Abstract
Terminally differentiated/non-dividing macrophages contain extremely low cellular dNTP concentrations (20–40 nm), compared with activated CD4+ T cells (2–5 μm). However, our LC-MS/MS study revealed that the non-canonical dUTP concentration (2.9 μm) is ∼60 times higher than TTP in macrophages, whereas the concentrations of dUTP and TTP in dividing human primary lymphocytes are very similar. Specifically, we evaluated the contribution of HIV-1 reverse transcriptase to proviral DNA uracilation under the physiological conditions found in HIV-1 target cells. Indeed, biochemical simulation of HIV-1 reverse transcription demonstrates that HIV-1 RT efficiently incorporates dUTP in the macrophage nucleotide pools but not in the T cell nucleotide pools. Measurement of both pre-steady state and steady state kinetic parameters of dUTP incorporation reveals minimal selectivity of HIV-1 RT for TTP over dUTP, implying that the cellular dUTP/TTP ratio determines the frequency of HIV-1 RT-mediated dUTP incorporation. The RT of another lentivirus, simian immunodeficiency virus, also displays efficient dUTP incorporation in the dNTP/dUTP pools found in macrophages but not in T cells. Finally, 2′,3′-dideoxyuridine was inhibitory to HIV-1 proviral DNA synthesis in macrophages but not in T cells. The data presented demonstrates that the non-canonical dUTP was abundant relative to TTP, and efficiently incorporated during HIV-1 reverse transcription, particularly in non-dividing macrophages.
Keywords: DNA Polymerase, Enzyme Kinetics, Mutagenesis Mechanisms, Nucleotide, Reverse Transcription
Introduction
2′-Deoxyuridine triphosphate (dUTP) is one of the key metabolic intermediates during cellular pyrimidine nucleotide biosynthesis. dUTP incorporation into chromosomal DNA by cellular DNA polymerases is a well characterized mutagenic event (1, 2), and cells are equipped with repair mechanisms that efficiently remove the incorporated dUMP from DNA (3). This cellular non-canonical dUTP also has a similar mutagenic impact on viruses that synthesize their genomic DNA in those cells. In fact, it is well established that viruses synthesizing viral DNA either encode their own repair systems or hijack the host repair system. These systems can replace the uracil resulting from either misincorporated dUTP or deaminated dCMP (resulting from ABOBEC3G in HIV-1), which have both been shown to be distinctly mutagenic (reviewed in Ref. 4) with dTMP.
Human immunodeficiency virus type 1 (HIV-1) uniquely infects terminally differentiated/non-dividing macrophages as well as dividing, activated CD4+ T cells. Infected macrophages are known to be a long lived cellular viral reservoir, especially in the CNS (5–7). Upon infection, the single-stranded viral genomic RNA is reverse transcribed into double-stranded proviral DNA by a virally encoded DNA polymerase, reverse transcriptase (RT). Although the cellular dNTP concentrations in various types of dividing cells, including cancer cells, has been extensively studied, the dNTP concentrations of human primary non-dividing macrophages were only recently determined (8, 9). These studies revealed that macrophages harbor strikingly low cellular dNTP concentrations (20–40 nm), compared to human primary activated CD4+ T cells (1–4 μm). Due to these limited cellular dNTP pools, viruses infecting macrophages may encounter difficulties in replicating their DNA genomes unless they are equipped with their own dNTP biosynthetic machinery. Because HIV-1 lacks this nucleotide biosynthesis machinery, it was of interest to determine how HIV-1 is able to efficiently synthesize proviral DNA in macrophages with such a limited dNTP substrate pool. Our kinetic studies revealed that HIV-1 RT is uniquely able to synthesize DNA even at low dNTP concentrations due to its high affinity for dNTP, enabling HIV-1 to overcome the limited dNTP pools in macrophages (8, 10).
