag might influence processivity. Nor can we eliminate the possibility that an AID molecule bound to DNA might recruit other AID proteins to the same DNA. However, experiments with native APOBEC3G show that this untagged ssDNA C deaminase is highly processive (36). Moreover, a mutant of APOBEC3G that exists predominantly as a monomer, alone and in the presence of ssDNA, exhibits processive deamination behavior similar to the native enzyme (47).
Notably, although the enzyme remained bound to its substrate for a matter of minutes, the deaminations accumulated with an extremely slow average rate, ∼1 per min. Yet, while remaining bound to a single ssDNA, AID is able to scan the entire ssDNA gap region (∼200 nt for double cassettes and ∼300 nt for triple cassettes); this is based on many independent occurrences of individual clones containing deaminations near the 3′- and 5′-ends of a cassette and between the two ends. Therefore, it appears that AID is spending most of its time scanning ssDNA but not deaminating a large majority of WRC hot motifs.
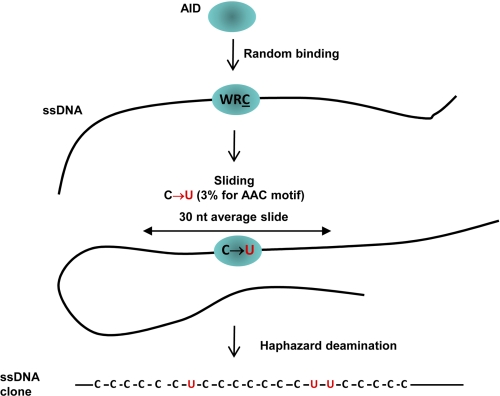
We have proposed a simple stochastic model to analyze the data, which we call the modified random walk model (Fig. 6). The first model parameter is the probability of deamination when AID encounters a motif. This probability depends on the sequence of the motif. The ratio of probabilities for the different motifs is fixed by the ratio of mutations in the different motifs observed in the data. In the model, AID traverses ssDNA by sliding. The second model parameter is the average length of each slide obtained from a geometric distribution (shorter slides are more common than longer ones). When one slide ends, AID slides again, in either direction with equal probability, and continues to randomly deaminate encountered motifs.
FIGURE 6.
Random walk model depicting AID scanning and catalyzing haphazard and inefficient deamination of C to U on ssDNA. AID binds at a random location on ssDNA. AID scans in accord with a modified random walk model (see text) in which the length of a slide in either direction is determined by a geometric distribution, i.e. shorter slides are favored over longer slides. An excellent fit of the model to the data shows that AID only rarely deaminates C to U, at about 3% efficiency for even “preferred” AAC hot motifs, thus ensuring mutational diversity. Deaminations occur predominantly during sliding. Intramolecular jumping/hopping (Fig. 3d) or inter-segmental transfer can also occur to reposition AID to another location on the same ssDNA molecule without altering the deamination patterns (see text). Deaminations occur mostly as singletons, although deamination clusters arise less frequently during repeated sliding over the same region of ssDNA. The average length of a random slide is 30 nt (10 trinucleotide motifs).
With just two parameters, this model provides an excellent fit to the data (Tables 1 and 2). The parameter values are a deamination efficiency of 3% in AAC hot motifs (1.7% for the AGC hot′ motif), and an average sliding distance of 10 motifs (30 nt) (Fig. 6). Thus, when encountering a favored WRC motif, AID appears to ignore this motif more often than 9 times out of 10. Because deaminations occur infrequently, clustered mutations occur when AID revisits ssDNA sites multiple times (supplemental movie 1). We have also added small amounts of jumping to the model (Fig. 3d and supplemental Tables S1 and S2) without adversely affecting the quality of the fit. Independent of the modeling, the relatively low number of consecutively mutated motifs in both the combined (AAC AGC)15 cassettes (Fig. 1, b–d, left-hand cassettes) and the transcription experiment (Fig. 5) confirms that the deamination efficiency is low (<10%).
Another important aspect of the data is the unexpectedly large influence of sequences on AID-catalyzed deamination. AID behaves as expected for two sequence contexts, AAC hot and AGC hot′ motifs and for AAC hot and GTC cold motifs. AID deaminates AAC 1.8-fold more efficiently than AGC and deaminates AAC 4-fold more efficiently than GTC, while maintaining an alternative high-low-high-low pattern over each cassette (Figs. 3, a and b, and 4a). In contrast, AID behaves decidedly differently when AAC is located near GAC (Fig. 4, b and c). Here, the AAC hot motifs undergo a 2-fold reduction in efficiency (Fig. 4, b and c). This reduction does not require that the AAC and GAC alternate (Fig. 4c) nor does it require that AAC be adjacent to GAC (Fig. 4c).
