Abstract
Acid-sensing ion channels (ASICs) are known to be primarily activated by extracellular protons. Recently, we characterized a novel nonproton ligand (2-guanidine-4-methylquinazoline, GMQ), which activates the ASIC3 channel subtype at neutral pH. Using an interactive computational-experimental approach, here we extend our investigation to delineate the architecture of the GMQ-sensing domain in the ASIC3 channels. We first established a GMQ binding mode and revealed that residues Glu-423, Glu-79, Leu-77, Arg-376, Gln-271, and Gln-269 play key roles in forming the GMQ-sensing domain. We then verified the GMQ binding mode using ab initio calculation and mutagenesis and demonstrated the critical role of the above GMQ-binding residues in the interplay among GMQ, proton, and Ca2+ in regulating the function of ASIC3. Additionally, we showed that the same residues involved in coordinating GMQ responses are also critical for activation of the ASIC3E79C mutant by thiol-reactive compound DTNB. Thus, a range of complementary techniques provide independent evidence for the structural details of the GMQ-sensing domain at atomic level, laying the foundation for further investigations of endogenous nonproton ligands and gating mechanisms of the ASIC3 channels.
Keywords: Acid-sensing Ion Channels (ASIC), Computer Modeling, Ion Channels, Ligand-binding Protein, Membrane Biophysics, Neuroscience, Atomic Level, Channel Gating, Homology Modeling, Nonproton Ligand Sensor
Introduction
Acid-sensing ion channels (ASICs)4 are proton-gated cation channels (1–3), opening in response to extracellular pH reduction. To date, at least six ASIC subunits encoded by four distinctive genes (asic1, asic2, asic3, and asic4) have been identified (1, 2): 1a, 1b, 2a, 2b, 3, and 4. The first low pH crystal structure of chicken ASIC1 has been resolved (4, 5), revealing significant insights into many fundamental issues about these trimeric ion channels (6). Recently, we discovered a nonproton ligand, 2-guanidine-4-methylquinazoline (GMQ) (see Fig. 1A), which activates ASIC3 channels under neutral pH conditions (7). Remarkably, GMQ- and acid-induced (pH 5.0) currents exhibit distinct kinetics. The acid-induced currents desensitized rapidly, whereas the GMQ-evoked currents showed no desensitization when GMQ was continuously present. Using electrophysiological analysis of a series of ASIC3 channel mutants, we demonstrated that the carboxyl-carboxylate interaction pair, Glu-79–Glu-423, is crucial for sustained activation of ASIC3 channels at neutral pH. Furthermore, using cysteine substitution mutants, we showed that covalent modification at E79C by DTNB, an Ellman's reagent used to modify and quantitatively detect sulfhydryl groups in proteins, was able to activate the channel, suggesting the importance of the Glu-79 residue in channel gating (7). Although the key role of Glu-79 and Glu-423 in GMQ-ASIC3 interactions has been elucidated, the makeup of this nonproton ligand-sensing domain remains largely unknown. In this study, taking advantage of computational approaches (8) and the available high resolution three-dimensional structure of ASIC channels (4, 5), we aimed to delineate the architecture of the nonproton ligand-sensing domain as well as the GMQ binding mode in ASIC3 channels. To achieve this, we employed diverse approaches including electrophysiological recording, comprehensive mutagenesis, homology modeling, in silico docking, and ab initio calculations. We uncovered the structural details at the atomic level, which well explain the GMQ-ASIC3 interactions and shed new light on the mechanisms of ASIC3 activation by nonproton ligands.
FIGURE 1.
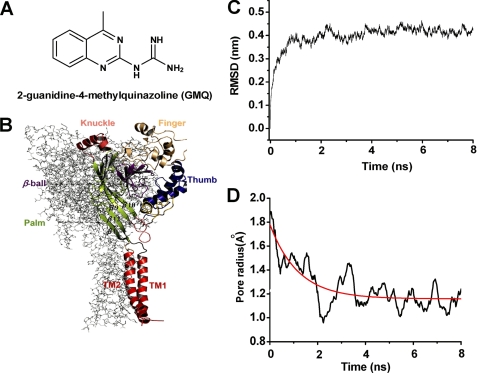
Structure of GMQ and homology model of ASIC3 channels. A, chemical structure of GMQ. B, stereo view of the three-dimensional structural model of ASIC3 channels. The structure model is constructed on the basis of a homology model of ASIC3 (see “Experimental Procedures”) and shown in parallel to the membrane layer. Domain organization of subunit A is highlighted with colors, whereas subunits B and C are displayed as gray lines. This figure and other structural images in Figs. 2, 3, 4, 6, and 8 are made using PyMol. C and D, MD simulations of structural model of ASIC3. C, time dependence of the root mean square deviation (RMSD) of the Cα atoms from the initial homology model of ASIC3 in the MD simulation. D, time dependence of the channel pore radius during the MD simulation.
EXPERIMENTAL PROCEDURES
Solutions, Drugs, Cell Culture, and Transfection
All of the solutions and drugs were purchased and prepared as described previously (7). All of the constructs were expressed in CHO cells as described previously (7). In brief, CHO cells were cultured in F-12 medium (1 mm l-glutamine, 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin) at 37 °C in a humidified atmosphere (5% CO2 and 95% air). Plasmid transfections were carried out using LipofectamineTM 2000.
Site-directed Mutagenesis
As described previously (7), the cDNA of ASIC3 was subcloned into the pEGFPC3 vector. Each mutant was generated with a QuikChange® mutagenesis kit. Individual mutations were verified by DNA sequencing.
Electrophysiology
Electrophysiological recordings were performed using the conventional whole cell configuration under voltage clamp as described previously (9). Briefly, membrane currents were measured using a patch clamp amplifier (Axon 700A, Axon Instruments, Foster City, CA) and were sampled and analyzed using a Digidata 1320A interface and a computer running the Clampex and Clamp-fit software. The membrane potential was held at −60 mV throughout the experiment under voltage clamp conditions. All of the experiments were carried out at room temperature (23 ± 2 °C). Except where otherwise indicated, all of the GMQ responses were recorded at conditional pH of 7.4.