Cellular dUTP can be synthesized either directly by the deamination of dCTP or phosphorylation of dUDP produced from UDP by ribonucleotide reductase (11). Mammalian cells lack the enzyme catalyzing the former reaction in the triphosphate form (12), and the latter is the major pathway for dUTP biosynthesis. However, dividing cells are also known to actively reduce levels of dUTP by dUTPase (1, 13). Although the cellular concentration of dUTP in dividing cells was previously reported, the dUTP concentrations vary vastly among cell types (14–16). One possible reason for these variations could result from technical difficulties in reliably differentiating dUTP from two other common cellular chemical analogs, UTP and TTP, in those methods employed.
It has been suggested that because dUTP incorporation into DNA is a genomic mutagenic event, cellular dUTP functions as a cellular defense mechanism (17) that can induce lethal hypermutagenesis upon incorporation into the DNA genomes of infecting cellular parasites, bacteria, and viruses, including HIV-1. HIV-1, however, efficiently escapes from this biochemical cellular defense mechanism by co-packaging a cellular enzyme, UNG2 (uracil DNA glycosylase 2), which repairs the dUMP molecules incorporated into proviral DNA by HIV-1 RT (18). An HIV accessory protein, viral protein R, has been considered as a viral effector that co-packages UNG2 from the infected cells (19, 20).
Here we employed quantitative LC-MS/MS technology for precise measurements of cellular dUTP in both human peripheral blood mononuclear cells (PBMCs)2 and monocyte-derived macrophages. With the determined dUTP concentration, we simulated HIV-1 reverse transcription in vitro and then confirmed the biochemical findings virologically. Indeed, we observed unexpectedly abundant cellular dUTP levels in non-dividing macrophages, which can result in efficient dUTP incorporation into HIV-1 proviral DNA replicated therein.
EXPERIMENTAL PROCEDURES
Preparation and Culture of Human Primary Macrophages and PBMCs for LC-MS/MS dUTP/TTP Assay
Human monocytes were isolated from buffy coats of HIV-1-negative, hepatitis B virus/hepatitis C virus-negative donors with density gradient centrifugation coupled with enrichment for CD14+ monocytes with Rosette Sep antibody mixture (Stem Cell Technologies, Vancouver, Canada). Cells were seeded at a concentration of 1.0 × 106 cells/well for 1 hr at 37 °C, 5% CO2 to allow plastic adherence prior to repeated washes with 1× PBS. Monocytes were allowed to differentiate for 7 days in RPMI medium (Hyclone, Logan, UT) containing heat-inactivated 20% fetal calf serum (FCS) (Atlanta Biologicals, Lawrenceville, GA), 1% penicillin/streptomyocin (Invitrogen), supplemented with 100 units/ml m-CSF (R&D Systems, Minneapolis, MN) at 37 °C, 5% CO2. For all conditions, macrophages were stained with CD11b-APC (Miltenyi Biotec, Auburn, CA) and subjected to FACS to determine purity of >99%. Human PBMCs were also isolated from buffy coats derived from healthy donors. Activated PBMCs were maintained in RPMI medium supplemented with heat-inactivated 20% FCS, 1% penicillin/streptomyocin, and 2% l-glutamine (Cellgro/Mediatech, Inc., Manassas, VA); 6 μg/ml PHA (J-Oils Mills, Inc., Tokyo, Japan) was added to the cells 72 h prior to experiments in order to activate them.
Extraction of Intracellular Nucleotide Fraction and LC-MS/MS Analysis
For both macrophages and PBMCs, the isolated cells were washed twice with ice-cold 1× PBS to remove any residual medium. Cells were resuspended in 70% CH3OH overnight, and extracts were centrifuged at 13,000 × g for 10 min (Thermo Electron Corp., Marietta, OH). Supernatants were subsequently dried, and the resulting samples were reconstituted in HPLC mobile phase for LC-MS/MS analysis as described previously (21). For dUTP, the MS/MS transition 469 → 81 was applied, and [13C15N]TTP was used for calibration.
