Abstract
Retrograde vesicular transport from the Golgi to the ER requires the Dsl1 tethering complex, which consists of the three subunits Dsl1, Dsl3, and Tip20. It forms a stable complex with the SNAREs Ufe1, Use1, and Sec20 to mediate fusion of COPI vesicles with the endoplasmic reticulum. Here, we analyze molecular interactions between five SNAREs of the ER (Ufe1, Use1, Sec20, Sec22, and Ykt6) and the Dsl1 complex in vitro and in vivo. Of the two R-SNAREs, Sec22 is preferred over Ykt6 in the Dsl-SNARE complex. The NSF homolog Sec18 can displace Ykt6 but not Sec22, suggesting a regulatory function for Ykt6. In addition, our data also reveal that subunits of the Dsl1 complex (Dsl1, Dsl3, and Tip20), as well as the SNAREs Ufe1 and Sec20, are ER-resident proteins that do not seem to move into COPII vesicles. Our data support a model, in which a tethering complex is stabilized at the organelle membrane by binding to SNAREs, recognizes the incoming vesicle via its coat and then promotes its SNARE-mediated fusion.
Keywords: Endoplasmic Reticulum (ER), Endosomes, Fusion Protein, Membrane Fusion, Reconstitution Of Membrane Transporters, Dsl1 Complex, SNARE, Ykt6, Retrograde Transport, Tethering
Introduction
Vesicles transport biosynthetic cargo and lipids between different compartments of the endomembrane system. Formation of the transport vesicles requires adaptors, coat proteins, and regulatory GTPases of the Arf1/Sar1 family. The initial contact between a vesicle and its target membrane requires Rab GTPases and tethers, which are in most cases multisubunit complexes. Rab GTPases, which cycle between an inactive GDP- and active GTP-bound state, and tethers coordinate the assembly of SNARE proteins on vesicle and target membrane into a four-helix bundle, which ultimately drives bilayer fusion (1).
The Dsl1 tethering complex functions in fusion of Golgi-derived vesicles at the ER4 membrane and consists of the three subunits Dsl1, Dsl3/Sec39, and Tip20. It forms a stable complex with the ER SNAREs Sec20, Ufe1, and Use1 (2, 3). In addition, Dsl1 and Tip20 are linked to the coatomer, which implies a role in the recognition and/or uncoating of the COPI vesicle (4–7). In agreement with this, Dsl1 depletion leads to a massive accumulation of COPI-coated vesicles (8).
The Dsl1 complex is closely linked to the SNAREs Ufe1, Use1/Slt1, Sec20, and Sec22, which are required for fusion at the ER membrane (9–12). The R-SNARE Sec22 is generally accepted as the v-SNARE on COPI vesicles. However, Sec22 has not been previously identified as part of the Dsl1 complex and is dispensable for yeast survival. In addition, it can be functionally replaced by the R-SNARE Ykt6 in anterograde (13) and potentially also in retrograde transport. Ykt6, which lacks a transmembrane domain and thus is unlikely to function as the sole v-SNARE, is found in multiple SNARE complexes at the Golgi, endosomes, and the vacuole (10, 14).
Here, we present additional insights into the interactions and functions of the Dsl1 complex. We show that the two R-SNAREs Sec22 and Ykt6 are associated with the Dsl1 complex, with Sec22 being the preferred subunit. Only Ykt6 is sensitive to Sec18/NSF, whereas the remaining interaction between SNAREs and the Dsl1 complex is unaffected. Reconstitution approaches reveal that the Dsl1 complex contains several interfaces for SNAREs, and in vivo studies suggest that subunits of the Dsl1 complex and the Q-SNAREs are ER-resident proteins. Our data support a model of tethering via coat recognition, followed by SNARE assembly and fusion.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmid Construction
Yeast strains used in this study are listed in supplemental Table S1. These were either generated by homologous recombination of PCR-amplified fragments or by transformation with plasmids (see below). For yeast two-hybrid analysis, full-length or truncated ORFs were cloned into both pACT2 (Clontech) and pFBT9 (15) plasmids and co-transformed into yeast strain PJ69–4A (as shown in supplemental Table S2). Transformants were selected on synthetic dextrose complete-Leu-Trp plates, and four clones of each tested interaction were restreaked to quadruple drop-out plates (synthetic dextrose complete-Leu-Trp-His-Ade). Growth was assayed after 4–7 days. Plasmids for purification of recombinant proteins were cloned either into pGEX-2TK (GE Healthcare), pETHIS (14), or pET32c(-Trx) (modified from pET32c(+), Novagen, by removal of the thioredoxin tag) and are listed in supplemental Table S3. The SNAREs were cloned without their transmembrane domain. PCR amplification was performed using Pfu polymerase (Fermentas GmbH), and all Y2H plasmids were sequenced (GATC Biotech AG). Restriction digest and cloning were performed according to the manufacturer's instructions (Fermentas GmbH).
