Abstract
Intracellular transport is largely dependent on vesicles that bud off from one compartment and fuse with the target compartment. The first contact of an incoming vesicle with the target membrane is mediated by tethering factors. The tethering factor responsible for recruiting Golgi-derived vesicles to the ER is the Dsl1 tethering complex, which is comprised of the essential proteins Dsl1p, Dsl3p, and Tip20p. We investigated the role of the Tip20p subunit at the ER by analyzing two mutants, tip20-5 and tip20-8. Both mutants contained multiple mutations that were scattered throughout the TIP20 sequence. Individual mutations could not reproduce the temperature-sensitive phenotype of tip20-5 and tip20-8, indicating that the overall structure of Tip20p might be altered in the mutants. Using molecular dynamics simulations comparing Tip20p and Tip20-8p revealed that some regions, particularly the N-terminal domain and parts of the stalk region, were more flexible in the mutant protein, consistent with its increased susceptibility to proteolysis. Both Tip20-5p and Tip20-8p mutants prevented proper ER trans-SNARE complex assembly in vitro. Moreover, Tip20p mutant proteins disturbed the interaction between Dsl1p and the coatomer coat complex, indicating that the Dsl1p-coatomer interaction could be stabilized or regulated by Tip20p. We provide evidence for a direct role of the Dsl1 complex, in particular Tip20p, in the formation and stabilization of ER SNARE complexes.
Keywords: Endoplasmic Reticulum (ER), Membrane Fusion, Membrane Trafficking, Vesicles, Yeast, Dsl1 Complex, SNARE Complex, Retrograde Transport, Vesicle Fusion, Vesicle Tethering
Introduction
The correct targeting and delivery of proteins and lipids to various organelles, including the cell membrane, is an essential process in eukaryotic cells. Vectorial transport ensures directionality and provides the order in which proteins travel through organelles along the secretory pathway. Traffic between different membrane-bound compartments is mediated mostly by transport vesicles. The basic principles for generation and consumption of COPII and COPI vesicles that operate in the ER-Golgi3 shuttle and within the Golgi complex are very similar. In either case, a small GTPase (Sar1p for COPII and Arf1p for COPI) is recruited to the donor membrane and activated. The GTPase in turn recruits cargo, SNAREs, which are important for the subsequent fusion event, and additional coat proteins (1–3). In addition to Sar1p, the COPII coat consists of the Sec23/34 and Sec13/31 subcomplexes, whereas COPI vesicles require Arf1p, an ArfGAP, and the heptameric coatomer complex (4–8). The polymerization of coat proteins deforms the membrane, and ultimately, transport vesicles are released. The exact timing for uncoating of vesicles remains elusive, but some partial uncoating may occur already during the budding process and could be completed after tethering to the target membrane (3, 9–11). The first contact of the vesicle with the acceptor membrane is mediated by tethering factors bringing the vesicle closer to the membrane at which a vesicle SNARE (v-SNARE) and the SNAREs on the target membrane (t-SNAREs) form a four-helix bundle (trans-SNARE complex), thereby promoting fusion of the lipid bilayers (12–15). Some SNAREs can participate in multiple fusion events at different compartments, and these can act redundantly to other SNAREs (16–21). Therefore, additional factors are needed to provide specificity in the fusion process. Rab/Ypt-GTPases, tethering factors as well as Sec1/Munc18 proteins have been shown to orchestrate, stabilize, and proofread the assembly of cognate v-t-SNARE complexes (22–29).
In yeast, the SNARE complexes involved in the fusion of Golgi-derived COPI vesicles with the ER are the v-SNARE Sec22p and the three t-SNAREs Sec20p, Ufe1p, and Use1p (24, 30–32). In addition, another v-SNARE, Bet1p, could also participate in the fusion of retrograde transport carriers with the ER (33). In contrast, the trans-SNARE complexes formed during the fusion of COPII vesicles at the Golgi contain the t-SNARE Sed5p and the v-SNAREs Bos1p, Bet1p, and Sec22p or Ykt6p, which seem to be functionally redundant in this process in vivo (18, 24); in this case, the v-SNAREs seem to provide three helices and the t-SNARE only one.
The Dsl1 tethering complex consisting of Dsl1p, Dsl3p, and Tip20p, is essential for the fusion of retrograde Golgi-derived COPI vesicles with the ER (27, 34–39). The Dsl1 complex binds to the ER through interaction of Dsl3p and Tip20p with Use1p and Sec20p, respectively, whereas Dsl1p interacts with the coatomer complex (27, 34–38). Recent evidence suggests that the complex also accelerates the formation of ER trans-SNARE complexes (27). Thus, the Dsl1 complex appears to have two functions: one is tethering COPI vesicles through Dsl1p and the second is increasing the efficiency of the fusion process through acceleration of SNARE complex assembly.
Previously, we showed that a temperature-sensitive allele of TIP20, tip20-8, caused the back-fusion of COPII vesicles to the ER (40), a process that normally does not occur in the cell. In contrast, another allele, tip20-5, or dsl1-1did not show this phenotype. In this study, we aimed to understand the function of Tip20p in the Dsl1 complex. We found that tip20-8 and tip20-5 contained multiple mutations that were not clustered to a specific part of the protein. No single point mutation showed a significant phenotype. However, molecular dynamics simulations revealed that Tip20-8p is generally more flexible than the wild-type protein. In particular, the N-terminal hinge region, which is in immediate vicinity of the Dsl1p interactions site and several residues within the long α-helical stalk region that also includes the binding site for Sec20p, showed increased fluctuations. The Tip20 mutant proteins were defective in Dsl1 complex formation, and, as a consequence, SNARE complex assembly was strongly reduced. In addition, Tip20p mutant proteins inhibited binding of coatomer to Dsl1p. We provide evidence that Tip20p plays two different regulatory roles in the Dsl1 complex: first controlling the interaction with coatomer and second driving SNARE complex assembly at the ER.
EXPERIMENTAL PROCEDURES
Alignment and Evolutionary Conserved Residues
The alignment and evaluation of evolutionarily conserved residues were performed using the ConSurf database (41). For the alignment, the algorithm used PSI-Blast to extract in total 56 related sequences from the UniProt database and aligned these using standard methods. The evolutionary conservation of each amino acid position was calculated using the Rate4Site algorithm (42). The conservation scores were normalized and translated to nine color codes, which represent the grade of conservation, one is maximum variability, and nine is maximum conservation.
Mapping of Mutations in Crystal Structure
The x-ray structure for Tip20p was downloaded from the Protein Data Bank (Protein Data Bank code 3FHN). This structure has a number of missing loops, which were rebuilt using the ModLoop server (43) for automated modeling of loops in protein structures. The tip20-5 and the tip20-8 mutations were incorporated into the structure using the mutation tool in the Swiss-pdb Viewer (44). The side chain conformations of the mutated residues were regenerated from the backbone structure of 3FHN using the program SCWRL (45).
Strain Construction
Standard techniques for DNA manipulation (46) and standard yeast genetic techniques and media (47) were used throughout. Yeast strains used in this study are listed in supplemental Table S1, sequences of the primers are listed in supplemental Table S2, and constructs are listed in supplemental Table S3.
Yeast strains that express variants of Tip20p containing only one of the mutations identified in tip20-8 or tip20(Δ1–81), tip20(I10D,L28E) or tip20(V17E), were constructed as follows. Expression plasmids (kindly provided by F. M. Hughson) of the corresponding constructs or wild-type TIP20 were subcloned into a LEU2 plasmid carrying a fusion construct of the 5′- and 3′-UTR of TIP20. These plasmids then were transformed into a yeast strain in which TIP20 was chromosomally deleted. TIP20 is essential, therefore a URA3 plasmid with a wild-type copy of TIP20 was present in the Δtip20 strain to keep it viable. After transformation of the LEU2 plasmids, the URA3 plasmid was shuffled out of the strains using 5-fluoroorotic acid, leaving a tip20 variant as the sole source of Tip20p.