Homology Modeling for the Three-dimensional Model of ASIC3 and ASIC1a
The recent crystal structure of ASIC1mfc (Protein Data Bank code 3HGC; resolution, 3.0 Å) (5) was solved by molecular replacement using the EC domain of a chicken ΔASIC1 (Protein Data Bank code 2QTS; resolution, 1.9 Å) promoter as a search probe, and the EC domains of two crystal structures (4, 5) are essentially identical. Thus, only the high resolution x-ray crystal structure of ΔASIC1 (4) was used as the template to construct the three-dimensional model of the rat ASIC3. The sequence of rat ASIC3 was retrieved from UniProt (entry O35240). The three-dimensional model of ASIC3 (see Fig. 1B) was constructed using the homology module encoded in Insight II (Molecular Simulations, Inc., San Diego, CA) (10, 11). Briefly, the alignment of the target sequence with the template was performed mainly using the method reported by Jasti et al. (4). According to the secondary structure information of the template, the sequence alignment was adjusted manually to obtain a more reasonable alignment. Then the coordinates of the structurally conserved regions of the template were assigned to ASIC3. The loops that connect the structurally conserved regions were generated de novo and selected so as to avoid clashes with other atoms. Refined routines in the homology module of Insight II were used to adjust the positions of the side chains. Finally, the constructed model was checked and validated by the program Procheck (12). A similar approach was used to construct the three-dimensional structure of the ASIC1a.
The model was created at neutral pH and optimized by molecular dynamics (MD) simulations (13, 14). Because the crystal structure of ASIC1 (Protein Data Bank code 2QTS) represents a desensitized conformation under acidic pH condition, the homology model of ASIC3 based on this template may be also desensitized. To obtain a stable and closed conformation of ASIC3 at neutral pH, we employed an 8-ns MD simulation to optimize the ASIC3 homology model. In this simulation, the root mean square deviation profile of the protein tends to be stable at 1 ns (see Fig. 1C). We also monitored the radius of the channel pore. The radius fluctuation along the MD trajectory also suggests that the channel tends to be in a closed state (see Fig. 1D). In particular, in the initial conformation, the radius is ∼1.8 Å (a desensitized pore radius), and then it decreases gradually to 1.0 Å at 2 ns. Thereafter, the pore radius fluctuates ∼1.2 Å. Thus, the snapshot at 2.5 ns on the MD trajectory was extracted as the structural model because the channel reached a closed and stable state after such time scale MD simulations.
In Silico Docking
The docking program Glide (version 4.0; Schrödinger, LLC, New York, NY) (15) was applied to dock GMQ into the potential binding site of ASIC3. For Glide docking, the protein structure was preprocessed using the protein preparation and refinement components in the Glide docking package. Then the grid for the protein was defined as an enclosing box with 15 Å in all three dimensions, which included all the residues involved in the binding of the ligand. For the docking runs, the standard precision docking mode was selected. All of the procedures including protein preparation, refinement, grid generation, and docking were performed using the default Glide parameters. The top 10 poses with the best Glide-Score for GMQ were saved as candidates. The tool of Glide pose viewer was used to check and select the candidate poses. The poses that interact with the correct residues were regarded as reasonable binding conformations. The GMQ-ASIC1a complexes were also prepared by using this docking paradigm.
Ab Initio and QM/MM Interaction Energy Calculations
All of the ab initio calculations were carried out using the density functional theory, a computational quantum mechanics method (Gaussian 03; Gaussian, Inc., Pittsburgh, PA) (16), at the level of B3LYP/6–31G(d). The interaction energy (ΔEcp) was obtained from the difference between the total energy of the residue-ligand complexes and the sum of total energies of the two monomers. The basis set superposition error was eliminated by means of the counterpoise method of Boys and Bernardi (17). All of these calculations were carried out using the Gaussian 03 suite of programs (16). All of the QM/MM calculations were carried out using the QM/MM protocol encoded in Dmol3 of Discovery Studio, version 2.1, with a BLYP functional parameter and an ultrafine basis of Dmol3. A natural division had defined the ligand as the QM region and the ASIC3 channel as the MM zone.
Construction of the Three-dimensional Models of DTNB-modified ASIC3
Covalent docking of DTNB to ASIC3 was carried out using the Gold docking program (version 3.1; Cambridge Crystallographic Data Center, Cambridge, UK) (18). A covalent linkage between the sulfur atom of the TNB molecule and the sulfur atom of residue Cys-79 in the mutated ASIC3 (E79C) was used to constrain the docking. Three TNB molecules were covalently docked into the three chains of the mutated ASIC3, respectively. The pose with the best Gold score was retained for each chain. After alignment of the three chains with covalently docked ligands, the predicted conformation of DTNB-modified ASIC3 was constructed.
Molecular Dynamics Simulations
The constructed structure of ASIC3 was used as the starting structure for MD simulations. A large dimyristoylphosphatidylcholine bilayer (19) containing 512 lipid molecules was constructed by replicating an equilibrated block with 128 dimyristoylphosphatidylcholine lipids four times. Thirty-six lipids were subsequently removed from the constructed bilayer to generate a suitable membrane system into which the transmembrane (TM) domain of the ASIC3 could be embedded. The protein/dimyristoylphosphatidylcholine system was then solvated in a bath of simple point charge water molecules (20). Counter ions were subsequently added to compensate for the net negative charge of the system.
The MD simulations were performed using the GROMACS package (version 3.2.1) (13, 14) with constant number, pressure, and temperature and periodic boundary conditions. A modified GROMOS87 force field (21) was employed for the protein. To maintain the system at a constant temperature of 310 K, the Berendsen algorithm (22) was applied to couple protein and other molecules separately with a coupling time of 0.1 ps. All of the bond lengths including hydrogen atoms were constrained by the Linear Constraint Solver algorithm. Electrostatic interactions between charged groups at a distance less than 10 Å were calculated explicitly; long range electrostatic interactions were calculated using the particle mesh Ewald method (23) with a grid width of 1.2 Å and a fourth order spline interpolation. A cutoff distance of 10 Å was applied for the Lennard-Jones interactions. The whole system was minimized using the steepest descent algorithm to reach the convergence criterion. Afterward, the first 160-ps MD simulation was carried out to heat the solvent molecules and ions to 310 K with protein and ligand fixed. Then the second 20-ps MD simulations were performed with the protein main chain and ligand fixed. Subsequently, with the whole system relaxed except for protein Cα atoms and ligand, the equilibration was completed after the third 20-ps MD simulations. Finally, 8-ns MD simulations were performed. All of the MD simulations were run on the DAWNING 4000A at the Shanghai Supercomputer Center (with 2,128 AMD OpteronTM CPUs). Preparation, analysis, and visualization were performed on a 128-CPU Silicon Graphics Origin3800 server.