Microscopy
Yeast cells expressing either Tip20-monomeric GFP, Dsl1-GFP, or Dsl3-monomeric GFP were grown to early log-phase, harvested, washed once with PBS, and mounted on object slides. Visualization was performed on a fluorescence microscope (Leica DM5500 B; Leica Microsystems GmbH) equipped with a GFP filter (excitation, D480/30; emission, D535/40, Beamsplitter 505dclp; Chroma Technology), captured with a digital camera (Spot Pursuit-4MP; Diagnostic Instruments, Inc.) and processed using Metamorph (Molecular Devices) and Autoquant X (Media Cybernetics). Strains bearing a temperature-sensitive allele were analyzed both after growth at 23 °C and after a temperature shift to 37 °C for 30 min. For Dsl1 and Ypt1 depletion experiments, strains were grown in YPG, washed once in PBS, and then grown in YPD for 9 h.
Tandem Affinity Purification
Tandem affinity purification (TAP) tag protein tandem affinity purification was performed as described in Refs. 16, 17 using the following buffer: 50 mm HEPES/KOH, pH 7.4, 150 mm NaCl, 0.15% Nonidet P-40 (Igepal CA-630; Sigma-Aldrich), and 1.5 mm MgCl2. The buffer was supplemented with 1 mm PMSF, 1 mm DTT, and 1× protease inhibitor mix FY (Serva). For washing of IgG-Sepharose (GE Healthcare), the buffer was supplemented with 0.5 mm DTT. Additional methods are provided in the supplemental data.
RESULTS
Isolation of Dsl1 Complex Identifies R-SNARE Ykt6
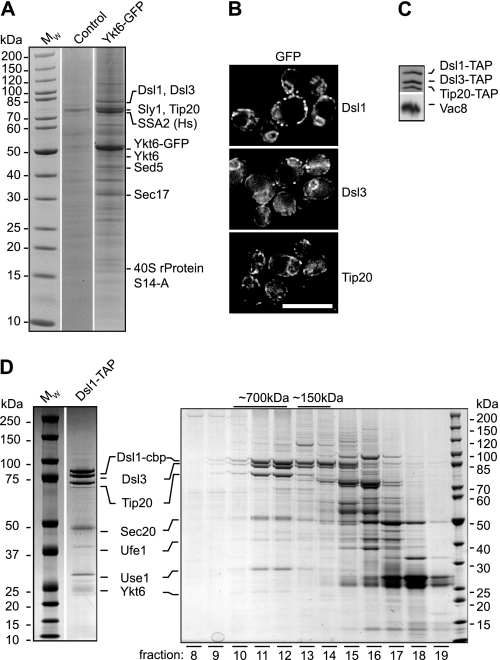
The SNARE Ykt6 is distributed between cytosol and membranes. To identify a potential receptor of Ykt6, we tagged Ykt6 with GFP in a loop contained within its N-terminal longin domain and isolated the protein using antibodies against GFP coupled to protein A-Sepharose. Proteins that co-eluted with Ykt6-GFP were identified by mass spectrometry (Fig. 1A). Besides known interaction partners (Sed5, Sec17, and Sly1), we were able to identify Dsl1, Dsl3, and Tip20 in the eluate. All three proteins belong to the Dsl1 complex at the ER, localize similarly to punctate structures at the cortical and perinuclear ER (Fig. 1B), and are equally abundant (Fig. 1C), as reported (2).
FIGURE 1.
Ykt6 interacts with the Dsl1 complex. A, components of the Dsl1 complex are co-purified with Ykt6-GFP. Ten thousand A600 units of the yeast strain overexpressing Ykt6-GFP (GAL1-YKT6-GFP) were lysed, processed as described under “Experimental Procedures,” and subjected to immunoprecipitation using affinity-purified GFP antibodies coupled to protein A-Sepharose. Eluates were applied to 4–12% SDS gels and prominent bands analyzed by mass spectrometry after colloidal Coomassie staining. B and C, in vivo localization of Dsl1 complex subunits. C-terminally tagged Dsl1 complex subunits were analyzed by fluorescence microscopy (B; scale bar indicates 10 μm). Subcellular fractionation of yeast cell lysates was done with TAP-tagged Dsl1, Dsl3, or Tip20. Proteins from membrane fraction were analyzed by SDS-PAGE and Western blotting using antibodies against the calmodulin binding peptide of the TAP tag or Vac8 (C). D, Ykt6 is found in association with the Dsl1 complex. Dsl1-TAP was purified either by tandem affinity purification or on IgG-Sepharose alone and subjected to gel filtration on a Superose 6 column as described (36). Aliquots of the resulting fractions were analyzed on SDS-PAGE (Coomassie-stained). Fraction numbers are indicated below the gel. Prominent bands of tandem affinity purification were cut for identification in mass spectrometry. cbp, calmodulin binding peptide.