RT Purification
The HXB2 HIV-1 RT gene was previously cloned into pET28a (Novagen), and the N-terminal hexahistidine-tagged p66/p66 homodimer HIV-1 RT was subsequently expressed in Escherichia coli BL21 (DE3) and purified with Ni2+-NTA chromatography followed by DEAE and SP anion exchange, as described previously (22), and a similar procedure was used for murine leukemia virus (MuLV) RT. SIVagm Sab1 RT was also previously cloned and purified (23). Foamy virus RT was described previously (24). These RT proteins were quantified and stored in 10% glycerol dialysis buffer as described previously (25, 26).
Primer Extension Assays
All DNA and RNA primers used in this study were purchased from Integrated DNA Technologies and Dharmacon Research, respectively. Assay mixtures (20 μl) contained 10 nm template-primer, the RT protein concentrations specified in the individual figure legends, four dNTPs (Amersham Biosciences) at the concentrations indicated in the figure legends, and 1× Reaction Buffer (50 mm Tris-HCl (pH 7.5), 50 mm NaCl 5 mm MgCl2). Reactions were initiated by adding the RT proteins and incubated at 37 °C for the defined times. Reactions were terminated with 10 μl of 40 mm EDTA, 99% formamide. Reaction products were immediately denatured by incubating at 95 °C for 5 min, and 4 μl of each 30-μl final reaction mixture was quantitated by PhosphorImager analysis (PerkinElmer Life Sciences) of 14% polyacrylamide-urea denaturing gels (SequaGel, National Diagnostics).
For the primer reaction described in the legend to Fig. 3C, a 5′-end 32P-labeled 17-mer A primer (5′-CGCGCCGAATTCCCGCT-3′) annealed to a 40-mer RNA (5′-AAGCUUGGCUGCAGAAUAUUGCUAGCGGGAAUUCGGCGCG-3′; template/primer ratio 2.5:1) was extended for various time points under the reaction conditions described above using the nucleotide pools specified in the figure legends.
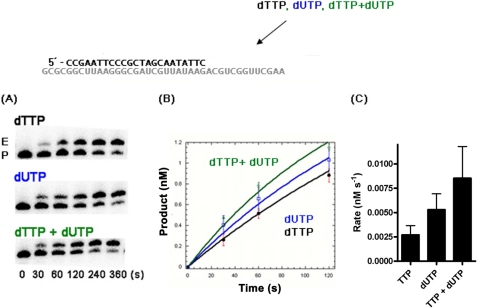
FIGURE 3.
TTP and dUTP incorporation rates of HIV-1 RT in macrophage conditions. A, single nucleotide extension reaction time-courses using the same template-primer (P) used in Fig. 2 were performed in vitro with the reconstituted TTP and dUTP concentrations found in macrophages. E, extended product. B, product formation for TTP (black line), dUTP (blue line), and both TTP and dUTP (green line) were plotted. C, the rates of the product formation (B) were determined with a double exponential equation as described under “Experimental Procedures.” A 20 nm final concentration of HIV-1 RT was used to initiate the reaction in which small aliquots were obtained over the time course indicated. The experiments were repeated in triplicate.
Single Nucleotide Incorporation Rates of HIV-1 RT in Cellular dUTP/TTP Pools
To assess the HIV-1 RT-mediated rates of TTP and dUTP incorporation in Fig. 3, a 5′-end 32P-labeled Ext-T 23-mer primer (5′-CCGAATTCCCGCTAGCAATATTC) was individually annealed to the 40-mer RNA template, which was used at a 2 nm final concentration in the reaction time course. These were then used to measure rates of product formation with macrophage levels of TTP, dUTP, or both (Table 1).
TABLE 1.