Antibodies
Polyclonal rabbit antibodies directed against Tip20p, Arf1p (33), Sec61p (generous gift from M. Spiess), Dsl1p, Dsl3p (both generous gifts from H. D. Schmitt), Ykt6p (generous gift from C. Ungermann), Bos1p and coatomer (48), mouse monoclonal anti-HA (Sigma), anti-His (AbD Serotec and GE Healthcare), anti-Pgk1p (Invitrogen) antibodies, and HRP-conjugated anti-His antibody (Sigma) were used in this study.
Growth Assays
For growth assays, cells of the indicated strains were grown to logarithmic phase in YPD medium, diluted to a cell density of 0.1 A600, followed by four serial dilutions of 10-fold each. Drops were spotted on YPD plates and incubated at indicated temperatures for appropriate times.
Preparation of Yeast Total Cell Extract
Of each of the indicated strains, cells from logarithmically growing cultures were harvested (8 A600), washed once with H2O and resuspended in 200 μl buffer B88 (20 mm HEPES, pH 6.8, 150 mm KAc, 5 mm Mg(Ac)2, 250 mm sorbitol) supplemented with 1 mm DTT, aprotinin, leupeptin, and pepstatin A. About 160 μl of glass beads were added. After vigorous vortexing for 15 min at 4 °C, cell debris and glass beads were pelleted (5 min, 300 × g, 4 °C), and the supernatant (= total cell extract) was transferred to a fresh reaction tube. For subsequent analysis by SDS-PAGE and immunoblotting, 30 ng of the total cell extracts were used.
Subcellular Fractionation
Overnight cultures were diluted to 0.1 A600 and grown at the permissive temperature (23 °C) to A600 0.4–0.6. Cells equivalent of 13–26 A600 were harvested by centrifugation at 1,800 × g for 5 min, washed once with water, resuspended to 5 A600/ml in buffer A (100 mm Tris-Cl, pH 9.4, 10 mm DTT), and incubated for 5 min at room temperature. Afterward, the cells were harvested by centrifugation at 1,800 × g for 5 min and converted into spheroplasts by incubation at 5 A600/ml in buffer B (0.75 × YP, 0.7 m sorbitol, 0.5% glucose, 50 mm Tris-Cl, pH 7.5) containing 25 μl/ml Zymolase T20 (25 mg/ml) for 30 min at 23 °C. The spheroplasts were collected by centrifugation at 200 × g for 3 min, resuspended in 170–340 μl B88* buffer (20 mm HEPES, pH 6.8, 250 mm sorbitol, 150 mm NaAc, 5 mm Mg(Ac)2) supplemented with 1 mm DTT, aprotinin, leupeptin, and pepstatin A, transferred to a microfuge tube, and disrupted with a Dounce homogenizer on ice. Unlysed spheroplasts and cell debris were removed by centrifugation at 2000 × g for 2 min at 4 °C. The supernatant was transferred to a fresh microfuge tube and centrifuged at 13,000 × g for 10 min at 4 °C. Afterward, the supernatant was centrifuged at 100,000 × g for 60 min at 2 °C. Pellets were solubilized in the starting volume of modified B88* buffer. Samples were analyzed by immunoblot.
Molecular Dynamics Simulations
The molecular dynamics simulations for the native and mutant protein were carried out using the software package GROMACS (49). The protein structures were immersed into a water box of dimension 112.40 Å × 106.23 Å × 164.92 Å with periodic boundary conditions. A steepest descent minimization was performed to minimize the energy of each system and to relax the water molecules. Then, the systems were equilibrated to 300 K, and a production simulation was performed for 6 ns. The program Gromacs was used for the subsequent analysis of root mean square deviation (r.m.s.d.) and root mean square fluctuation (r.m.s.f.) with the modules g_rms and g_rmsf. For the principal component analysis the g_covar module was used to calculate and diagonalize the covariance matrix. The corresponding eigenvectors were analyzed with the g_anaeig module.
Protein Purification
The C-terminal GST-tagged cytoplasmic region of Ufe1p was purified from Escherichia coli cell lysates via glutathione (GSH)-agarose (Sigma-Aldrich). STE buffer (25% (w/v) sucrose, 50 mm Tris-Cl, pH 8.0, and 40 mm EDTA) served as lysis buffer. For washes, PBS containing 15% glycerol was used, and the elution was performed with 150 mm Tris-Cl, pH 8.0, 120 mm NaCl, 50 mm glutathione, 5 mm DTT, 1 mm EDTA, and 1 mm PMSF. The N-terminal GST-tagged cytoplasmic tail of Sec20p and the N-terminal GST-tagged Dsl1p were purified via GSH-agarose. B88 buffer (20 mm HEPES, pH 6.8, 150 mm KAc, 5 mm Mg(Ac)2, 250 mm sorbitol) with 0.5% Triton X-100 was used for lysis and washes. For elution, 20 mm HEPES, pH 6.8, 150 mm KAc, 5 mm Mg(Ac)2, 250 mm sorbitol, 50 mm glutathione, 0.5% Triton X-100 was used. The N-terminal His6-tagged Dsl1p, the N-terminal His6-tagged Tip20p, the N-terminal His6-tagged cytoplasmic region of Use1p coexpressed with Dsl3p, and the N-terminal His6-tagged cytoplasmic region of Sec20p were purified via nickel-nitrilotriacetic acid (Ni-NTA)-agarose (Qiagen). Lysis and washes were performed in 50 mm Tris-Cl, pH 8.0, 200 mm NaCl, 20 mm imidazole, and 2 mm β-mercaptoethanol. Proteins were eluted with 50 mm HEPES, pH 7.5, 150 mm KCl, 270 mm imidazole, 10% glycerol, and 2 mm β-mercaptoethanol. N-terminal His6-tagged Tip20-5p and Tip20-8p were purified via Ni-NTA-agarose (Qiagen). The lysis and washes were performed in 50 mm Tris-Cl, pH 7.5, 500 mm NaCl, 20 mm imidazole, 1% Triton X-100, 10% glycerol, and 1 mm TCEP. Protein was eluted with 50 mm Tris-Cl, pH 7.5, 500 mm NaCl, 300 mm imidazole, 1% Triton X-100, 10% glycerol, 1 mm TCEP. The C-terminal His6-tagged cytoplasmic regions of Sec22p, Ykt6p, Bet1p, Bos1p, and Snc1p were purified via Ni-NTA-agarose (Qiagen) according to manufacturer's instructions. Coatomer was purified as described previously (50).
GST Pulldown Assay
GST fusion proteins (2.5 μg) were immobilized onto 25 μl of 50% glutathione-agarose slurry (GE Healthcare) for 60 min at 4 °C. Unbound proteins were removed by three washes with Buffer C (25 mm Tris-Cl, pH 7.5, 150 mm KCl, 10% glycerol, 1% Triton X-100, 2 mm β-mercaptoethanol). The beads were incubated with recombinant His6-tagged proteins (5 μg or 100 μg) in Buffer C at 4 °C on a rotating wheel. The total reaction volume was 200–800 μl. After binding, beads were washed three times with Buffer C, transferred to a fresh tube, washed once with 25 mm Tris-Cl, pH 7.5, and heated to 65 °C for 10 min in sample buffer. Eluted proteins were analyzed by SDS-PAGE followed by Coomassie Blue staining or immunoblotting.