To confirm the interaction between the Dsl1 complex and Ykt6, we tagged Dsl1 and Dsl3 with a C-terminal TAP tag and performed a tandem affinity purification via IgG Sepharose and CaM beads (Fig. 1D and supplemental Fig. S1A). Using mass spectrometry, we identified not only the three Dsl1 complex subunits, but also all of the components of the ER SNARE complex Ufe1, Use1, and Sec20. This stable complex of tethers and SNAREs was reported previously but lacked the appropriate R-SNARE (2). When we subjected the eluate of the IgG beads to gel filtration, we recovered the Dsl1 complex together with the previously identified Q-SNAREs and Ykt6 in a high molecular mass complex of ∼700 kDa in fractions 11, 12 (Fig. 1D and supplemental Fig. S1A), and a subcomplex of Dsl1 and Dsl3 in fractions 13 and 14 (Fig. 1D), in agreement with recent structural work (3).
Sec22 and Ykt6 Interaction with Dsl1 Complex Differs
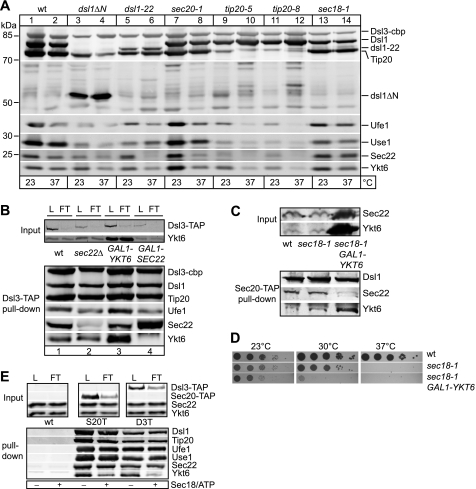
Sec22 was previously described to be the R-/v-SNARE in transport between the Golgi and ER (11, 18, 19), though we found Ykt6 (Fig. 1). However, we detected both Sec22 and Ykt6 in association with Dsl3-TAP and the other subunits of the Dsl1 complex on Western blots (supplemental Fig. S1B), whereas control beads did not precipitate any SNARE (Fig. 2E). We therefore wondered whether we could detect differences in the composition of the Dsl1 tether complex with SNAREs and thus determine the precise interactions. We tested different temperature-sensitive mutants of the Dsl proteins, Dsl1 and Tip20, the SNARE Sec20 and Sec18, the ATPase required for SNARE complex disassembly. The composition of the Dsl1 complex and its interaction with SNAREs were unchanged in the sec18-1 mutant, which is blocked in SNARE disassembly at the restrictive temperature (Fig. 2A, lane 14, supplemental Fig. S1B). In contrast, mutants in dsl1 (lanes 2–6), or tip20 (lanes 9–12) strongly affected the stability of the Dsl1 complex (Fig. 2A). This indicates that the interaction of the Dsl1 complex with all five SNAREs is affected by Dsl1 complex mutations, but not by alterations in the SNARE chaperone Sec18.
FIGURE 2.
Differential binding of R-SNAREs to the Dsl1 complex. A, analysis of temperature-sensitive strains for the interaction between SNAREs and the Dsl1 complex. Cells were grown overnight at 23 °C and heat-shocked at 37 °C for 2 h. Lysates of 10,000 A600 units of cells were prepared as described under “Experimental Procedures” for TAP. After purification and tobacco etch virus elution, 10% of the tobacco etch virus eluate was loaded on a 4–12% SDS-PAGE gel, and proteins were analyzed by Western blotting using the indicated antibodies. B, overexpression of Sec22 displaces Ykt6 from the Dsl1 complex. Control cells (WT), sec22Δ cells, or cells overexpressing Ykt6 (GAL1-YKT6) or Sec22 (GAL1-SEC22) were grown overnight in YPG. Dsl3-TAP was purified from 1500 A600 units via IgG-Sepharose and tobacco etch virus cleavage. Eluates were analyzed as in described in A. L, load; FT, flowthrough; cbp, calmodulin binding peptide. C, in sec18-1 background, overexpression of Ykt6 displaces Sec22 from the Dsl1 complex. Cells were grown in YPG at 23 °C and shifted to 37 °C for 3 h. Sec20-TAP pull down was made from indicated strains as in B. Note that Sec22 is not visible in the Ykt6 overexpression lane to the Ykt6 signal in the Western blot. D, Ykt6 overexpression in the sec18-1 strain enhances the temperature-sensitive phenotype. Dilution series from 0.25 to 0.000025 A600 of indicated strains were grown at 23, 30, and 37 °C and photographed. E, Sec18 displaces Ykt6 from the Dsl1 complex. A membrane fraction (P10) was prepared from 3000 A600 units of the indicated cells as described under “Experimental Procedures” and was incubated in reaction buffer with or without Sec18 and ATP. Membranes were reisolated for 10 min at 20,000 × g at 4 °C and resuspended in lysis buffer, and the TAP-tagged proteins were isolated as before. Tobacco etch virus-eluted proteins analyzed as in A. S20T, Sec20-TAP; D3T, Dsl3-TAP.