The modified random walk model gives an excellent fit to the ensemble of data for all motifs (supplemental Fig. S1) by extending the recognition motif for AID from three to six nucleotides. A 6-nt “extended motif” has been shown for APOBEC3G, where deamination of a 5′YCC canonical hot motif is diminished by 20-fold in the presence of a neighboring NCC (5′NCCYCC) (36). However, to date AID has only been show to depend on 3-nt motifs (14, 15). Whether or not AID is found to have a more extensive motif recognition region, it is clear that sequence context can alter hot motif deamination efficiency to a significant extent.
Finally, we find that AAC hot and AGC hot′ motifs are dealt with differently than GAC and GTC non-hot motifs when clones with exactly one mutation are compared with multiply mutated clones. The WRC motif deamination efficiencies are ∼7–14-fold greater than the non-WRC motifs for clones with ≥2 mutations, but remarkably for clones with 1 mutation, hot motif deaminations are favored by less than 2-fold (Table 3). We speculate that perhaps AID is much more selective in distinguishing hot- from non-hot motifs while sliding than when jumping.
Clearly, there is much to be learned about ssDNA-scanning enzymes in general, and specifically APOBEC family members. We view the modified random walk model as an Occam's razor view of an AID scanning and deamination mechanism. The most basic properties of AID, which involve the scanning and deamination of naked ssDNA and the nontranscribed strand of actively transcribed dsDNA, are important to establish an underpinning for understanding V-gene targeting (48). We have taken an initial step in this direction by using AID to generate large numbers of independently derived DNA clones having widely diverse mutational patterns that suggest that AID behaves in a haphazard and inefficient manner. Many different V-region mutations must first be “tested” in naive B cells, so that only those that arise by chance that enhance antibody-antigen binding are selected for subsequent clonal expansion. Perhaps the best way to ensure a wide variety of mutations to achieve antigenic diversity is if AID works as a haphazard and catalytically inefficient enzyme.
Supplementary Material
Acknowledgments
We thank Minnie Chan and Catherine Khoo for technical assistance in data collection and Ian Slaymaker for making the AID scanning and C deamination movie.
This work was supported, in whole or in part, by National Institutes of Health Grants GM21422 and ES013192.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1, Tables S1–S4, and a movie.
- AID
- activation-induced deoxycytidine deaminase
- ssDNA
- single-stranded DNA
- nt
- nucleotide
- V
- variable
- S
- switch.
REFERENCES
- 1. Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) Annu. Rev. Immunol. 26, 481–511 [DOI] [PubMed] [Google Scholar]
- 2. Stavnezer J., Guikema J. E., Schrader C. E. (2008) Annu. Rev. Immunol. 26, 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Di Noia J. M., Neuberger M. S. (2007) Annu. Rev. Biochem. 76, 1–22 [DOI] [PubMed] [Google Scholar]
- 4. MacDuff D. A., Demorest Z. L., Harris R. S. (2009) Nucleic Acids Res. 37, 1854–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vartanian J. P., Henry M., Marchio A., Suspène R., Aynaud M. M., Guétard D., Cervantes-Gonzalez M., Battiston C., Mazzaferro V., Pineau P., Dejean A., Wain-Hobson S. (2010) PLoS Pathog. 6, e1000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhutani N., Brady J. J., Damian M., Sacco A., Corbel S. Y., Blau H. M. (2010) Nature 463, 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Popp C., Dean W., Feng S., Cokus S. J., Andrews S., Pellegrini M., Jacobsen S. E., Reik W. (2010) Nature 463, 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo J. U., Su Y., Zhong C., Ming G. L., Song H. (2011) Cell 145, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiricny J., Menigatti M. (2008) Cell 135, 1167–1169 [DOI] [PubMed] [Google Scholar]
- 11. Chelico L., Pham P., Petruska J., Goodman M. F. (2009) J. Biol. Chem. 284, 27761–27765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chahwan R., Wontakal S. N., Roa S. (2010) Trends Genet. 26, 443–448 [DOI] [PubMed] [Google Scholar]
- 13. Fritz E. L., Papavasiliou F. N. (2010) Genes Dev. 24, 2107–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003) Nature 424, 103–107 [DOI] [PubMed] [Google Scholar]