RESULTS
Mutations in tip20-8 and tip20-5 Are Scattered throughout the Gene
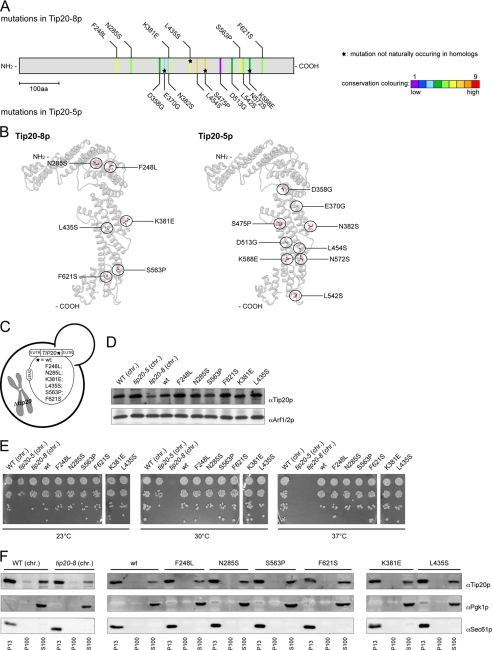
To better understand the role of Tip20p in the Dsl1 complex, we first sequenced two temperature-sensitive mutant alleles, tip20-5 and tip20-8. The tip20 mutant alleles were generated by error-prone PCR (51), suggesting that more than a single point mutation should be found in the mutants. Indeed, tip20-5 and tip20-8 contain nine and six mutations, respectively, that cause amino acid changes (Fig. 1A and supplemental Fig. S1). All mutations, except for L435S in tip20-8 and E370G, S475P, and K588E in tip20-5 also occur naturally in sequences of TIP20 homologues in other species (Fig. 1A and supplemental Fig. S1). It is likely that the conserved changes may not be the major cause of the tip20-8 phenotype. Because the mutations found in tip20-5 and tip20-8 did not cluster on the linear sequence, we determined whether they would cluster in the three-dimensional space by introducing the mutations into the model of the crystal structure (Fig. 1B). Still, the mutations in Tip20-8p and Tip20-5p were more or less evenly distributed throughout the protein, with some enrichment along the long helical axis. Taken together, by comparing the distribution of the mutations within the two alleles, we could not identify a specific region in Tip20p that would be responsible for the growth phenotype of tip20-5 and tip20-8 cells.
FIGURE 1.
Analysis of Tip20-5 and Tip20-8 mutants. A, sequencing of the tip20-5 and the tip20-8 alleles revealed nine and six amino acid (aa) changes, respectively. An alignment and evaluation of evolutionarily conserved residues for Tip20p was performed using the ConSurf database (41). The conservation scores were normalized and translated to nine color codes, which represent the grade of conservation; 1 is maximum variability and 9 is maximum conservation. The mutations occurring in Tip20-8p (top) and in Tip20-5p (bottom) were mapped onto the linear sequence. Stars indicate mutations that do not occur naturally in sequences of TIP20 homologues in other species. Neither the mutations found in tip20-8 nor the ones identified in tip20-5 cluster on the linear sequence. B, the mutations in Tip20-8p and Tip20-5p are relatively evenly distributed throughout the protein, with some enrichment along the α-helical stalk region of the protein. Mutations occurring in Tip20-8p (left) and in Tip20-5p (right) were incorporated into the x-ray crystal structure of Tip20p (3FHN) using the mutation tool in the Swiss-pdb Viewer (44). The side chain conformations of the mutated residues were regenerated from the backbone structure using the program SCWRL (45). C, schematic drawing of yeast strains expressing variants of Tip20p that contain only one of the mutations identified in tip20-8. D, all single point mutation constructs express Tip20p to a similar extent than the wild-type constructs. Immunoblots of protein extracts from the single point mutation were performed. Detection of Arf1/2p was used as loading control. chr., the chromosomally encoded TIP20 versions. E, none of the single point mutations showed any growth defect at any tested temperature. Growth assays were performed at the indicated temperatures to test the tip20 mutant strains. The tip20-8 strain displays a growth defect at 30 °C and above, whereas the tip20-5 strain only ceases to grow at 37 °C. F, in all strains, most of Tip20p was found in the P13 fraction, which contains mostly ER membranes. A smaller portion of Tip20p was found in the S100 fraction. Subcellular fractionations of the indicated strains were performed and analyzed by immunoblots. Pgk1p was used as a marker for cytosolic proteins, whereas Sec61p served as a marker for ER membranes.
Individual Point Mutations Do Not Recapitulate the Growth Phenotype of tip20-8
Because the mutations did not cluster in a particular part of TIP20, we constructed yeast strains that express variants of Tip20p containing only one of the mutations identified in tip20-8 (Fig. 1C). For this purpose, we cloned the single point mutations occurring in tip20-8 or wild-type TIP20 into a LEU2 plasmid carrying a fusion construct of the 5′- and 3′-UTR of TIP20. These plasmids were then transformed into a yeast strain in which TIP20 was chromosomally deleted. As TIP20 is essential, a URA3 plasmid with a wild-type copy of TIP20 kept the cells alive. After transformation of the point mutation containing LEU2 plasmid, the URA3 plasmid was shuffled out of the strains using 5-fluoroorotic acid, leaving the single point mutation-constructs as the sole source of Tip20p.
First, we had to ensure that all the single point mutation constructs were expressed. To this end, we prepared native lysates of the different yeast strains and checked for Tip20p expression by immunoblot (Fig. 1D). All single point mutation-construct strains expressed Tip20p to a similar extent than the wild-type constructs, indicating that none of the single point mutations caused severe protein instability. Next, we assayed the tip20 mutant strains for temperature-sensitive growth. The tip20-8 strain ceased to grow at 30 °C (36, 40, 51). In contrast, none of the single point mutations showed any growth defect at any tested temperature (Fig. 1E). The data show that none of the individual mutations severely affects Tip20p function and suggest that combinations of the mutations must be responsible for the tip20-8 phenotypes.
Membrane Association of Tip20p Is Not Altered in tip20 Mutants
Tip20p acts as part of the Dsl1 complex at the ER membrane (27, 34–39), and a more subtle effect, which may not lead to a growth defect, could be a less efficient recruitment of Tip20p variants to the ER. Possible changes in the distribution of the protein could contribute to the perturbed protein function in the tip20-8 strain. Therefore, we analyzed the membrane association of Tip20p variants by differential centrifugation (Fig. 1F) and found that in comparison with the wild-type strain neither the tip20-8 strain nor any of the single point mutation strains showed any changes in the membrane association of Tip20p. In all of the cases, most of the protein was present in the pellet fraction after a 13,000 × g spin (P13), which contains most of the ER as shown by the presence of the ER resident Sec61p protein. A smaller portion of Tip20p was detected in the S100 cytoplasmic pool. This finding is in agreement with Tip20p being a peripheral membrane protein. The membrane association remained largely unchanged when Tip20, tip20-5, and tip20-8 strains were shifted to 37 °C for 1 h prior to the differential centrifugation (supplemental Fig. S2). More importantly, the level of Tip20p membrane association was indistinguishable between the three Tip20 variants. Our data indicate that protein mislocalization or reduced ER association may not be the cause for the tip20-8 phenotype.