Data Analysis
The results are expressed as the means ± S.E. Statistical comparisons were made using Student's t test, where p < 0.05 (*) or p < 0.001 (**) was considered significantly different. Concentration-response relationships for GMQ activation of WT or mutated channels were obtained by measuring currents in response to different concentrations of GMQ as described previously (7). Briefly, a reference pH 5.0 was applied first to each CHO cell, and 2 min later a single test concentration of GMQ was applied, and the response was normalized to the reference pH 5.0-induced peak current. Each concentration was tested on at least three CHO cells, and all of the results used to generate a concentration-response relationship were from the same group. The data were fit to the Hill equation: I/Imax = 1/[1 + (EC50/[Ligand])n, where I is the normalized current at a given concentration of GMQ, Imax is the maximum normalized current, EC50 is the concentration of GMQ yielding a current that is half of the maximum, and n is the Hill coefficient.
RESULTS
Constructing Homology Model of ASIC3 at Neural pH
To explore the structural basis underlying GMQ-ASIC3 interactions, we first constructed a three-dimensional model of ASIC3 (Fig. 1B) at neutral pH based on the crystal structure of the chicken ASIC1 channel (4) (Protein Data Bank code 2QTS) using homology modeling and MD simulation approaches (see “Experimental Procedures”). The quality of this homology model was verified by the Ramachandran plot, in which 99.4% of the residues are in the most favored region with no residues in the disallowed region. The result of MD simulations indicated that the three-dimensional model of the ASIC3 channel became stable after 2.5 ns of simulation time, because the root mean square deviation profile for carbon atoms exhibited a plateau (Fig. 1C), and the pore radius converged to a low value (Fig. 1D). Accordingly, we used this optimized structural model of ASIC3 for the following studies.
Specific Interactions between GMQ and the Cavity Lined by Leu-77, Glu-79, Gln-269, Gln-271, Glu-423, and Arg-376
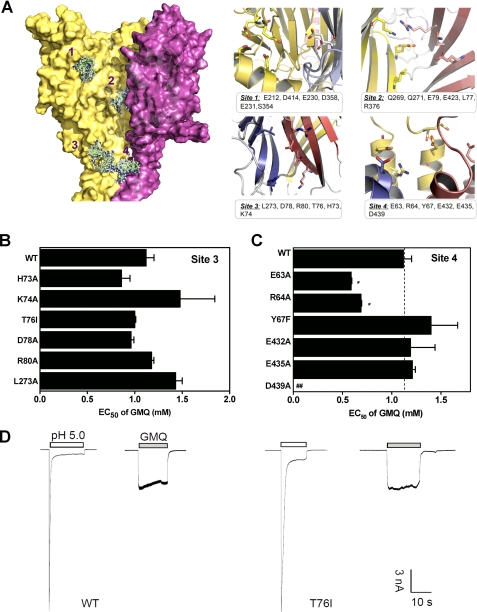
Using the three-dimensional structural model, we systemically examined potential GMQ-binding sites in ASIC3 by combining computational analysis, mutagenesis, and functional studies. First, we identified four putative GMQ-binding sites using the cavity searching algorithm as well as in silico (ligand-protein) docking (Fig. 2A). Site 1 has been previously implicated in pH sensing in ASIC1 (4) and ASIC3 (7) and is located in the cleft between the “thumb” and “finger” domains; site 2 is in the interface of the three subunits, namely the cavity lined by Glu-79 and Glu-423 in the palm domain (Figs. 2A and 3A); site 3 is in the interface of any two subunits; and site 4 is formed by a cluster of acidic residues in the post-TM1 and pre-TM2 regions (Fig. 2A). Using mutagenesis followed by functional analysis, we found that many residues in these sites are crucial for the pH-dependent gating of ASIC3 channels (7) (data not shown), consistent with previous mutational studies in the ASIC1a channel (4, 24). However, for GMQ-induced currents, amino acid mutations in site 1 (7), site 3, and site 4 only slightly affected EC50 value (Fig. 2, B and C; E63A and R64A even slightly decreased EC50 values) and exerted little effects on the kinetics of GMQ-induced currents (Fig. 2D). In contrast, most mutations in site 2 significantly affected the EC50 values of GMQ (Fig. 3B), suggesting key roles of these residues in GMQ binding.
FIGURE 2.
Potential binding sites of GMQ in the three-dimensional ASIC3 model. A, structure and key residues (displayed in sticks for emphasis) of potential binding sites of GMQ. The potential binding sites were detected using cavity searching algorithm encoded in Discovery Studio®, version 2.1. GMQ was docked into the potential binding site of ASIC3 using docking program Glide (version 4.0; Schrödinger). B and C, EC50 values of GMQ (means ± S.E., n = 3–10) for point mutations in Sites 3 (B) and 4 (C). In C, the WT EC50, taken from B, is regraphed again for comparison. *, p < 0.05 versus WT ASIC3 (dashed line). D, examples of GMQ-evoked (1 mm) currents at neutral pH in WT and mutated ASIC3 channels (as exemplified by T76I). Similar results were obtained in four other experiments.
FIGURE 3.
The action of GMQ is determined by residues in the central cavity of the palm domain. A, structure and key residues in the palm domain. The palm domains of subunits A, B, and C are colored yellow, pink, and blue (left panel), respectively. Only two of the three subunits are shown for clarity (right panel). B, EC50 values and maximal currents of GMQ (means ± S.E., n = 4–10) for point mutations. The values of E423A, E423D, E423L, E423Q, E423R, and E79A were derived from previously published data (7), shown here as white bars for comparison. *, p < 0.05; **, p < 0.001 versus WT ASIC3 (dashed line). #, maximal currents of GMQ were not normalized to IpH5.0 because of the markedly reduced IpH5.0 resulting from the mutation-induced steady-state desensitization at the neutral pH. The WT EC50, taken from Fig. 2B, is regraphed again for comparison.