Intracellular dUTP concentrations of primary human monocyte-derived macrophages and activated PBM cells as determined by LC-MS/MS
| TTPa | dUTP | |
|---|---|---|
| Concentration (μm) | ||
| Activated PBMCs | 16.0 ± 5.3 | 12.0 ± 1.7 |
| Macrophages | 0.05 ± 0.04 | 2.9 ± 1.3 |
| pmol/106cells | ||
| Activated PBMCs | 4.9 ± 1.6 | 3.8 ± 0.5 |
| Macrophages | 0.13 ± 0.1 | 7.7 ± 3.6 |
a Previously published (9).
Steady State Kinetics of HIV-1 RT with dUTP/TTP
The single nucleotide incorporation reactions were conducted using the Ext-T 23-mer described above annealed to RNA 40-mer in the 1× reaction buffer for 5 min at least in triplicate at the ranges of TTP and dUTP concentrations sufficient to assure proper non-linear regression fits for both phases of product formation. The individual reverse transcriptases discussed were titrated onto saturating TTP to obtain product formation within the linear range within the reaction parameters described. The Km and Kcat values were determined as described previously (9, 27) by non-linear regression.
Pre-steady State Kinetic Analysis of HIV-1 RT Protein
To determine the concentration of active purified WT RT, single-turnover burst assays were performed as described previously (10). A 5′-end 32P-labeled (100 nm) and cold (200 nm) 23-mer T primer annealed to 38-mer RNA template was extended with 100 nm WT RT in the presence of 800 μm dTTP for single round incorporation and rapidly quenched with EDTA at different time points using the Kintek RFQ3. The samples were then analyzed on a 14% sequencing gel under denaturing conditions, and the percentage of extended product was quantified (Quantity One 1-D analysis software, Bio-Rad). The product formation at each time point was plotted and fitted to Equation 1, which determined the concentration of active RT (Amp) to be 1.9 μm. The pre-steady state reactions with 200 nm active proteins and 50 nm template-primer complexes were performed to assess the observed rates of product formation (kobs) at six time points in duplicate for different dTTP or dUTP concentrations. The kobs values were then fitted to Equation 2 to yield Kd and kpol. Curve fitting was performed (Equations 1 and 2) with KaleidaGraph as described previously (10).
Coupled Primer Extension and UNG Digestion Assay
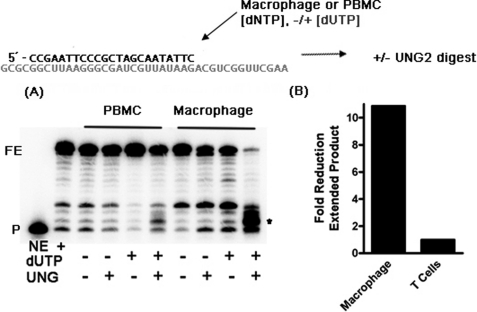
Reactions (5 min) with the conditions indicated in Fig. 2 (with or without dUTP and/or UNG2) were initiated with 40 nm RT at either macrophage or PBMC dNTP concentrations with the primer and template described above. These reactions were then quenched with 10 mm EDTA alone and divided and incubated with either double-distilled H2O or 2 units of E. coli UDG (New England Biolabs) for 60 min at 37 °C. The stop dye described above was then added, and these reactions were incubated at 95 °C for 10 min and analyzed by urea PAGE electrophoresis as mentioned above. Fig. 2 was analyzed by comparing background-subtracted, total product-normalized, and completely extended product from macrophage and T cells quantified with Bio-Rad Quantity One. These data were then plotted as -fold reduction in extended product directly due to dUTP incorporation and subsequent UNG2 digest.
FIGURE 2.
dUTP incorporation by HIV-1 RT with nucleotide substrate pools found in primary human macrophages and PBMCs. An RT extension reaction coupled with a UNG2 digestion assay is shown in A. A 5′-end 32P-labeled 23-mer DNA primer annealed to the 40-mer RNA template was extended in vitro by HXB2 HIV-1 RT with the reconstituted dNTP concentrations found in macrophages or PBMCs in the presence and absence of dUTP concentration found in those cell types, and then these reactions were quenched with EDTA and subjected to E. coli UNG2 digestion, followed by 95 °C incubation and analyzed by urea denaturing PAGE. The cleavage products induced by the dUTP incorporation are marked with an asterisk. P, primer. B, total fully extended products (FE) after heat treatment were determined, and the -fold difference between the fully extended product levels of the reactions under the two cell type conditions is shown.