Mutations Occurring in Tip20-8p Lead to Increased Flexibility of N-terminal Hinge Region
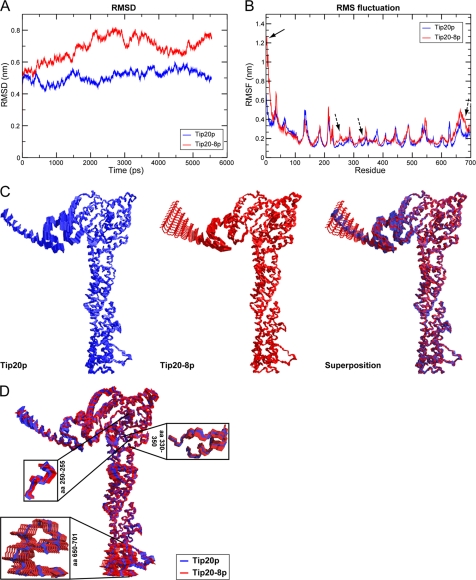
Since none of the point mutations in tip20-8 gave a noticeable phenotype, we analyzed to which extent the overall structure is affected in Tip20-8p compared with wild-type. To this end, we performed molecular dynamics simulations using the software package Gromacs. The Tip20-8p mutations were modeled onto the Tip20p structure (38). Then, both structures were subjected to identical conditions, and their molecular dynamics trajectories were calculated for 6 ns each. To estimate the quality and convergence of the molecular dynamics trajectory, the backbone r.m.s.d. values of each protein structure relative to their starting structures were calculated (Fig. 2A). We found that although Tip20p is quite stable during the simulation, the Tip20-8p simulation showed dramatic changes in the r.m.s.d. as an effect of the mutations. To further probe the source of these differences, r.m.s.f., which measures the movement of each residue in the system with respect to the average position of that residue, was calculated for both structures (Fig. 2B). These calculations showed a striking difference between the Tip20p and Tip20-8p for the first 25–30 residues and indicated that the N terminus in Tip20-8p is very flexible. This result is interesting because no mutation in Tip20-8p is located in this part of the molecule. However, it is not uncommon that amino acid mutations have an effect on protein structure, even in a remote position of the mutated site (52–54). In addition, further differences in the regions of residues 250–260, 330–350, and at the C terminus (residues 650–701) could be observed. To determine the exact range and location of the motions and the difference between Tip20p and Tip20-8p, we performed a principle component analysis on both of the structures. This technique is used to discriminate the background atomic fluctuations from larger more relevant movements of the protein. The principle component analysis confirmed the findings of the r.m.s.f. calculation. Analysis of the first two largest components showed very large movements in the first 30 N-terminal amino acids of Tip20-8p (Fig. 2C) (component 2), and some further smaller movement, particularly in regions of residues 250–255, 330–350, and 650–701 (Fig. 2D) (component 1). Taken together, the molecular dynamics simulations showed that Tip20-8p is in general more flexible compared with wild-type. This flexibility is most apparent in the N-terminal hinge region, which is in close vicinity of the Dsl1p binding site and in three parts along the long α-helical stalk region. Interestingly, the affected residues in the stalk region include the binding area (amino acids 82–356) of the ER t-SNARE Sec20p (27, 38). The increase in flexibility could make the Tip20-8p more susceptible to degradation by proteases. Indeed, we observed reduced Tip20p levels in tip20-8 compared with wild-type (Fig. 1D).
FIGURE 2.
Tip20-8p is more flexible than wild-type Tip20p. A, although Tip20p (blue) behaves rather stably during the molecular dynamics simulation, Tip20-8p (red) as an effect of the mutations shows dramatic changes in the r.m.s.d.. The backbone r.m.s.d. values of each protein structure relative to their starting structures were calculated to estimate the quality and convergence of the molecular dynamics trajectory. B, a striking difference between the Tip20p (blue) and Tip20-8p (red) for the first 25–30 residues (indicated by an arrow) and further differences in the regions of residues 250–260, 330–350, and the C terminus (residues 650–701) (indicated by dashed arrows) could be detected. The sources of the observed differences in r.m.s.d. were determined by computation of the r.m.s.f. Thereby, the movement of each residue in the system with respect to the average position of that residue was calculated for both structures. C, component 2 of the principal component analysis reflects the very large movements in the first 30 N-terminal amino acids. The maximal range as well as intermediate states of the movements for the wild-type Tip20p (left, blue) and the Tip20-8p (middle, red) is shown. On the right, a superposition (Tip20p in blue, Tip20-8p in red) is displayed. D, component 1 of the principle component analysis mirrors the observed fluctuations in the regions of the long α-helical stalk. A superposition of the maximal range as well as intermediate states of the movements for wild-type Tip20p (blue) and Tip20-8p (red) is shown. The boxes represent an enlargement of the regions (amino acids 250–255, 330–350, and 650–701) that displayed the biggest amplitude in movement.
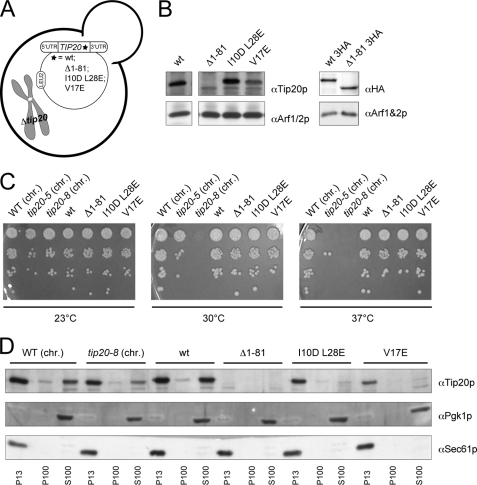
N Terminus of Tip20p Is Not Required for Growth or ER Localization
The increased flexibility of the hinge region connecting the N-terminal finger to the downstream α-helices in Tip20-8p suggested that the N terminus might be critical for Tip20p function. The N terminus of Tip20p (amino acids 1–81) appears to be necessary and sufficient for interaction with Dsl1p and thus for the correct assembly of the Dsl1 complex (27, 38). We used the same strategy as described above to construct yeast strains that contain either a version of Tip20p that is lacking the amino acids 1–81 (Δ1–81), or contains two (I10D,L28E) or one point mutation(s) (V17E), respectively, within the N-terminal region of the protein (Fig. 3A). These point mutant constructs were shown to abolish the interaction with Dsl1p similarly to the Δ1–81 construct (38). Extracts from the different tip20 variant strains revealed that the expression level of the point mutations was comparable with wild-type Tip20p (Fig. 3B). In contrast, the signal for the Δ1–81 construct was very low and appeared to co-migrate with another band, which could be a degradation product of Tip20p. To ascertain that the lower molecular weight band in Fig. 3B corresponded to Tip20Δ1–81p, we constructed yeast strains in which a HA3 tag was added to the C terminus of wild-type and the N-terminal deletion construct. The HA antibody recognized a band for wild-type Tip20p-HA and Tip20Δ1–81p-HA at the same height as the Tip20p antibody (Fig. 3B). Moreover, the band intensities for both constructs were comparable, indicating that Tip20Δ1–81p is not less stable than wild-type Tip20p and that the main epitope, which is recognized by our polyclonal antibodies resides in the N-terminal part of Tip20p.
FIGURE 3.
The N terminus of Tip20p is not required for growth or membrane localization. A, schematic drawing of yeast strains expressing variants of Tip20p that contain either a version of Tip20p that is lacking the amino acids 1–81 (Δ1–81) or containing two (I10D,L28E) or one point mutation (V17E), respectively. B, all constructs of N-terminal tip20 variants express Tip20p to a similar extent than the wild-type constructs. Immunoblots of protein extracts from the N-terminal tip20 variants were performed. Detection of Arf1/2p was used as loading control. C, none of the N-terminal tip20 variants showed a growth phenotype. Growth assays were performed at the indicated temperatures using the indicated tip20 variant strains. D, none of the N-terminal tip20 variants showed an aberrant localization of Tip20p. Subcellular fractionations of the indicated strains were performed and analyzed by immunoblots. chr., the chromosomally encoded versions of TIP20.
Next, we checked whether the N-terminal mutation constructs are essential for growth at various temperatures (Fig. 3C). None of the N-terminal mutations showed a growth phenotype, indicating that the direct interaction between Dsl1p and Tip20p is not essential for the function of the Dsl1 complex. One explanation for this observation could be that the two proteins do not need to interact for their proper localization. To test this possibility, we performed differential centrifugation experiments (Fig. 3D). All Tip20p constructs were membrane-associated to the same extent as wild-type, suggesting that Tip20p localizes to membranes independent of its interaction with Dsl1p. This finding is supported by data from Ren et al. (27), that show that the Dsl3p and the Tip20p subunits of the Dsl1 complex bind independently to the ER t-SNAREs Use1p and Sec20p, respectively.