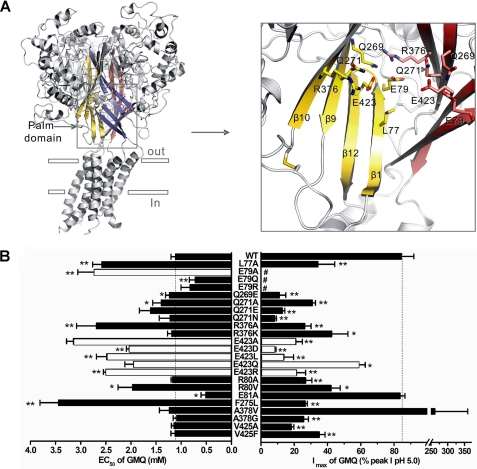
A closer examination of site 2 revealed that it is made up of a series of charged, polar, and hydrophobic residues, including Glu-79, Glu-423, Arg-376, Gln-271, Gln-269, and Leu-77 (Fig. 3A). Mutations at these key residues resulted in increased EC50 values and decreased maximal GMQ responses in mutated channels (Fig. 3B). To establish the interaction specificity, we performed more extensive mutations at residues (Fig. 4A, i.e. Asp-78, Arg-80, Glu-81, Leu-273, Lys-379, Glu-380, Glu-418, and Val-425) adjacent to the core of site 2 cavity. These mutations exerted less effects on the EC50 of GMQ (Fig 4B) when compared with E423A, E79A, L77A, and R376A mutations (Fig. 3B). We noted that R80V and E418A mutations caused noticeable increases, whereas E81A caused noticeable decreases of EC50 values of GMQ, which may be explained by allosteric effects because of their proximity to Glu-79 or Glu-423. Therefore, it is likely that residues Glu-423, Glu-79, Leu-77, and Arg-376 make up the core of the GMQ-binding site, whereas the surrounding residues play a role in stabilizing the conformations of this site. These data, together with the previous finding that modification of the introduced cysteine residue by DTNB or covalently linked GMQ (a GMQ dimer) is able to activate the ASIC3E79C channels (7) at neutral pH, collectively suggest that specific interactions at the site 2 cavity lined by Glu-79, Glu-423, Leu-77, Arg-376, Gln-269, and Gln-271 are important for the GMQ-induced channel gating.
FIGURE 4.
Residues surrounding the putative GMQ-binding site core. A, structure and key residues (displayed in colorful sticks for emphasis) around the putative GMQ-binding site core (displayed in gray sticks with mesh surrounding). Only two of three subunits are shown for clarity. The palm domains of subunit A and B are colored yellow and pink, respectively. B, EC50 values of GMQ (means ± S.E., n = 3–23) for point mutations. *, p < 0.05; **, p < 0.001 versus WT ASIC3 (dashed line). The maximal current values were obtained at the saturating concentrations (3–5 mm GMQ). The green bars represent data derived from Fig. 3B for comparison. The WT EC50, taken from Fig. 2B, is regraphed again for comparison.
Constructing GMQ Binding Mode Using in Silico Docking
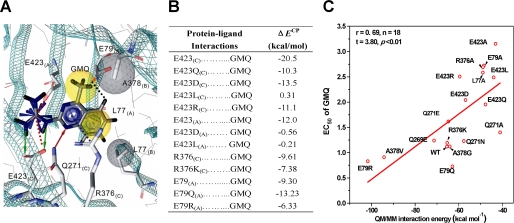
Next, a putative GMQ binding mode in the site 2 cavity lined by residues around Glu-79 and Glu-423 of the palm domain was formulated by using in silico (ligand-protein) docking simulation (see “Experimental Procedures”). This GMQ binding mode predicts that the guanidinium group of GMQ binds to a small pocket consisting of Glu-423(C), Gln-269(C), Glu-E423(A), Gln-271(A), and Arg-376(A), and the 4-methylquinazoline group resides in a relatively large pocket composed of Arg-376(C), Arg-376(B), Gln-271(C), Gln-271(B), Leu-77(A), Leu-77(B), and Glu-79(A) (Fig. 5A, where (A), (B) and (C) indicate the three subunits). In particular, the guanidinium group of GMQ interacts with the side chains of Glu-423(C) and Glu-423(A) through H-bonding and/or electrostatic interaction, respectively; the side chains of Arg-376(C) form typical cation-π interactions with the quinazoline ring; the side chains of Leu-77(A) and Leu-77(B) interact with the benzene ring of GMQ through hydrophobic interactions; the methyl group of GMQ contacts with the side chain of A378(B) through hydrophobic interaction; and the side chain of Glu-79(A) forms a charge-assisted H-bond (–C=O····H H-bond) (25) with a hydrogen atom of the benzene ring.
FIGURE 5.
GMQ binding mode. A, residues contacting GMQ. Residues interacting with GMQ are displayed in sticks for emphasis. The green and red arrows indicate H-bond contact between GMQ and Glu-423 or Gln-271, respectively. The black, red, and blue dashed lines indicate charge-assisted H-bond, electrostatic, and cation-π interactions between residues and GMQ, respectively. The residues of ASIC3 (gray sphere) and groups of GMQ (yellow sphere) implicated in the hydrophobic interactions are displayed with a surface sphere. All ligand-ASIC3 interactions were autodetected by Ligandscout 2.02 (trail version), except for the charge-assisted H-bond. B, summary of the ab initio calculations of ΔECP based on the proposed GMQ binding mode. ΔECP represents interaction energy with basis set superposition error correction. C, correlation between the GMQ-ASIC3 interaction energies and their corresponding EC50 values of GMQ in the WT and mutated ASIC3 channels. The interaction energies were calculated using QM/MM hybrid methods encoded in Dmol3 of Discovery Studio®, version 2.1. A natural division had defined the ligand as the QM region and the ASIC3 channel as the MM zone.