Visualization, Quantification, and Kinetic Analysis of the Primer Extension Reactions
All primer extension reactions in this study were analyzed with urea denaturing 16% polyacrylamide gels and scanned with a PMI phosphor imager (Bio-Rad) and quantified with the Bio-Rad Quantity One software. Nonlinear regression analysis was done with Kaleida Graph, rates were determined with a single exponential, and Michaelis-Menten fits were completed as described previously (27).
Assays for Inhibition of HIV-1 Infection and Proviral DNA Synthesis by ddUTP in Macrophages and T Cells
The purified human monocytes from multiple donors were differentiated to macrophages as described previously, and the D3HIV-GFP vector system expressing GFP was also prepared as described previously (8, 10, 28, 29). Equal p24 level of the produced vector was used to transduce human macrophages preincubated with different concentrations of 2′,3′-dideoxyuridine (ddU) (Sigma) for at least 2 h. The harvested cells were used for determining the percentages of GFP-expressing and/or propidium iodide-stained cells by FACS at 7 days postinfection. All cells used to assess transduction efficiency were gated or stained with propidium iodide to assess cell death. 2LTR quantitative RT-PCR assays were conducted identically, with genomic DNA purified with Promega Genomic DNA kit on day 7. Quantitative 2LTR circle PCR assay using real-time PCR was normalized by total genomic DNA (Bio-Rad). The primers and amplification protocol have been used in our previous studies (28, 30). ddUTP was tested in T cells as follows. Human primary activated CD4+ T cells (2.5 × 105) isolated from two donors were pretreated with 0, 0.1, and 1 mm ddUTP for at least 2 h, and then the cells were transduced with DHIV-GFP. The transduced cells were prepared 48 h post-transduction and analyzed by FACS for determining the percentage of GFP+ cells. The analysis was conducted in triplicate per donor, and the means and S.D. values were derived from all data obtained.
Cellular Viability Assays
CHME5 cells (24 h), MRC5 primary HLF cells (48 h), primary CD4+ T cells (48 h), and primary macrophages (5 days) were incubated with the ddU concentrations indicated for the time period in parenthesis. These cells were then stained with propidium iodide (Sigma) and analyzed with flow cytometry.
RESULTS
LC-MS/MS-based Quantification of Cellular dUTP from Human Primary Macrophages and Activated PBMC
Cellular dUTP concentrations in human primary cells have been previously reported, whereas the dUTP concentration in human primary terminally differentiated/non-dividing macrophages, which is one of the key target cell types of HIV-1, has not been determined definitively. We previously observed that the TTP concentration is extremely low in macrophages (0.05 μm) compared with that in activated PBMCs (16 μm; Table 1 and Fig. 1A). Because both TTP and dUTP are the substrates for chromosomal DNA replication, which is absent in macrophages, it is a reasonable assumption that the dUTP concentration in macrophages would also be much lower than that in the activated PBMCs. In this study, first, we measured the dUTP concentration of human primary macrophages and activated PBMC whole cell extracts by employing the quantitative LC-MS/MS technology, which not only can accurately differentiate dUTP from other chemically analogous compounds, such as UTP and TTP, by their molecular weights, but also can sensitively detect and quantify dUTP and TTP extracted from human primary macrophages and activated PBMCs isolated from multiple donors (see supplemental Fig. 1 for the separation profiles for dUTP, dTTP, and UTP).