Tip20-8p and Tip20-5p Binding to Dsl1p and Sec20p Is Reduced in Vivo and in Vitro
Because the interaction between Dsl1p and Tip20p did not seem to be essential for Dsl1 complex function, we wanted to determine whether the interaction of Tip20p with other proteins was impaired in tip20-5 and tip20-8 mutants. For this purpose, we chromosomally tagged TIP20, tip20-5, and tip20-8 with a Strep-tag and performed affinity purifications under native conditions followed by LC/MS analysis and immunoblotting in parallel. We identified, as reported previously (36), Sec22p and Ufe1p as interactors of Tip20p, and found that these interactions were strongly reduced in the mutant affinity purifications (data not shown). However, we could not detect the previously reported interaction with the ER t-SNARE Sec20p (36, 38, 55). To overcome this shortcoming, we performed purifications under denaturing conditions and included a cross-linking step using chromosomally tagged SEC20 with a His6-biotinylation sequence-His6 (HBH) tag in Tip20p wild-type and mutant strains. We found that less Tip20-5p and Tip20-8p were associated with Sec20p, when compared with wild-type (data not shown). These results are consistent with the wide distribution of the mutations over the entire sequence of TIP20 in the mutants and an increase of flexibility in various parts of at least Tip20-8p. We conclude that the interaction with all known Tip20p binding proteins was strongly reduced in both Tip20-5p and Tip20-8p in vivo.
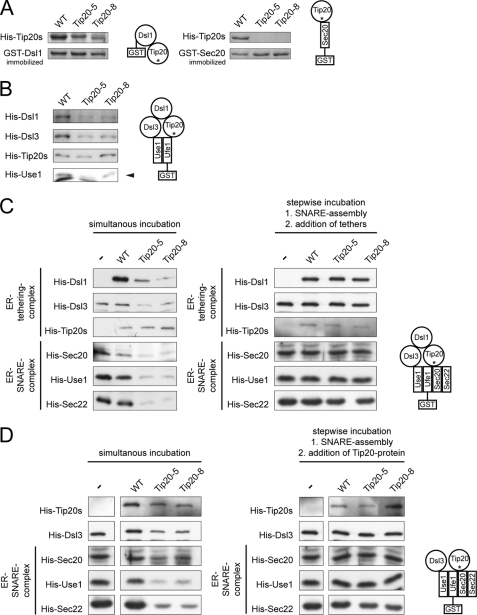
To confirm our results, we performed in vitro pulldown assays using Dsl1p-GST and His6-tagged Tip20p variants. The affinity of Tip20-8p and Tip20-5p for Dsl1p was significantly decreased compared with wild-type Tip20p in vitro (Fig. 4A). Because Tip20p interacts directly with Sec20p, we tested next the binding of Tip20p variants to GST-Sec20p. Similarly to the GST-Dsl1p experiment, the binding ability of the mutants was strongly reduced (Fig. 4A). Taken together, these experiments demonstrate a reduced binding capacity of Tip20p mutant proteins to Sec20p and Dsl1p in vivo and in vitro and are consistent with previously published data (36).
FIGURE 4.
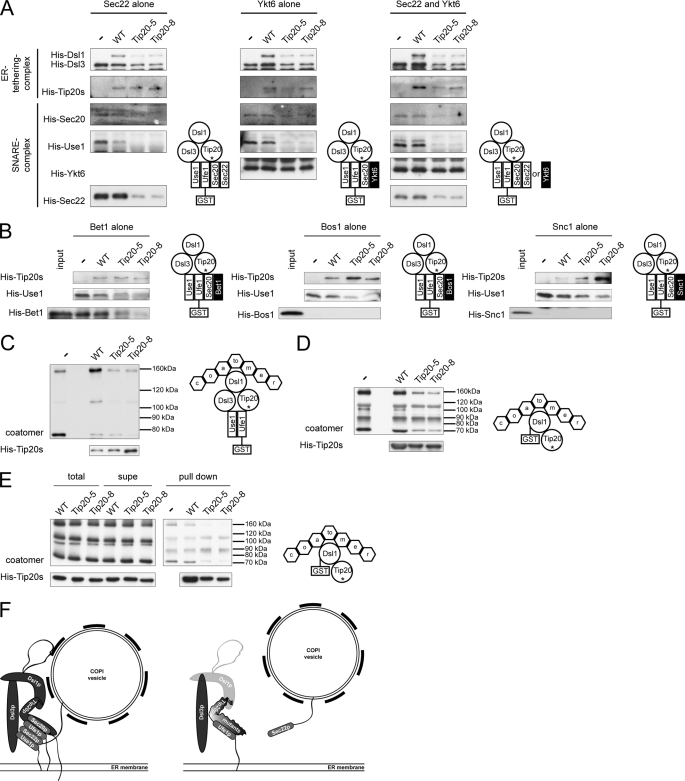
In vitro assembly of Dsl1 tethering complexes and ER trans-SNARE complexes is affected by Tip20p mutants. A, Tip20p mutants have a lower affinity for Sec20 and Dsl1p than wild-type Tip20p. A GST pulldown was performed with GST-Ds1p or GST-Sec20p and Tip20p variants. B, Dsl1p binding to Tip20-5p or Tip20-8p is drastically decreased. To reconstitute the Dsl1 tethering complex in vitro, pulldown assays were performed as indicated in the graphical representation and analyzed by immunoblotting. Note that Dsl3p and Use1p were added as a complex. Dsl1p and Dsl3p were detected with protein specific antibodies, whereas an anti-His antibody was used to detect Tip20p and Use1p. The arrowhead in the Use1p blot points to the upper band, which is the specific one. C, Tip20p is required for SNARE complex assembly. ER SNARE complexes were reconstituted either in the presence or absence of Dsl1 complex members. Mutant Tip20p proteins strongly reduced the amount of SNARE complexes formed. No effect was observed when the Dsl1 complex members were added after SNARE assembly had occurred. D, Dsl1p does not influence SNARE complex assembly. In vitro pulldown assay were performed as in C, but Dsl1p was omitted. The asterisk under Tip20 in the drawings is meant to illustrate the usage of wild type and mutant versions in the pulldowns.
Tip20-8p and Tip20-5p Block Dsl1 Complex Assembly in Vitro
Because the Tip20p-Dsl1p interaction was reduced in the presence of Tip20p mutant proteins, we asked next whether the mutants would generally affect Dsl1 complex assembly. Although it had been shown previously that Dsl1p interacts directly with Dsl3p (27), our attempts to pull down His6-Dsl3p with immobilized GST-Dsl1p failed, probably due to nonfunctional Dsl3p. Dsl3p directly interacts with the ER t-SNARE Use1p (27, 36), and Use1p could be only efficiently purified when coexpressed with Dsl3p (27). Thus, we used as a source of Dsl3p the complex of His6-Use1p/Dsl3p. Given this slight complication, we decided to build up the Dsl1 complex from the SNARE site and used a GST fusion to the ER t-SNARE, Ufe1p, (32), which has been shown to interact with both Use1p and the Dsl1 complex (30, 31, 36) and performed GST pulldowns (Fig. 4B). Interestingly, Tip20-5p and Tip20-8p bound to Ufe1p with similar efficiencies than wild-type, indicating that Tip20p can bind directly to Ufe1p (supplemental Fig. S3), independent of Sec20p, and that the mutations do not destabilize this interaction, whereas Dsl1p binding was strongly impaired. The affinity of Tip20p to Ufe1p appears to be relatively low (supplemental Fig. S3) and may be stabilized by the interaction with other proteins in vivo. Because Dsl3p is expressed in a complex with Use1p, and Use1p can bind directly to Ufe1p, we refrained from drawing any conclusions about Dsl3p directly. However, Dsl3p was unable to efficiently immobilize Dsl1p, indicating that either this interaction is too weak to stably recruit Dsl1p or that the Dsl3p-Use1p complex was also recruited inefficiently to Ufe1p-GST. The interaction of the Tip20p mutant proteins with Ufe1p explains why Tip20p mutants are still ER localized, despite the loss of interaction with Sec20p. Moreover, these data provide strong evidence that the Dsl1 complex is destabilized in tip20-5 and tip20-8 mutants, probably also in vivo.