Testing GMQ Binding Mode Using ab Initio Calculation and Mutagenesis
We then tested the GMQ binding mode in the ASIC3 channels using a combination of site-directed mutagenesis, electrophysiological assays, and ab initio calculations of residue-GMQ interaction energies (ΔECP; see “Experimental Procedures”). Consistent with the mode prediction, the EC50 values of GMQ-induced currents of E423A, E423D, E423L, E423Q, and E423R mutant channels were all increased, because these mutations abolished the H-bonding and/or electrostatic interactions of GMQ with Glu-423(C) and/or Glu-423(A) (Figs. 3B and 5A) (7). Moreover, the ΔECP between Glu-423(C) and GMQ decreased in E423Q, E423D, E423L, and E423R mutations, as compared with the WT channel (Fig. 5B). Similarly, the interaction between Glu-423(A) and GMQ was fully abolished by E423D and E423L mutations as indicated by the ΔECP values (Fig. 5B). Additionally, L77A mutation increased the EC50 value, probably because of the reduced hydrophobic interaction between L77A and GMQ. Also, interrupting the cation-π interaction by R376A mutation increased the EC50, while maintaining the cation-π interaction in R376K mutation had no effect on EC50 (Figs. 3B and 5A), which is consistent with the ab initio calculated ΔECP values (Fig. 5B). Furthermore, because the E79A mutation abolishes the -C=O····H H-bond, it increased the EC50 value dramatically (Fig. 3B). However, the E79Q mutation decreased the EC50, whereas E79R had no significant effect on EC50 (Fig. 3B). The effect of E79Q may be attributed to the stabilization of the -C=O····H H-bond, whereas the E79R mutation may have created a new cation-π interaction between the cationic end of arginine and the benzene ring of GMQ. In support of these explanations, ΔECP between GMQ and the channel was increased or only slightly decreased in the E79Q or E79R mutant, respectively, when compared with the WT channel (Fig. 5B). In addition, the contact of GMQ with Gln-271, Gln-269, and Ala-378 may not be as important as the contact with Glu-423, Leu-77, Arg-376, or Glu-E79 in determining the apparent GMQ affinity, because mutations at these positions (Q269E, Q271A, Q271N, Q271E, A378G, and A378V) only slightly increased or had no effect on the EC50 values (Fig. 3B). However, mutations at these positions significantly altered the amplitude of GMQ-induced maximal currents (Fig. 3B), suggesting an allosteric role of contacts between GMQ and these residues in GMQ-induced channel gating (see below and Fig. 6).
FIGURE 6.
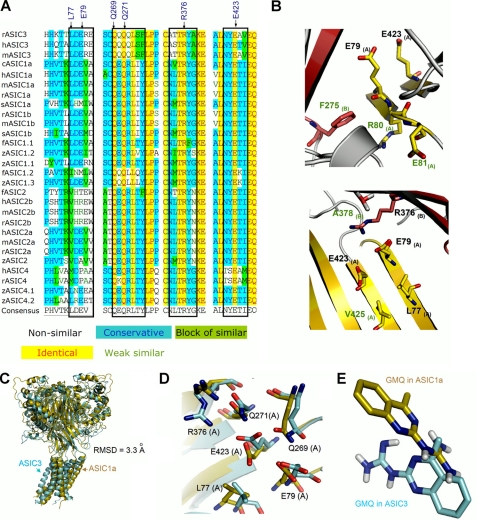
Analysis of GMQ-interacting residues in different ASIC subunits. A, amino acid sequence alignment of different ASIC subunits of the region corresponding to residues in ASIC3 that interact with GMQ. The key amino acids involved in forming the GMQ-binding site in ASIC3 are marked by black arrows. The black boxes indicate that although the key residues involved in shaping the GMQ-binding site are highly conserved, the amino acids adjacent to these residues are not conserved among various ASIC subunits. B, conserved residues (black labels) involved in shaping the GMQ-binding site and the adjacent nonconserved residues (green labels). Residues are displayed as sticks for emphasis. C, structural superimposition of ASIC3 (azure) with ASIC1a (golden) after 2.5 ns of MD simulations at neutral pH. D, same as C, highlighting variations in the positions of key residues involved in shaping the putative GMQ-binding site in ASIC3 (azure) as compared with that of ASIC1a (golden). E, distinctive orientation of GMQ docked into the snapshots of ASIC3 (azure) and ASIC1a (golden) after 2.5 ns of MD simulations at neutral pH.
Although the agreement between ab initio calculations and mutational analyses is encouraging, it may also be somewhat fortuitous, because the calculated ΔECP values can only be used to detect changes in the interaction energy between GMQ and a single amino acid. To further quantify the correlation between the EC50 values and interaction energies, we performed an additional QM/MM hybrid analysis (see “Experimental Procedures”) for all mutations. Correlation analysis suggests that the predicted interaction energy changes for the mutations correlate with the observed changes in EC50 values (r = 0.69, n = 18, t = 3.80, p < 0.01) (Fig. 5C), which further supports the proposed GMQ binding mode (Fig. 5A).
GMQ Binding Mode Explains ASIC3 Subunit Specificity
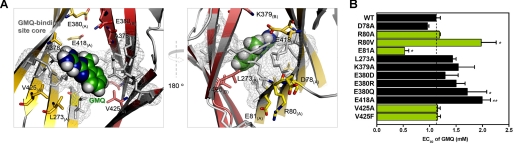
The key residues forming the GMQ-binding site are highly conserved among various ASIC subunits (Fig. 6A). Therefore, the observed specificity of GMQ to ASIC3 over ASIC1a (7) cannot be simply explained by the proposed GMQ binding mode. To address this issue, we performed multi-sequence alignment and MD simulations on both ASIC1a and ASIC3 channels at neutral pH (see “Experimental Procedures”) and followed up with mutational analysis. We noted that although the key residues involved in GMQ binding are highly conserved, the amino acids adjacent to these residues are unique in ASIC3 (e.g. Arg-80, Glu-81, Phe-275, Ala-378, and Val-425; Fig. 6, A and B). MD simulations revealed that ASIC1 and ASIC3 may adopt different conformational transition pathways, because the structural superposition between ASIC3 and ASIC1a showed that after 2.5-ns simulations, the conformation of ASIC3 was distinct from that of ASIC1a (Fig. 6C), especially the steric dispositions of the corresponding residues of the GMQ-binding site in ASIC3 (Fig. 6D). After docking GMQ into the optimized channels (see “Experimental Procedures”), we found distinct interaction patterns for GMQ-ASIC3 and GMQ-ASIC1a (Fig. 6E). These analyses led us to speculate that the subtle difference in the amino acid sequence could confer the specificity of GMQ to ASIC3 over ASIC1a. We chose to study six mutations (R80V, E81A, F275L, A378G, A378V, and V425A) that we expected to affect the GMQ effect. For most of these mutations, the changes in the GMQ-induced maximal currents (Fig. 3B) were in good agreement with model prediction. Notably, F275L markedly affected both the maximal current and EC50 value of GMQ (Fig. 3B), probably because of the significantly reduced constraint to the shape of GMQ-binding site (Fig. 6B). Thus, given their close positions to the core of GMQ-binding site, these nonconserved residues may play a part in defining subunit specificity of GMQ through stabilizing the spatial conformation of the GMQ-binding site.