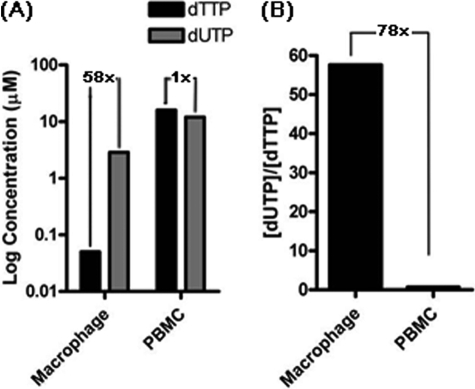
FIGURE 1.
Comparison of dUTP and TTP concentrations of human primary macrophages and activated PBMCs. A, the dUTP concentrations of macrophages and activated PBMCs, which were determined by LC-MS/MS (summarized in Table 1), are plotted with gray bars. The black bars represent TTP concentrations of human primary macrophages and activated CD4+ T cells previously determined by LC-MS/MS (9). The -fold differences between dTTP and dUTP in each cell type are shown. B, the dUTP/TTP concentration ratio difference between the two cell types calculated from the LC-MS/MS analysis data.
As shown in Table 1 and Fig. 1A, our LC-MS/MS analysis demonstrated that human PBMCs have a high dUTP concentration (12.0 μm) almost equal to the TTP concentration (16.0 μm). Surprisingly, however, unlike canonical TTP, human primary macrophages significantly maintain high dUTP concentrations, and thus macrophages appear to have an extreme imbalance between dUTP and TTP with a [dUTP]/[TTP] ratio of 58 (Fig. 1A). Indeed, macrophages harbor a 78-fold more imbalanced ratio of dUTP over TTP, compared with the activated PBMCs (Fig. 1B). This uniquely large dUTP/TTP imbalance in macrophages implies that any viruses that synthesize DNA in macrophages during their life cycle encounter a great kinetic stress to incorporate highly mutagenic dUTPs into their DNA genomic materials. More importantly, we reasoned that this unique abundant dUTP and severe dUTP/TTP imbalance may force HIV-1 RT to incorporate dUTP much more frequently during HIV-1 proviral DNA synthesis, particularly in macrophages. However, it is also highly plausible that this high dUTP pool environment can be counteracted by the efficient enzymatic selectivity of HIV-1 RT against dUTP.
Efficient dUTP Incorporation by HIV-1 RT with the Macrophage Nucleotide Pools but Not with the Ones with Activated PBMCs
Because the LC-MS/MS data in Fig. 1 revealed that non-canonical dUTP is abundant and a greater disparity between TTP and dUTP exists in macrophages, compared with the activated PBMCs, we next biochemically investigated how HIV-1 RT synthesizes DNA under the dNTP/dUTP pools found in these cells. For this, we simulated the HIV proviral DNA synthesis by employing a primer extension reaction with a 5′-end 32P-labeled 23-mer DNA primer annealed to a 40-mer RNA template (template-primer). In this biochemical simulation, the template-primer complex was extended by HIV-1 RT with either dNTP alone or dNTP and dUTP mixtures at the concentrations found in macrophages and activated PBMCs, as illustrated in Fig. 2 (top). In order to monitor the dUTP incorporation during DNA synthesis in this reaction, we incubated the reaction products with E. coli UNG followed by heat treatment, which has been used extensively to detect the dUMP embedded in DNA (31, 32). In this treatment, UNG cleaves the glycosidic bond of uracil in DNA containing dUMP, generating the abasic sites that heat treatment then hydrolyzes at the phosphodiester bond, resulting in dUMP-specific DNA fragments. As shown in Fig. 2, HIV-1 RT efficiently extended the primer in dNTP alone pools or dNTP/dUTP mixture pools found in both cell types, generating the 40-mer products (FE). However, upon UNG treatment, no smaller cleavage products were observed in the reactions with both PBMCs and macrophage dNTPs alone, implying the absence of dUTP incorporation and nonspecific cleavage reactions under this experimental condition. However, a significant amount of the full-length product was degraded to small cleavage products in the reactions using the dNTP/dUTP pools of macrophages (see the asterisk in Fig. 2A). In contrast, during the DNA polymerization reaction with the PBMCs, dNTP/dUTP pools displayed little degradation of the full-length product and minimal cleavage product. When the cleavage products were quantified (Fig. 2B), it was clear that the macrophage nucleotide pool generated 10-fold more cleavage products than the PBMC nucleotide pool. Therefore, the data shown in Fig. 2, A and B, clearly support the efficient incorporation of dUTP during HIV-1 RT-mediated DNA synthesis under the macrophage nucleotide pools but not the PBMC nucleotide pools.