Tip20p Mutants Inhibit Trans-SNARE Complex Assembly in Vitro
Formation of a SNARE complex consisting of the ER-localized SNAREs Sec20p, Ufe1p, and Use1p with the v-SNAREs Sec22p and/or Bet1p is necessary for the fusion of COPI vesicles with the ER (24, 30–33). Moreover, a recent study suggests that the Dsl1 complex accelerates SNARE complex assembly at the ER (27). Because Tip20-5p and Tip20-8p failed to bind to Sec20p, but not to Ufe1p, we asked whether Tip20-8p and Tip20-5p would influence ER SNARE complex assembly. To this end, GST-Ufe1p was immobilized and incubated with the remaining ER t-SNAREs, the v-SNARE Sec22p and Dsl1 complex members (Fig. 4C). As expected, SNARE complex assembly occurred in the presence of wild-type Tip20p. In contrast, in the presence of the Tip20p mutant proteins, ER SNARE complex assembly was severely perturbed, and none of the other SNARE proteins was efficiently incorporated into the SNARE complex. Tip20p must have a regulatory function during SNARE complex assembly at the ER because SNARE complexes containing Sec20p, Use1p, and Sec22p were formed properly, when first SNARE complexes were formed, and then Dsl1 complex members were added in a second incubation step (Fig. 4C). Dsl1p was recruited efficiently to these SNARE complexes. However, this binding was most likely dependent on the Dsl3p-Use1p complex, which was present during the SNARE complex assembly step. These data are consistent with the observation that ER SNARE complex assembly is accelerated in vitro by the presence of the Dsl1 complex (27). Taken together, our data indicate that Tip20p is required for proper SNARE complex assembly.
Tip20p and Not Dsl1p Is Required for Proper SNARE Complex Assembly
So far, the defect in SNARE complex assembly could be due to the reduced interaction of Tip20-5p and Tip20-8p with Dsl1p. Alternatively, the phenotype could be completely independent of Dsl1p. To distinguish between those possibilities, we repeated the above experiment in the absence of Dsl1p (Fig. 4D). Again, SNARE complexes assembled in the absence of Dsl1p when wild-type Tip20p was present, and this assembly was reduced when mutant Tip20p was added to the incubation mixture. Interestingly, the reduction in SNARE complex assembly was independent of the presence of Dsl1p in the assay. Therefore, we conclude that Tip20p may have a more prominent role in SNARE complex assembly than Dsl1p.
ER SNARE Complex Assembly Is Not Rescued by Alternative v-SNAREs in tip20 Mutants
Sec22p is not the only v-SNARE that could potentially engage in a trans-SNARE complex at the ER. Bet1p and Ykt6p have been shown to be substitutes for Sec22p (18, 33). In addition, it has been shown that SNARE-SNARE interactions under some circumstances are promiscuous and that the formations of nonphysiological SNARE complexes can take place (20, 21, 56, 57). Therefore, we tested whether in the presence of Tip20-5p and Tip20-8p, ER SNARE complexes would become more promiscuous. First, we decided to look at Ykt6p, which seems to be able to substitute for v-SNAREs in more than one type of SNARE complexes (18, 24, 58). Ykt6p interacted with Ufe1p equally well, independent of the presence of wild-type or a mutant form of Tip20p in the assay (Fig. 5A). However, this interaction did not improve the recruitment of Use1p or Sec20p into the complex. Therefore, binding of Ykt6p to Ufe1p is independent of the other ER t-SNAREs. Adding Ykt6p and Sec22p simultaneously did not improve the SNARE complex assembly at the ER. Because neither Sec20p nor Use1p were efficiently recruited to Ufe1p in the presence of Ykt6p, Ykt6p does not drive SNARE assembly and cannot overcome the defects introduced by the Tip20p mutant proteins.
FIGURE 5.
The assembly of ER SNARE complexes in tip20 mutants is not rescued by alternative v-SNAREs and coatomer binding is affected in the presence of Tip20-5p or Tip20-8p. A, Ykt6p binds to GST-Ufe1p but does not promote SNARE complex assembly. Pulldown assays were performed as indicated in the graphical representation. B, Bet1p, Bos1p, and Snc1p do not improve SNARE complex assembly in the presence of the Tip20p mutants. Pulldown assays were performed as indicated in the graphical representation. C, coatomer binding to mutant Dsl1 complexes is reduced in vitro. To assess whether the coatomer binding function of the Dsl1 complex is affected by the Tip20p mutants, Dsl1 complexes were assembled, and coatomer was added either at the same time or only after Dsl1 complex preassembly. D, Tip20-5 and Tip20-8 negatively regulate the interaction between coatomer and Dsl1p. Tip20p variants and coatomer were incubated with GST-Dsl1p and the binding of the proteins to GST-Dsl1p was determined by immunoblot. E, GST-Dsl1p and Tip20p do not compete for the same binding site on coatomer. Preincubation of Tip20p proteins with coatomer did not change the binding behavior. A pulldown experiment was performed as described in D except that coatomer and the Tip20p proteins were preincubated at 4 °C. To eliminate possible aggregation, the reaction mixture was spun for 15 min at 20,000 × g at 4 °C, and the supernatant was used for the binding reaction to GST-Dsl1p. F, scheme describing the results. For an explanation, see text.
Next, we tested whether Bet1p was efficiently recruited into SNARE complexes. Bet1p behaved similarly to Sec22p and was only incorporated into SNARE complexes in the presence of wild-type Tip20p (Fig. 5B). Tip20p mutants caused a strong reduction of Bet1p binding, and less SNARE complexes were formed. The effects for the ER v-SNAREs were specific because noncognate SNAREs as the v-SNARE at the Golgi, Bos1p, or the plasma membrane v-SNARE Snc1p could not be recruited at all to engage into SNARE complex formation (Fig. 5B). Our data indicate that in the presence of Tip20p mutant proteins, SNARE complex assembly at ER is severely altered but proper recognition of cognate v-SNAREs is not affected, as noncognate v-SNAREs were not recruited into ER SNARE complexes under any conditions tested in these assays.
Tip20p Regulates Binding of Dsl1p to Coatomer
So far, we have shown that Tip20p mutant proteins disturb the formation of ER SNARE complexes. We next wanted to test whether the upstream function the Dsl1 complex, namely the tethering of COPI vesicles through direct interaction with Dsl1p is also perturbed by Tip20p mutant proteins (27, 34–38). First, we performed a pulldown with GST-Ufe1p and the members of the Dsl1 complex and coatomer. Indeed, less coatomer was precipitated in the Tip20p mutant incubations compared with wild-type (Fig. 5C). This result could be explained by either less Dsl1p binding to mutant Tip20p and hence less coatomer recruitment or a more direct role of Tip20p in coatomer binding. To distinguish between these two possibilities, we performed a pulldown with GST-Dsl1p, coatomer, and Tip20p variants. Interestingly, the presence of Tip20p mutants reduced the amount of immobilized coaotmer, suggesting that Tip20p may play a more active role in Dsl1p binding to coatomer by either stabilizing the interaction between Dsl1p and coatomer or providing an additional coatomer binding site (Fig. 5D). Moreover, preincubation of Tip20p with coatomer did not change the binding efficiency of Tip20p or coatomer to Dsl1p, indicating that Tip20p and Dsl1p are not competing for the same binding site (Fig. 5E). Taken together, our results suggest a prominent role for Tip20p in the recognition of incoming COPI vesicles and in the assembly of trans-SNARE complexes at the ER.