GMQ Binding Mode Explains Interplay among H+, Ca2+, and GMQ
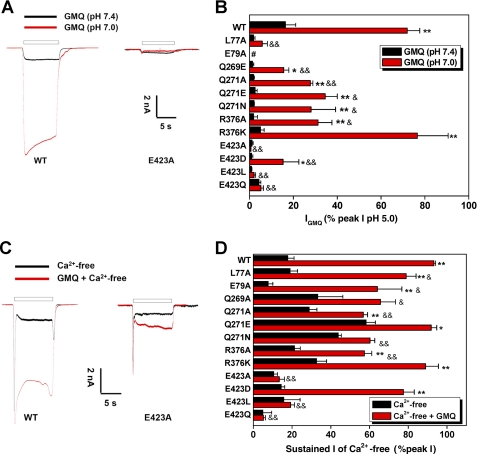
In a previous study, we demonstrated a dynamic interplay between GMQ and extracellular protons and Ca2+ in regulating ASIC3 channel activation (7) and suggested that the GMQ-binding domain may constitute a multifunctional sensor (26) that integrates diverse cellular signals (e.g. mild acidosis or reduction of extracellular Ca2+), which may be attained under pathophysiological conditions (27–31). As an additional step to validate the GMQ binding mode, we measured GMQ responses under pH 7.0 or Ca2+-free conditions and compared the channel activities among WT and various ASIC3 mutants with altered GMQ-binding residues. As expected, most mutations at the GMQ-binding residues, especially the alterations at the Glu-423 position, markedly reduced the enhancing effect of mild acidosis or reduced extracellular Ca2+ (Fig. 7). We noted that amino acid mutations on some residues of the nonproton ligand-sensing domain resulted in either increased (e.g. Q271A, Q271E, Q271N, Q269A, and R376K) or decreased (e.g. E79A, E423A, and E423Q) currents in the absence of Ca2+ (Fig. 7D), suggesting that the nonproton ligand-sensing domain is also involved in mediating Ca2+-free solution-induced sustained currents. Together, these results support that the residues involved in GMQ binding can also sense mild acidosis or directly interact with extracellular Ca2+ (4). Thus, changes of extracellular pH and Ca2+ may modulate the GMQ response through altering the conformation of the nonproton ligand-sensing domain in ASIC3 channels.
FIGURE 7.
Potentiation of GMQ-evoked current by mild acidosis (pH 7.0) or removal of extracellular Ca2+ is determined by GMQ-interacting residues. A, representative current traces showing the GMQ-induced current at pH 7.0 or 7.4 recorded from WT or the E423A mutant of ASIC3 channels. B, GMQ-induced (1 mm) currents (means ± S.E., n = 4–6) at pH 7.0 or 7.4 in CHO cells expressing various mutant channels. *, p < 0.05; **, p < 0.001; GMQ (pH 7.0) versus GMQ (pH 7.4). &, p < 0.05; &&, p < 0.001; GMQ responses (pH 7.0) in mutated versus WT channels. C, representative current traces showing the GMQ-induced (1 mm) sustained current in a Ca2+-free standard solution (pH 7.4) recorded from WT or the E423A mutant of ASIC3 channels. D, GMQ-induced (1 mm) currents (means ± S.E., n = 5–9) in Ca2+-free solution (with 5 mm EGTA) in CHO cells expressing various mutant channels. *, p < 0.05; **, p < 0.001; GMQ + Ca2+-free versus Ca2+-free. &, p < 0.05; &&, p < 0.001; GMQ responses (Ca2+-free) in mutated versus WT channels. For data normalization in B and D, a reference acidic solution (pH 5.0) was applied first to each CHO cell, and 2 min later, a test mixed solution composed of GMQ and pH 7.0 or Ca2+-free solution was applied, and the sustained current was normalized to the reference pH 5.0-induced peak current. GMQ-induced currents in ASIC3E79A mutant were normalized to the currents induced by pH 5.0 following a preconditioning pH 9.0 (to restore steady-state desensitization of ASIC3E79A mutant at pH 7.4).
GMQ Binding Mode Explains DTNB-induced ASIC3 Activation
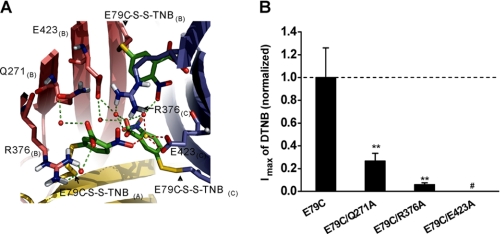
Finally, we tested the GMQ binding mode by using DTNB, which has been previously shown to lock the mutant ASIC3E79C channels in an open state (7). If residues involved in shaping the GMQ-binding site are essential for activating ASIC3 channels independent of acidosis, mutations at these residues would affect the DTNB-induced channel gating. Indeed, a further molecular covalent-docking test (see “Experimental Procedures”) suggested that Glu-423, Arg-376, and Gln-271 (Fig. 8A), which bind GMQ, might be important for coordinating the docking of DTNB to ASIC3E79C. Mutations at these three sites significantly reduced DTNB-induced ASIC3E79C activation (Fig. 8B), strengthening the notion that covalent activation of ASIC3E79C channels requires specific interactions between side chains of TNB and residues involved in forming the nonproton ligand-sensing domain (7).
FIGURE 8.
DTNB-induced ASIC3E79C channel activation at neutral pH is determined by GMQ-interacting residues. A, residues involved in DTNB-induced ASIC3E79C channel activation. Water-mediated (red balls, green dashed line) and Na+-mediated (gold ball, red dashed line) contacts with Arg-376, Glu-423, and Gln-271 established by the E79C-S-S-TNB complex. B, normalized maximal currents induced by DTNB in mutant channels (means ± S.E., n = 4–6). **, p < 0.001 versus ASIC3E79C channels (dashed line); #, little or no response to DTNB at up to 4 mm for 5 min.