To unravel whether the two R-SNAREs compete for the same binding site on the complex, we overexpressed either Sec22 or Ykt6 and purified the Dsl1 complex via Dsl3. Overexpression of Ykt6 did not have any significant effect on Sec22 binding to the Dsl1 complex, whereas Sec22 overexpression completely abolishes binding of Ykt6 (Fig. 2B, lane 3 versus 4). We then asked whether the competition of Ykt6 and Sec22 may be enhanced if the turnover of the SNARE complex was impaired. Indeed, overexpression of Ykt6 in the sec18-1 mutant was sufficient to displace Sec22 from the Dsl1 complex (Fig. 2C), indicating that Ykt6 can compete with Sec22 under these conditions. Moreover, these cells grew more slowly at higher temperatures than cells with the sec18-1 allele alone (Fig. 2D). It was demonstrated previously that Ykt6 functionally replaces Sec22 in transport between the Golgi and the ER (13). We therefore reasoned that Ykt6 might be enriched in the Dsl1 complex if Sec22 is lacking. Contrary to our assumption, the pulldown of Dsl3-TAP in a sec22Δ strain did not show a significant accumulation of Ykt6 in the Dsl1 complex (Fig. 2B, lane 2). We conclude that association of Ykt6 with the Dsl1 complex occurs upstream of Sec22 binding and the latter prevents Ykt6 from re-entering the complex.
We then asked whether the interaction of Sec22 and Ykt6 with the Dsl1 complex is altered by the addition of purified Sec18, which can disassemble SNARE complexes (20). When ER-enriched membranes were incubated with Sec18 and ATP, only Ykt6 was selectively displaced from Sec20 or Dsl3, whereas the remaining complex stayed intact (Fig. 2E and supplemental Fig. S1C, lane 4). Our data suggest that Ykt6 occupies a Sec18-sensitive binding site on the assembled SNARE-Dsl1 complex.
Dsl1 Complex Binds Q-SNAREs Sec20 and Use1 via Their N-terminal Domains
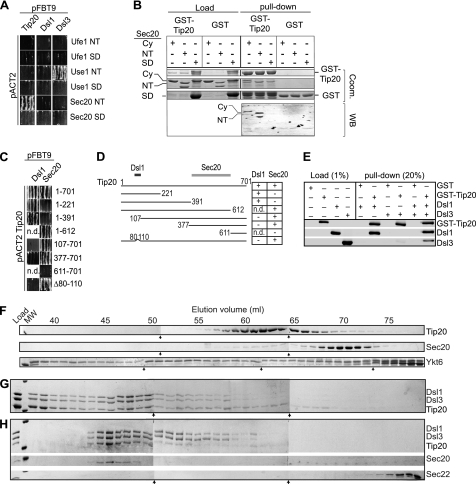
To map the interactions between components of the Dsl1 complex and SNAREs, we employed yeast two-hybrid analysis. Interestingly, several subunits like Use1 and Dsl1 showed multiple interactions, suggesting that they occur in the context of the partially assembled Q-SNARE-Dsl1 complex (supplemental Fig. S1D). We therefore focused on the N-terminal and the SNARE domains of each Q-SNARE (Fig. 3A). Our data indicate that the Dsl1 complex recognizes the N-terminal domains of the SNARE Use1 (residues 1–141) and Sec20 (residues 1–196) via its subunits Dsl3 and Tip20, in agreement with Hughson and colleagues (Fig. 3, A and B) (21). For Tip20, we mapped the binding site for Sec20 to residues 377–611 (Fig. 3, C and D), C-terminal of the proposed binding site by Ren et al. (21). Whereas Tripathi et al. (3) mapped the interaction site of Tip20 to Dsl1 to the first 41 residues, an internal deletion of residues 80–110 also abolished the binding to Dsl1, which may be due to changes in secondary structure. Alternatively, the binding site between Tip20 and Dsl1 includes additional Tip20 segments.
FIGURE 3.