DISCUSSION
In this paper, we investigated the function of the Dsl1 complex member Tip20p by analyzing the phenotypes of tip20 mutants and found that they interfered with the proper assembly of trans-SNARE complexes at the ER (Fig. 5F). Trans-SNARE complexes consisting of the t-SNAREs Sec20p, Ufe1p, and Use1p and the v-SNARE Sec22p and/or Bet1p promote fusion of Golgi-derived COPI-coated vesicles with the ER (24, 30–32). Ufe1p, Sec20p, Use1p, and Sec22p/Bet1p could not be efficiently assembled into ER SNARE complexes in the presence of mutant Tip20p in vitro. This defect was not compensated for by the third vesicle SNARE of the ER-Golgi shuttle, Bos1p, which was not incorporated into the ER SNARE complex under any conditions tested. Whereas Sec22p and Bet1p can participate in SNARE complex formation at both the ER and the Golgi (18, 33, 59), Bos1p appears only to act in the fusion process of COPII vesicles at the Golgi (18, 24, 33, 59, 60). The noncognate v-SNARE Snc1p, which acts at the plasma membrane, could also not be engaged in SNARE complex formation. Interestingly, Ykt6p, another v-SNARE, which can functionally replace Sec22p in the fusion of ER-derived COPII vesicles with the Golgi (18) binds efficiently to Ufe1p, irrespective of the presence of wild-type or Tip20 mutants, and even when added together with Sec22p. Yet, the presence of Ykt6p did not improve the incorporation of Sec20p, Use1p, or Sec22p in vitro, and hence, no functional SNARE complexes were formed. Our in vitro data suggest that Tip20-5p and Tip20-8p could act as dominant negative inhibitors of SNARE complex assembly. However, we cannot exclude that in vivo, when e.g. other factors like the Sec1/Munc18 protein Sly1p are present, Ykt6p could potentially assist in the incorporation of cognate SNAREs in the trans-SNARE complex at the ER. Our data are consistent with findings by Meiringer et al. (61), that Ykt6p can bind to the ER t-SNARE-Dsl1 complex but that it does not act as v-SNARE in this scenario. We propose that Tip20p mutant proteins do not change the specificity for cognate SNARE complex assembly but rather prevent trans-SNARE complex assembly. Consistent with this notion, mutant Tip20p already disturbed the interaction of the ER t-SNAREs with each other. However, when SNARE complexes were assembled in vitro prior to the addition of Tip20p variants, no effect on SNARE complex assembly was observed, indicating that Tip20p mutant proteins may have a decreased off-rate from the SNAREs. A plausible scenario is that the Dsl1 complex may associate with ER t-SNAREs and hand over the COPI vesicle to the ER SNAREs, which may be in a partially preassembled state. Sec22p interacts with the ER t-SNAREs and forms a trans-SNARE complex and allows fusion. In the presence of Tip20p mutant proteins, less COPI vesicles would be brought into close proximity to the ER because, although the mutant Tip20p are still ER-associated, the binding to Dsl1p, which interacts with coatomer, is reduced (27, 34–39). Tip20p seems to play also a regulatory role in the binding of Dsl1p to coatomer, the levels of which were drastically reduced in the presence of Tip20p mutant proteins. Alternatively, Tip20p could possess a cryptic coatomer-binding site, which would become more important in the mutant situation, and the mutant proteins would displace coatomer from Dsl1p. Such a cryptic interaction site is thought to be located in the N terminus of Tip20p of Schizosaccharomyces species (62). Thus, in principle, it is conceivable that the N-terminal region, which contains the Dsl1p binding site (27, 38) and which becomes very flexible in Tip20-8p, could adopt a conformation that is unable to recognize Dsl1p and becomes instead a coatomer-binding site. Yet, SNARE complex formation might not only be reduced because of the lower amount of COPI vesicles at the ER membrane but also because Tip20p mutant proteins would block efficient ER SNARE assembly by interacting with Ufe1p. The finding that defective Dsl1 complexes interfere with a proper assembly of cognate ER trans-SNARE complexes and the observation that the Dsl1 complex accelerates SNARE complex formation (27) provide evidence for a novel function of the Dsl1 complex, namely a role in actively contributing to SNARE complex assembly. Such a function has been suggested before for other tethering complexes. Uso1p, an essential tethering factor at the Golgi in yeast, is required for the assembly of the v-SNARE-t-SNARE complexes (63). In mammalian cells, defects in the function of the intra-Golgi-tethering complex, the conserved oligomeric Golgi (COG) complex, led to a significant decrease in Golgi SNARE mobility and in the steady-state level of intra-Golgi SNARE complexes, accompanied by an accumulation of uncomplexed syntaxin 5 (64). Furthermore, the trans-Golgi located mammalian tethering complex GARP interacted directly with SNAREs that participate in the endosome-to-trans-Golgi network (TGN) retrograde route. Further functional analyses placed the Golgi-associated retrograde protein (GARP) complex upstream of the SNAREs, regulating their localization and assembly into SNARE complexes (26). Finally, the HOPS complex, the tethering complex present at the yeast vacuole, proofreads SNARE domain and N-terminal domain structures of vacuolar SNAREs and regulates the fusion capacity of trans-SNARE complexes, only allowing full function for wild-type SNARE configurations (28).
How mutant Tip20-5p and Tip20-8p prevent assembly of the SNARE complexes remains unclear. At least Tip20-8p is more flexible than wild-type Tip20p, and this flexibility may render Tip20p more susceptible to degradation. Indeed, Tip20-8 levels are slightly reduced in yeast, and recombinant Tip20-8p has more degradation products than wild-type. More importantly, parts of the helical stalk region become more flexible, and these regions comprise the binding site of Sec20p. The binding to Sec20p seems to be of regulatory importance because Tip20-8p and Tip20-5p are still efficiently recruited to membranes, albeit the interaction with Sec20p is severely reduced. Although we find direct binding of Tip20p to Ufe1p in vitro, the interaction of Tip20p with Sec20p might be the more important one in vivo and could potentially be the driving force for SNARE complex assembly. A plausible scenario is that the arrival of a vesicle is signaled via the Tip20p-Sec20p interaction, leading to an efficient recruitment of cognate SNAREs. The role of other players, like Sec1/Munc18 proteins, known to have a role in orchestrating and stabilizing fusion events at the ER, needs to be further examined in this context.
Supplementary Material
Acknowledgments
We are grateful to M. Spiess, F. Hughson, H. D. Schmitt, C. Ungermann, D. Banfield, J. Gerst, R.-W. Peng, P. Cosson, and R. Schekman for reagents. We thank C. Ungermann for sharing results prior to publication. Members of the Spang laboratory are acknowledged for discussions and critical comments.
This work was supported by the Swiss National Science Foundation and the University of Basel.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
- ER
- endoplasmic reticulum
- v-SNARE
- vesicle SNARE
- t-SNARE
- SNARE on the target membrane
- r.m.s.d.
- root mean square deviation
- r.m.s.f.
- root mean square fluctuation
- Ni-NTA
- nickel-nitrilotriacetic acid
- COP
- coat protein complex
- 5-FOA
- 5′ fluoroorotic acid.