DISCUSSION
This study combines computational analyses and mutagenesis/functional evaluations of ASIC3 channels to reveal the architecture of the nonproton ligand-sensing domain and detailed structural information about the ligand-ASIC3 interactions. Using homology modeling and MD simulations followed by functional testing of selected mutants, we show that GMQ binds specifically to the cavity made up of residues Glu-423, Glu-79, Leu-77, Arg-376, Gln-271, and Gln-269, to enable sustained activation of ASIC3 channels at neutral pH. Then using in silico docking and ab initio calculation, combined with mutational analysis, we proposed and characterized the GMQ-ASIC3 interaction mode. Whereas our data identified key determinants essential for mediating GMQ-ASIC3 interactions, there are at least three alternative explanations for the data observed here. First, GMQ most likely binds specifically to a cavity around Glu-79 and Glu-423 to activate the ASIC3 channel. Second, mutation-induced reduction of the actions of GMQ could result from allosteric effects if the cavity lined by Glu-79 and Glu-423 is proximate to the “real” GMQ-binding site. Third, it is also possible that the cavity around Glu-79 and Glu-423 represents just one of multiple GMQ-binding sites.
We favor a direct and specific interaction between the GMQ and the cavity lined by Glu-79 and Glu-423 for the following reasons. First, a covalent linkage between GMQ or TNB and Cys-79 is sufficient to activate ASIC3E79C channels, whereas GMQ dimer or DTNB treatment on WT or ASIC3E423C channels does not induce channel gating (7). Here, we extend this previous finding by showing that additional mutations at a cavity around Glu-79 and Glu-423 reduce the potency of covalent modification-induced channel gating (Fig. 8B). These results strongly suggest that stimulation of the cavity around Glu-79 and Glu-423 is both necessary and sufficient for ASIC3 channel gating. Second, we found that mutations at the residues located at the core of the putative GMQ-binding site reduced the apparent affinity of GMQ and abolished the maximal potency of GMQ on ASIC3 channels (Fig. 3B). However, mutations at the known pH sensors or residues adjacent to the putative GMQ-binding core did not affect or only slightly reduced the action of GMQ (Fig. 4B) (7). Finally, the GMQ binding mode as well as the makeup of the binding site well explains ASIC subunit selectivity (Fig. 6) and sensitivity of the GMQ response to mild acidosis and extracellular Ca2+ decrease (Fig. 7). The previous demonstration of a synthetic nonproton ligand (7) and the present elucidation of the specific binding domain involved in the nonproton ligand sensing should shed new light on future investigations of the endogenous nonproton ligands and gating mechanisms of ASIC3 channels.
ASICs are membrane-embedded trimeric complexes (4, 5) activated by the reduction of extracellular pH (1–3, 32). As the simplest ligand possible, whether H+ gates ASICs by titration of a single or multiple polar residues remains controversial (24). In recent years, multiple putative pH sensors have been proposed (4, 7, 8, 24, 33–43), e.g. the residues located in the cleft between the thumb and finger domains (4); residues around Glu-79 and Glu-423 in the palm domain (4, 7, 40); a cluster of acidic residues in the post-TM1 and pre-TM2 regions (Glu-63, His-72/His-73, and Asp-78 of ASIC1a) (24, 41). Because multiple proton-binding sites and multiple domains are involved in ASIC gating during acidosis, elucidation of a pore-opening mechanism of ASICs is substantially challenging. In addition to the putative pH sensors located on the extracellular domain, a number of studies provide evidence for the existence of at least three additional protonation sites involving residues in post-TM1 and pre-TM2 regions (24, 41), TM domains, and even intracellular domains (37, 42, 43). For all these known pH-sensing residues, only residues located in the cavity lined by Glu-79 and Glu-423 in the palm domain participate in the GMQ-induced channel gating (Fig. 3) (7), arguing for more specific roles of the palm domain in addition to its role as a pivotal module for transiting proton-induced conformational changes in the ASIC channel (4, 40). Furthermore, it is interesting to know whether the proposed GMQ-binding site also plays a dominant role in the acid-induced channel gating, if so, and how GMQ interaction with just a single pH sensor causes ASIC3 activation.
In summary, the present identification of the detailed atomic level GMQ binding model, together with the identification of nonproton ligands (7, 26), the availability of high resolution of crystallographic structures (4, 5), and the MD simulation methodology (44–47), provide a unique opportunity to refine our understanding of the structure and function of ASICs. Although the complexity of the ASICs presents a formidable challenge to theoretical studies, it is encouraging to note that many simulation studies from other families of ion channels have also yielded valuable information consistent with high resolution structural data (8, 45–47). Thus, the delineation of the architecture of nonproton ligand-sensing domain in ASIC3 channels at the atomic level represents a significant advancement in understanding the gating mechanisms of ASICs. Furthermore, given the emerging implications of ASIC3 channels in multiple sensory dysfunctions including nociception (48), this detailed structural information lay the foundation for conducting rational drug design and developing effective analgesics.
Acknowledgments
We thank all groups that provided us with ASIC cDNAs. We also thank Dr. M. X. Zhu for helpful comments on the manuscript.
This work was supported by National Natural Science Foundation of China Grants 30830035 and 9091300, National Basic Research Program of China Grant 2011CBA00408, the Shanghai Municipal Government Grant 09XD1404900, and the Sanofi-Aventis-Shanghai Institutes for Biological Sciences scholarship program 2009.
- ASIC
- acid-sensing ion channel
- GMQ
- 2-guanidine-4-methylquinazoline
- DTNB
- 5,5′-dithiobis(nitrobenzoic acid)
- MD
- molecular dynamics
- QM
- quantum mechanics
- MM
- molecular mechanics
- TM
- transmembrane
- TNB
- 5-thiol-2-nitrobenzonic acid.