Subunit interactions between the Dsl1 complex and SNARE proteins. A and B, Q-SNARE domain interaction with Dsl1, Dsl3, and Tip20. A, interactions between fragments of the N-terminal regulatory domain (NT) and SNARE domain (SD) of Ufe1, Use1, and Sec20 and full-length Dsl1, Dsl3, and Tip20 were analyzed by yeast two hybrid. B, pulldown with GST-Tip20 against cytosolic domains of Sec20. Full-length Tip20 bound to GSH beads was incubated with the cytosolic part of Sec20 (Cy, without Trx-tag), only N terminus or SNARE domain (both with Trx tag). GSH beads were eluted by boiling in Laemmli buffer, and the samples were analyzed by SDS-PAGE followed by Coomassie staining (Coom., upper panel). In addition, a Western blot (WB) was decorated with His-tag antibody (lower panel). C and D, mapping of Tip20 interactions. C, interactions between truncations of Tip20 and full-length Dsl1 and Sec20 were analyzed by yeast two-hybrid. Residues are listed as numbers. D, graphic depiction of C. E, reconstitution of the Dsl1 complex. Purified Dsl1 and Dsl3 were added to GSH beads carrying GST or GST-Tip20, and the pulldown assay was performed as described under “Experimental Procedures” and analyzed by SDS-PAGE and Western blotting. F, migration of proteins of the Dsl1 complex and indicated SNAREs on gel filtration. The indicated purified proteins were analyzed by gel filtration on a Sephacryl S-300 HR column (GE Healthcare). One-ml fractions were collected, TCA-precipitated, and analyzed by SDS-PAGE and Coomassie staining. Arrows indicate the single gels. G, reconstitution of the Dsl1 complex. Equimolar amounts of the Dsl3, Dsl1, and Tip20 lacking the GST were incubated at 4 °C and analyzed by gel filtration as in F. H, reconstitution of the Dsl1 complex interaction with Sec22 and Sec20. Fractions between 45 and 50 ml of reconstituted Dsl1 complex as in G were concentrated, incubated with 2-fold molar excess of Sec20 and Sec22, and analyzed by gel filtration. One-ml fractions were collected and analyzed as in F.
To test the interactions directly, we purified the components of the Dsl1 complex and reconstituted the complex assembly. We detected direct binding of Dsl1 to Dsl3 and Tip20 but not for Tip20 to Dsl3 (Fig. 3E). This is in agreement with previously published structural findings (3, 21). By gel filtration, we showed that monomeric Tip20 (Fig. 3F) assembled into the large Dsl1 complex (fractions 45–50), if incubated with Dsl1 and Dsl3 (Fig. 3G) and some large complex in the void volume. When the concentrated Dsl1 complex was incubated with an excess of either Sec20 or Sec22, only Sec20 bound to the Dsl1 complex, presumably via Tip20 (Fig. 3H). The reconstitution of the Dsl1 complex with all SNAREs, including the R-SNARE, was very inefficient (data not shown), and the interaction may occur only transiently in vivo. Our data reveal that the Dsl1 complex has binding sites for the N-terminal domains of Sec20 and Use1 and plays a role in the assembly of the SNARE complex, consistent with our initial observation (Fig. 1).
Evidence for ER-resident Dsl1 Complex
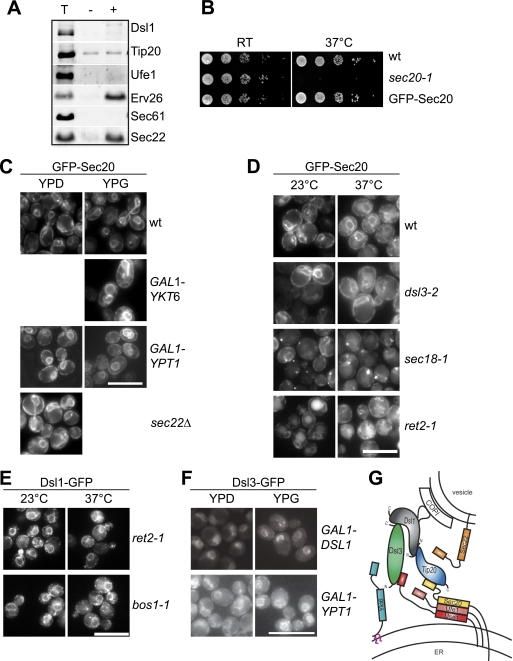
If Sec22 were the retrograde v-SNARE, it appears unlikely that it is replaced by acylated Ykt6, which lacks the transmembrane domain that is required to drive fusion of the lipid bilayers. Moreover, the interaction of Dsl1 and COPI suggested that Dsl1 could be recruited to retrograde vesicles. We therefore took advantage of a COPII budding assay (22) to monitor the incorporation of selected subunits of the Dsl1 complex and SNAREs (Fig. 4A). Vesicles generated in the presence of COPII components contained Sec22 and the cargo receptor Erv26, but lacked Dsl1, Tip20, and Ufe1, consistent with the predominant ER-localization of the Dsl1 complex in vivo (Fig. 1B). As we lacked an antibody to Sec20, we turned to an in vivo assay, using functional GFP-tagged Sec20 (Fig. 4B). Sec20 has been previously reported to contain a luminal HDEL motif, which might be responsible of its ER localization (23). However, neither sec22 deletion, Ykt6 overexpression, Ypt1 depletion, mutations in the δ-COP (ret2-1), nor alterations in Dsl3 functionality affected the steady-state localization of Sec20 to the ER (Fig. 4, C and D). In the sec18-1 mutant, we observed accumulations of GFP-Sec20, which increased in size at the restrictive temperature (Fig. 4D). These dots colocalized with the ER marker Sec63 (supplemental Fig. S2D). They might be the result of additional Ykt6 associated with the (Dsl1-)SNARE complex, which may impair transport between ER and Golgi, and subsequently growth (Fig. 2D). Our data therefore suggest that it is unlikely that Sec20 functions as a v-SNARE on retrograde vesicles.