- 15. Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) J. Biol. Chem. 279, 51612–51621 [DOI] [PubMed] [Google Scholar]
- 16. Yu K., Huang F. T., Lieber M. R. (2004) J. Biol. Chem. 279, 6496–6500 [DOI] [PubMed] [Google Scholar]
- 17. Wang M., Rada C., Neuberger M. S. (2010) J. Exp. Med. 207, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basu U., Meng F. L., Keim C., Grinstein V., Pefanis E., Eccleston J., Zhang T., Myers D., Wasserman C. R., Wesemann D. R., Januszyk K., Gregory R. I., Deng H., Lima C. D., Alt F. W. (2011) Cell 144, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., Nieland T. J., Root D. E., Casellas R., Nussenzweig M. C. (2010) Cell 143, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorman J., Chowdhury A., Surtees J. A., Shimada J., Reichman D. R., Alani E., Greene E. C. (2007) Mol. Cell 28, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorman J., Greene E. C. (2008) Nat. Struct. Mol. Biol. 15, 768–774 [DOI] [PubMed] [Google Scholar]
- 22. Komazin-Meredith G., Mirchev R., Golan D. E., van Oijen A. M., Coen D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10721–10726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blainey P. C., van Oijen A. M., Banerjee A., Verdine G. L., Xie X. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kochaniak A. B., Habuchi S., Loparo J. J., Chang D. J., Cimprich K. A., Walter J. C., van Oijen A. M. (2009) J. Biol. Chem. 284, 17700–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonnet I., Biebricher A., Porté P. L., Loverdo C., Bénichou O., Voituriez R., Escudé C., Wende W., Pingoud A., Desbiolles P. (2008) Nucleic Acids Res. 36, 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blainey P. C., Luo G., Kou S. C., Mangel W. F., Verdine G. L., Bagchi B., Xie X. S. (2009) Nat. Struct. Mol. Biol. 16, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terry B. J., Jack W. E., Modrich P. (1985) J. Biol. Chem. 260, 13130–13137 [PubMed] [Google Scholar]
- 28. Gowers D. M., Wilson G. G., Halford S. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15883–15888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Porecha R. H., Stivers J. T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10791–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ross S. M. (1994) A First Course in Probability, 4th Ed, pp. 167–169 MacMillan Publishing Co., New York [Google Scholar]
- 32. Larijani M., Martin A. (2007) Mol. Cell. Biol. 27, 8038–8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berg O. G., Winter R. B., von Hippel P. H. (1981) Biochemistry 20, 6929–6948 [DOI] [PubMed] [Google Scholar]
- 34. Halford S. E. (2001) Biochem. Soc. Trans. 29, 363–374 [DOI] [PubMed] [Google Scholar]
- 35. Bennett S. E., Sanderson R. J., Mosbaugh D. W. (1995) Biochemistry 34, 6109–6119 [DOI] [PubMed] [Google Scholar]
- 36. Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 37. Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwat A. S. (2003) Nucleic Acids Res. 31, 2990–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. (2003) Nature 422, 726–730 [DOI] [PubMed] [Google Scholar]
- 39. Landick R. (2006) Biochem. Soc. Trans. 34, 1062–1066 [DOI] [PubMed] [Google Scholar]
- 40. Liu C., Martin C. T. (2001) J. Mol. Biol. 308, 465–475 [DOI] [PubMed] [Google Scholar]
- 41. Sohn Y., Kang C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Canugovi C., Samaranayake M., Bhagwat A. S. (2009) FASEB J. 23, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shen H. M., Ratnam S., Storb U. (2005) Mol. Cell. Biol. 25, 10815–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carey D. C., Strauss P. R. (1999) Biochemistry 38, 16553–16560 [DOI] [PubMed] [Google Scholar]
- 45. Dowd D. R., Lloyd R. S. (1990) J. Biol. Chem. 265, 3424–3431 [PubMed] [Google Scholar]
- 46. Lindahl T., Barnes D. E. (2000) Cold Spring Harbor Symp. Quant. Biol. 65, 127–133 [DOI] [PubMed] [Google Scholar]
- 47. Chelico L., Prochnow C., Erie D. A., Chen X. S., Goodman M. F. (2010) J. Biol. Chem. 285, 16195–16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. MacCarthy T., Kalis S. L., Roa S., Pham P., Goodman M. F., Scharff M. D., Bergman A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8629–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.