REFERENCES
- 1. Wemmie J. A., Price M. P., Welsh M. J. (2006) Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 2. Krishtal O. (2003) Trends Neurosci. 26, 477–483 [DOI] [PubMed] [Google Scholar]
- 3. Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 4. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 5. Gonzales E. B., Kawate T., Gouaux E. (2009) Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Canessa C. M. (2007) Nature 449, 293–294 [DOI] [PubMed] [Google Scholar]
- 7. Yu Y., Chen Z., Li W. G., Cao H., Feng E. G., Yu F., Liu H., Jiang H., Xu T. L. (2010) Neuron 68, 61–72 [DOI] [PubMed] [Google Scholar]
- 8. Yang H., Yu Y., Li W. G., Yu F., Cao H., Xu T. L., Jiang H. (2009) PLoS Biol. 7, e1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu L. J., Duan B., Mei Y. D., Gao J., Chen J. G., Zhuo M., Xu L., Wu M., Xu T. L. (2004) J. Biol. Chem. 279, 43716–43724 [DOI] [PubMed] [Google Scholar]
- 10. Mahajan S., Ghosh S., Sudbeck E. A., Zheng Y., Downs S., Hupke M., Uckun F. M. (1999) J. Biol. Chem. 274, 9587–9599 [DOI] [PubMed] [Google Scholar]
- 11. Wang Q., Halpert J. R. (2002) Drug Metab. Dispos. 30, 86–95 [DOI] [PubMed] [Google Scholar]
- 12. Laskowski R., MacArthur M., Moss D., Thornton J. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 13. Berendsen H. J. C., van der Spoel D., van Drunen R. (1995) Comput. Phys. Commun. 91, 43–56 [Google Scholar]
- 14. Lindahl E., Hess B., van der Spoel D. (2001) J. Mol. Model. 7, 306–317 [Google Scholar]
- 15. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 16. Frisch M. J., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Zakrzewski V. G., Montgomery J. A., Jr., Stratmann R. E., Burant J. C., Dapprich S., Millam J. M., Daniels A. D., Kudin K. N., Strain M. C., Farkas O., Tomasi J., Barone V., Cossi M., Cammi R., Mennucci B., Pomelli C., Adamo C., Clifford S., Ochterski J., Petersson G. A., Ayala P. Y., Cui Q., Morokuma K., Malick D. K., Rabuck A. D., Raghavachari K., Foresman J. B., Cioslowski J., Ortiz J. V., Baboul A. G., Stefanov B. B., Liu G., Liashenko A., Piskorz P., Komaromi I., Gomperts R., Martin R. L., Fox D. J., Keith T., Al-Laham M. A., Peng C. Y., Nanayakkara A., Challacombe M., Gill P. M., Johnson B., Chen W., Wong M. W., Andres J. L., Gonzalez C., Head-Gordon M., Replogle E. S., Pople J. A. (2003) Gaussian 03, Gaussian, Inc., Pittsburgh, PA [Google Scholar]
- 17. Boys S. F., Bernardi F. (1970) Mol. Phys 19, 553–566 [Google Scholar]
- 18. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 19. Gurtovenko A. A., Patra M., Karttunen M., Vattulainen I. (2004) Biophys. J. 86, 3461–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berendsen H. J., Postma J. P., van Gunsteren W. F., Hermans J. (1981) In Intermolecular Forces (Pullman B. ed) p. 331, Reidel, Dordrecht, The Netherlands [Google Scholar]
- 21. Vanbuuren A. R., Marrink S. J., Berendsen H. J. C. (1993) J. Phys. Chem. 97, 9206–9212 [Google Scholar]
- 22. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. (1984) J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 23. Darden T. A., York D., Pedersen L. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 24. Paukert M., Chen X., Polleichtner G., Schindelin H., Gründer S. (2008) J. Biol. Chem. 283, 572–581 [DOI] [PubMed] [Google Scholar]
- 25. Grabowski S. J. (2004) J. Phys. Org Chem. 17, 18–31 [Google Scholar]
- 26. Li W. G., Yu Y., Zhang Z. D., Cao H., Xu T. L. (2010) Mol. Pain 6, 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutherland S. P., Benson C. J., Adelman J. P., McCleskey E. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Issberner U., Reeh P. W., Steen K. H. (1996) Neurosci. Lett. 208, 191–194 [DOI] [PubMed] [Google Scholar]
- 29. Nicholson C., Bruggencate G. T., Steinberg R., Stöckle H. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 1287–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Immke D. C., McCleskey E. W. (2001) Nat. Neurosci. 4, 869–870 [DOI] [PubMed] [Google Scholar]
- 31. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 33. Coscoy S., de Weille J. R., Lingueglia E., Lazdunski M. (1999) J. Biol. Chem. 274, 10129–10132 [DOI] [PubMed] [Google Scholar]
- 34. Coric T., Zhang P., Todorovic N., Canessa C. M. (2003) J. Biol. Chem. 278, 45240–45247 [DOI] [PubMed] [Google Scholar]
- 35. Vukicevic M., Weder G., Boillat A., Boesch A., Kellenberger S. (2006) J. Biol. Chem. 281, 714–722 [DOI] [PubMed] [Google Scholar]
- 36. Coric T., Zheng D., Gerstein M., Canessa C. M. (2005) J. Physiol. 568, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfister Y., Gautschi I., Takeda A. N., van Bemmelen M., Kellenberger S., Schild L. (2006) J. Biol. Chem. 281, 11787–11791 [DOI] [PubMed] [Google Scholar]
- 38. Bargeton B., Kellenberger S. (2010) J. Biol. Chem. 285, 13816–13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liechti L. A., Bernèche S., Bargeton B., Iwaszkiewicz J., Roy S., Michielin O., Kellenberger S. (2010) J. Biol. Chem. 285, 16315–16329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cushman K. A., Marsh-Haffner J., Adelman J. P., McCleskey E. W. (2007) J. Gen Physiol. 129, 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paukert M., Babini E., Pusch M., Gründer S. (2004) J. Gen Physiol. 124, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen X., Gründer S. (2007) J. Physiol. 579, 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W. Z., Chu X. P., Li M. H., Seeds J., Simon R. P., Xiong Z. G. (2006) J. Biol. Chem. 281, 29369–29378 [DOI] [PubMed] [Google Scholar]
- 44. Klepeis J. L., Lindorff-Larsen K., Dror R. O., Shaw D. E. (2009) Curr. Opin Struct. Biol. 19, 120–127 [DOI] [PubMed] [Google Scholar]
- 45. Khalili-Araghi F., Gumbart J., Wen P. C., Sotomayor M., Tajkhorshid E., Schulten K. (2009) Curr. Opin Struct. Biol. 19, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klein M. L., Shinoda W. (2008) Science 321, 798–800 [DOI] [PubMed] [Google Scholar]
- 47. Shaw D. E., Maragakis P., Lindorff-Larsen K., Piana S., Dror R. O., Eastwood M. P., Bank J. A., Jumper J. M., Salmon J. K., Shan Y., Wriggers W. (2010) Science 330, 341–346 [DOI] [PubMed] [Google Scholar]
- 48. Li W. G., Xu T. L. (2011) ACS Chem. Neurosci. 2, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]