FIGURE 4.
Analysis of Q-SNARE and Dsl1 complex cycling between ER and Golgi. A, Dsl1, Tip20, and Ufe1 are not incorporated in COPII vesicles. Purified ER membranes were incubated with COPII subunits. Vesicles were then isolated, and proteins analyzed by SDS-PAGE and Western blotting using antibodies against the indicated proteins. 10% of a total reaction (T) was compared with budded vesicles produced in the absence (−) or presence (+) of COPII proteins. B, GFP-Sec20 is functional. Cells carrying a sec20-1 temperature-sensitive mutant or GFP-Sec20 were serially diluted, spotted on YPD plates, and incubated at the indicated temperatures. C, neither Ykt6 overexpression, SEC22 deletion, nor Ypt1 depletion affect Sec20 localization. D, localization of GFP-Sec20 in selected mutants. E, localization of Dsl3-GFP at the ER is not disturbed by depletion of Dsl1 or Ypt1. Western blots showing Ypt1 and Dsl1 depletion are in supplemental Figs. S1E and S2C. F, Dsl1-GFP localizes at the ER in temperature-sensitive mutants defective in ER to Golgi (bos1-1) or Golgi to ER (ret2-1) transport. For C–F, microscopy of GFP constructs was done as described under “Experimental Procedures.” Scale bar, 10 μm. G, model of the Dsl1 complex assembled with the three Q-SNAREs Sec20, Ufe1, and Use1 and docked COPI vesicle. Whereas Sec20 and Use1 bind directly to Tip20 and Dsl3 via their N-terminal domains, Ufe1 does not interact with the Dsl1 complex as well as the R-SNAREs Sec22 and Ykt6. Ykt6 is lacking a transmembrane domain and associates to membranes by a farnesyl and a palmitoyl anchors.
Furthermore, depletion of Ypt1 or Dsl1, the latter one causes a massive accumulation of COPI vesicles (8), did not affect Dsl3 localization to the ER (Fig. 4E). Upon depletion of Dsl1, Sec20 and Dsl3 showed colocalization with the ER marker Sec63 but not with the Golgi marker Mnn9 (supplemental Fig. S2, A and B). Also Dsl1 itself, which does not bind directly to a SNARE or any other ER transmembrane protein, seem to be stably localized at the ER. In ret2-1 cells or a temperature-sensitive mutant of the ER to Golgi v-SNARE bos1 (bos1-1), Dsl1 is found at the ER (Fig. 4F), similar to our observations upon Ypt1 depletion (data not shown). Our data are therefore consistent with a stable ER-resident Dsl1 complex, which is kept in place by binding ER-resident Q-SNAREs.
DISCUSSION
Fusion of Golgi-derived vesicles with the ER requires a close cooperation of the Dsl1 complex (2) with the SNAREs Ufe1, Use1, Sec20, and Sec22 (12, 19). Previous purification of the Dsl1 complex did not yield any R-SNARE but only the Q-SNAREs Use1, Ufe1, and Sec20 (2). However, Use1 interacts with Sec20, Ufe1, and the R-SNAREs Sec22 and Ykt6 (19). We now demonstrate that Sec22 and Ykt6 are found in substoichiometric amounts in association with the isolated Dsl1 complex, suggesting that Ykt6 may regulate vesicle fusion by binding not only the Q-SNAREs, but also the SNARE-Dsl1 complex. We could not identify a binding site between any Dsl1 complex subunit and Ufe1 or Sec22, indicating that both bind to the Dsl1 complex via the SNAREs Sec20 and Use1 (11), which bind Tip20 and Dsl3 via their N-terminal domains (Fig. 3, A and B) (21). Our data suggest that Sec22 is indeed the missing R-SNARE, as expected from previous studies (12, 19). Overexpression of Sec22 completely removes Ykt6 from its association with the Dsl1 complex, whereas Ykt6 could not do so in reverse. A similar association of Ykt6 with the Dsl1-SNARE complex was recently observed by Spang and colleagues (37). In addition, Sec18 addition displaced Ykt6, but not Sec22 or the Q-SNAREs, from the Dsl1 complex. It is therefore possible that Ykt6 acts as an acceptor for the preferred SNARE Sec22 at the ER. Our data are consistent with the view that Sec22 is the v-SNARE in vivo, whereas the Q-SNAREs seem to be resident proteins of the ER.