REFERENCES
- 1. Bonifacino J. S., Glick B. S. (2004) Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2. Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 3. Springer S., Spang A., Schekman R. (1999) Cell 97, 145–148 [DOI] [PubMed] [Google Scholar]
- 4. Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 5. Letourneur F., Gaynor E. C., Hennecke S., Démollière C., Duden R., Emr S. D., Riezman H., Cosson P. (1994) Cell 79, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 6. Waters M. G., Serafini T., Rothman J. E. (1991) Nature 349, 248–251 [DOI] [PubMed] [Google Scholar]
- 7. Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. (1991) Cell 67, 239–253 [DOI] [PubMed] [Google Scholar]
- 8. Yang J. S., Lee S. Y., Gao M., Bourgoin S., Randazzo P. A., Premont R. T., Hsu V. W. (2002) Journal of Cell Biology 159, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antonny B., Madden D., Hamamoto S., Orci L., Schekman R. (2001) Nat. Cell Biol. 3, 531–537 [DOI] [PubMed] [Google Scholar]
- 10. Spang A. (2009) Curr. Opin. Cell Biol. 21, 531–536 [DOI] [PubMed] [Google Scholar]
- 11. Tanigawa G., Orci L., Amherdt M., Ravazzola M., Helms J. B., Rothman J. E. (1993) J. Cell Biol. 123, 1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fasshauer D., Bruns D., Shen B., Jahn R., Brünger A. T. (1997) J. Biol. Chem. 272, 4582–4590 [DOI] [PubMed] [Google Scholar]
- 13. Fiebig K. M., Rice L. M., Pollock E., Brunger A. T. (1999) Nat. Struct. Biol. 6, 117–123 [DOI] [PubMed] [Google Scholar]
- 14. Poirier M. A., Xiao W., Macosko J. C., Chan C., Shin Y. K., Bennett M. K. (1998) Nat. Struct. Biol. 5, 765–769 [DOI] [PubMed] [Google Scholar]
- 15. Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 16. Borisovska M., Zhao Y., Tsytsyura Y., Glyvuk N., Takamori S., Matti U., Rettig J., Südhof T., Bruns D. (2005) EMBO J. 24, 2114–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Götte M., Gallwitz D. (1997) FEBS Lett. 411, 48–52 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y., Barlowe C. (2002) Mol. Biol. Cell 13, 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pelham H. R. (2001) Trends Cell Biol. 11, 99–101 [DOI] [PubMed] [Google Scholar]
- 20. Tsui M. M., Banfield D. K. (2000) J. Cell Sci. 113, 145–152 [DOI] [PubMed] [Google Scholar]
- 21. Fasshauer D., Antonin W., Margittai M., Pabst S., Jahn R. (1999) J. Biol. Chem. 274, 15440–15446 [DOI] [PubMed] [Google Scholar]
- 22. Cai H., Reinisch K., Ferro-Novick S. (2007) Dev. Cell 12, 671–682 [DOI] [PubMed] [Google Scholar]
- 23. Carr C. M., Rizo J. (2010) Curr. Opin. Cell Biol. 22, 488–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 25. Markgraf D. F., Peplowska K., Ungermann C. (2007) FEBS Lett. 581, 2125–2130 [DOI] [PubMed] [Google Scholar]
- 26. Pérez-Victoria F. J., Bonifacino J. S. (2009) Mol. Cell Biol. 29, 5251–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren Y., Yip C. K., Tripathi A., Huie D., Jeffrey P. D., Walz T., Hughson F. M. (2009) Cell 139, 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Starai V. J., Hickey C. M., Wickner W. (2008) Mol. Biol. Cell 19, 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ungermann C., Langosch D. (2005) J. Cell Sci. 118, 3819–3828 [DOI] [PubMed] [Google Scholar]
- 30. Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Söllner T. H., Lithgow T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9873–9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. (2003) EMBO J. 22, 3664–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewis M. J., Rayner J. C., Pelham H. R. (1997) EMBO J. 16, 3017–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spang A., Schekman R. (1998) J. Cell Biol. 143, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andag U., Neumann T., Schmitt H. D. (2001) J. Biol. Chem. 276, 39150–39160 [DOI] [PubMed] [Google Scholar]
- 35. Andag U., Schmitt H. D. (2003) J. Biol. Chem. 278, 51722–51734 [DOI] [PubMed] [Google Scholar]
- 36. Kraynack B. A., Chan A., Rosenthal E., Essid M., Umansky B., Waters M. G., Schmitt H. D. (2005) Mol. Biol. Cell 16, 3963–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reilly B. A., Kraynack B. A., VanRheenen S. M., Waters M. G. (2001) Mol. Biol. Cell 12, 3783–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tripathi A., Ren Y., Jeffrey P. D., Hughson F. M. (2009) Nat. Struct. Mol. Biol. 16, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zink S., Wenzel D., Wurm C. A., Schmitt H. D. (2009) Dev. Cell 17, 403–416 [DOI] [PubMed] [Google Scholar]
- 40. Kamena F., Spang A. (2004) Science 304, 286–289 [DOI] [PubMed] [Google Scholar]
- 41. Goldenberg O., Erez E., Nimrod G., Ben-Tal N. (2009) Nucleic Acids Res. 37, D323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pupko T., Bell R. E., Mayrose I., Glaser F., Ben-Tal N. (2002) Bioinformatics 18, S71–77 [DOI] [PubMed] [Google Scholar]
- 43. Fiser A., Sali A. (2003) Bioinformatics 19, 2500–2501 [DOI] [PubMed] [Google Scholar]
- 44. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 45. Canutescu A. A., Shelenkov A. A., Dunbrack R. L., Jr. (2003) Protein Sci. 12, 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 47. Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 48. Rexach M. F., Latterich M., Schekman R. W. (1994) J. Cell Biol. 126, 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 50. Hosobuchi M., Kreis T., Schekman R. (1992) Nature 360, 603–605 [DOI] [PubMed] [Google Scholar]
- 51. Cosson P., Schröder-Köhne S., Sweet D. S., Démollière C., Hennecke S., Frigerio G., Letourneur F. (1997) Eur J. Cell Biol. 73, 93–97 [PubMed] [Google Scholar]
- 52. Brzović P. S., Sawa Y., Hyde C. C., Miles E. W., Dunn M. F. (1992) J. Biol. Chem. 267, 13028–13038 [PubMed] [Google Scholar]
- 53. Galiano L., Ding F., Veloro A. M., Blackburn M. E., Simmerling C., Fanucci G. E. (2009) J. Am. Chem. Soc. 131, 430–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ingram V. M. (1956) Nature 178, 792–794 [DOI] [PubMed] [Google Scholar]
- 55. Sweet D. J., Pelham H. R. (1993) EMBO J. 12, 2831–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wendler F., Tooze S. (2001) Traffic 2, 606–611 [DOI] [PubMed] [Google Scholar]
- 57. Yang B., Gonzalez L., Jr., Prekeris R., Steegmaier M., Advani R. J., Scheller R. H. (1999) J. Biol. Chem. 274, 5649–5653 [DOI] [PubMed] [Google Scholar]
- 58. Fischer von Mollard G., Stevens T. H. (1999) Mol. Biol. Cell 10, 1719–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cao X., Barlowe C. (2000) J. Cell Biol. 149, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parlati F., McNew J. A., Fukuda R., Miller R., Söllner T. H., Rothman J. E. (2000) Nature 407, 194–198 [DOI] [PubMed] [Google Scholar]
- 61. Meiringer C., Rethmeier R., Auffarth K., Wilson J., Perz A., Barlowe C., Schmitt H. D., Ungermann C. (2011) J. Biol. Chem. 286, 25039–25046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmitt H. D. (2010) Trends Cell Biol. 20, 257–268 [DOI] [PubMed] [Google Scholar]
- 63. Sapperstein S. K., Lupashin V. V., Schmitt H. D., Waters M. G. (1996) J. Cell Biol. 132, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shestakova A., Suvorova E., Pavliv O., Khaidakova G., Lupashin V. (2007) J. Cell Biol. 179, 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.