While our manuscript was in preparation, two studies provided insight into the structure of the Dsl1 complex (3, 21). Dsl1 and Tip20 resemble known structures of the exocyst complex (3). In agreement with our results, Hughson and co-workers (21) demonstrate that Use1 and Sec20 bind via their N termini to Dsl3 and Tip20, respectively. Our data indicate that Ufe1 requires Sec20 for binding, whereas Sec22 only binds if all other SNAREs are present. This observation is consistent with our initial isolation of Sec22 and Ykt6 with the Dsl1 complex from yeast (Figs. 1 and 2). The purified Dsl1 complex is, however, very inefficient in promoting SNARE assembly (3), and it has not been tested whether the slight increase in complex formation correlates with increased fusion.
The Dsl1 complex is a critical factor involved in COPI vesicle recognition at the ER (8). Its subunit Dsl1 binds directly to the heterodimeric complex of ϵ-COP and the C terminus of α-COP (24), presumably to tether the COPI vesicles to the ER and promote uncoating. Our data agree with such a combined function and suggest that the ER resident Dsl1 complex then promotes assembly of Sec22 with the Dsl1 complex-bound Q-SNAREs.
At the Dsl1 complex, Ykt6 may occupy a binding site to facilitate the association of Sec22 with the Q-SNAREs. Indeed, Sec22 can displace Ykt6, and the reverse is also possible if Sec18 is impaired. Potentially, Ykt6 orients the Dsl1-Q-SNARE complex in such a way that is predominantly competent to bind Sec22 in trans. If this is the case, then Ykt6 would have a more active role in ensuring proper assembly. We consider it unlikely that Ykt6 functions as a v-SNARE on Golgi-derived vesicles because its lipid anchor does not support membrane fusion (25). However, we cannot exclude a function of Ykt6 on COPI vesicles if Sec22 is lacking. It is also possible that in sec22Δ cells, a small portion of Sec20, Ufe1, or Use1 are recycled via the Golgi back to the ER and thus act as a v-SNARE, though we did not observe incorporation of Ufe1 into COPII-coated vesicles or relocalization of Sec20 into Golgi-like structures, except some dot-like structures in sec18-1 mutants (Fig. 4, C and D). It should be noted that more Ykt6 enters COPII-coated vesicles in sec22Δ cells (13), suggesting that Ykt6 may compensate for some of the functions of Sec22. Alternatively, the Golgi SNARE Bet1, which interacts with Ufe1 and Use1 in vitro (37), may function in this process.
For the mammalian homolog of Dsl1, ZW10, it has been proposed that the protein is cycling between the ER and the Golgi (26). This model is based on the observations that in primate cells ZW10 localizes at the ER, whereas in rodent cells, it is found at the Golgi (27–29). Our data on the yeast Dsl1 complex support a model, in which the entire tethering complex resides on one organelle and recruits vesicles via their coat (30). Transport vesicles should then maintain their coat to ensure the recognition at the target organelle. Our interaction studies confirm that the Dsl1 complex binds the N-terminal domains of Use1 and Sec20, though it is possible that the complex has additional membrane binding sites. This would position the complex such that it directly couples tethering of COPI vesicles and SNARE-mediated fusion, by bringing together the SNARE on the vesicle with the assembled t-SNARE complex on the ER membrane (Fig. 4G). Interestingly, such a scenario is reminiscent of the proposed interaction of the HOPS complex with the AP-3 coat. Here, the AP-3 subunit Apl5 binds the HOPS subunit Vps41 (31–33), which is regulated by phosphorylation (34). HOPS might get stabilized similarly by SNAREs on the vacuole (35) and could then tether AP-3 vesicles by binding the coat. We therefore postulate that the tether-coat interaction might be of general importance to drive fusion reactions.
Supplementary Material
Acknowledgments
We thank Fred Hughson for the Dsl3 expression plasmid and all members of the Ungermann laboratory for fruitful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 431, 944, and UN111/4-2) and the Hans-Mühlenhoff Foundation (to C. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Tables S1–S3, Figs. S1 and S2, and additional references.
- ER
- endoplasmic reticulum
- TAP
- tandem affinity purification
- v-SNARE
- vesicle membrane associated SNARE
- Q-SNARE
- SNARE with glutamine in SNARE domain
- R-SNARE
- SNARE with arginine in SNARE domain
- HOPS
- homotypic vacuole fusion and protein sorting
- YPD
- yeast peptone dextrose
- YPG
- yeast peptone galactose.
REFERENCES
- 1. Cai H., Reinisch K., Ferro-Novick S. (2007) Dev. Cell 12, 671–682 [DOI] [PubMed] [Google Scholar]
- 2. Kraynack B. A., Chan A., Rosenthal E., Essid M., Umansky B., Waters M. G., Schmitt H. D. (2005) Mol. Biol. Cell 16, 3963–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tripathi A., Ren Y., Jeffrey P. D., Hughson F. M. (2009) Nat. Struct. Mol. Biol. 16, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frigerio G. (1998) Yeast 14, 633–646 [DOI] [PubMed] [Google Scholar]
- 5. Andag U., Neumann T., Schmitt H. D. (2001) J. Biol. Chem. 276, 39150–39160 [DOI] [PubMed] [Google Scholar]
- 6. Andag U., Schmitt H. D. (2003) J. Biol. Chem. 278, 51722–51734 [DOI] [PubMed] [Google Scholar]
- 7. Vanrheenen S. M., Reilly B. A., Chamberlain S. J., Waters M. G. (2001) Traffic 2, 212–231 [DOI] [PubMed] [Google Scholar]
- 8. Zink S., Wenzel D., Wurm C. A., Schmitt H. D. (2009) Dev. Cell 17, 403–416 [DOI] [PubMed] [Google Scholar]
- 9. Lewis M. J., Pelham H. R. (1996) Cell 85, 205–215 [DOI] [PubMed] [Google Scholar]
- 10. Burri L., Lithgow T. (2004) Traffic 5, 45–52 [DOI] [PubMed] [Google Scholar]
- 11. Lewis M. J., Rayner J. C., Pelham H. R. (1997) EMBO J. 16, 3017–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dilcher M., Veith B., Chidambaram S., Hartmann E., Schmitt H. D., Fischer von Mollard G. (2003) EMBO J. 22, 3664–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y., Barlowe C. (2002) Mol. Biol. Cell 13, 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meiringer C. T., Auffarth K., Hou H., Ungermann C. (2008) Traffic 9, 1510–1521 [DOI] [PubMed] [Google Scholar]
- 15. Haas A. K., Fuchs E., Kopajtich R., Barr F. A. (2005) Nat. Cell Biol. 7, 887–893 [DOI] [PubMed] [Google Scholar]
- 16. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 17. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 18. Ballensiefen W., Ossipov D., Schmitt H. D. (1998) J. Cell Sci. 111, 1507–1520 [DOI] [PubMed] [Google Scholar]
- 19. Burri L., Varlamov O., Doege C. A., Hofmann K., Beilharz T., Rothman J. E., Söllner T. H., Lithgow T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9873–9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ungermann C., Nichols B. J., Pelham H. R., Wickner W. (1998) J. Cell Biol. 140, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ren Y., Yip C. K., Tripathi A., Huie D., Jeffrey P. D., Walz T., Hughson F. M. (2009) Cell 139, 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otte S., Belden W. J., Heidtman M., Liu J., Jensen O. N., Barlowe C. (2001) J. Cell Biol. 152, 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sweet D. J., Pelham H. R. (1992) EMBO J. 11, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsia K. C., Hoelz A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11271–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McNew J. A., Weber T., Parlati F., Johnston R. J., Melia T. J., Söllner T. H., Rothman J. E. (2000) J. Cell Biol. 150, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmitt H. D. (2010) Trends Cell Biol. 20, 257–268 [DOI] [PubMed] [Google Scholar]
- 27. Arasaki K., Uemura T., Tani K., Tagaya M. (2007) Biochem. Biophys. Res. Commun. 359, 811–816 [DOI] [PubMed] [Google Scholar]
- 28. Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., Tagaya M. (2004) EMBO J. 23, 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varma D., Dujardin D. L., Stehman S. A., Vallee R. B. (2006) J. Cell Biol. 172, 655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Angers C. G., Merz A. J. (2011) Semin. Cell Dev. Biol. 22, 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rehling P., Darsow T., Katzmann D. J., Emr S. D. (1999) Nat. Cell Biol. 1, 346–353 [DOI] [PubMed] [Google Scholar]
- 32. Darsow T., Katzmann D. J., Cowles C. R., Emr S. D. (2001) Mol. Biol. Cell 12, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angers C. G., Merz A. J. (2009) Mol. Biol. Cell 20, 4563–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H. J., Reggiori F., Engelbrecht-Vandré S., Ungermann C. (2010) J. Cell Biol. 191, 845–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stroupe C., Hickey C. M., Mima J., Burfeind A. S., Wickner W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17626–17633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peplowska K., Markgraf D. F., Ostrowicz C. W., Bange G., Ungermann C. (2007) Dev. Cell 12, 739–750 [DOI] [PubMed] [Google Scholar]
- 37. Diefenbacher M., Thorstinsdottir H., Spang A. (2011) J. Biol. Chem. 286, 25027–25038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.