Next, we investigated the kinetic impact of dUTP on HIV-1 RT-mediated DNA synthesis reactions under cellular dNTP concentrations. For this, we employed the single nucleotide incorporation reaction with the same template-primer used in Fig. 2A. In this reaction, the primer was extended by HIV-1 RT with the macrophage concentrations of TTP alone, dUTP alone, and the mixture of TTP and dUTP. As shown in Fig. 3A, the rate of dUTP incorporation is equal or elevated as compared with TTP at their macrophage concentrations. However, as seen in Fig. 3B, upon the addition of both TTP and dUTP, the overall primer extension kinetics were additive as quantified in Fig. 3C when compared with TTP or dUTP alone. Indeed, the presence of dUTP elevates the primer extension rate by ∼3-fold compared with the dTTP alone at macrophage concentrations. Thus, these data, together with the data shown in Fig. 2A, indicate that not only does HIV-1 efficiently incorporate dUTP during DNA synthesis using the macrophage nucleotide pools, but also the presence of dUTP substrate improves HIV-1 RT-mediated DNA synthesis kinetics in macrophage dNTP concentrations.
Steady State and Pre-steady State Kinetic Comparison of TTP and dUTP Incorporation by HIV-1 RT
Next, we investigated the enzymatic selectivity of HIV-1 RT for dUTP and TTP by measuring their steady state incorporation parameters. We measured the steady state kinetic Km and kcat values of HIV-1 RT with TTP and dUTP using the single nucleotide incorporation assay. As shown in Table 2, HIV-1 RT displayed almost identical Km and kcat values for dUTP and TTP. This indicates that HIV-1 RT has an equal steady state incorporation capability (kcat/Km) for dUTP and TTP. In other words, HIV-1 RT has very poor capability to distinguish between TTP and dUTP during the steady state kinetic DNA synthesis.
TABLE 2.
Steady state kinetic parameters TTP/dUTP for HIV-1, MuLV, FIV, and SIVagm reverse transcriptases
| Km | Kcat | Kcat/Km | Selectivity (TTP/dUTP) | |
|---|---|---|---|---|
| μm | ||||
| HIV | ||||
| TTP | 0.022 ± 0.003 | 0.010 ± 0.003 | 0.44 | 1.30 |
| dUTP | 0.028 ± 0.007 | 0.009 ± 0.003 | 0.34 | |
| SIV | ||||
| TTP | 0.018 ± 0.004 | 0.001 ± 0.0002 | 0.054 | 0.84 |
| dUTP | 0.017 ± 0.004 | 0.001 ± 0.0001 | 0.065 | |
| FIV | ||||
| TTP | 0.059 ± 0.007 | 0.039 ± 0.0006 | 0.67 | 3.75 |
| dUTP | 0.180 ± 0.005 | 0.032 ± 0.0013 | 0.18 | |
| MuLV | ||||
| TTP | 0.050 ± 0.022 | 0.0008 ± 0.0001 | 0.016 | 4.97 |
| dUTP | 0.253 ± 0.052 | 0.0008 ± 0.0001 | 0.003 | |
To confirm the steady state results, we also performed pre-steady state quench flow titrations for both TTP and dUTP. As shown in Table 3, HIV-1 RT showed very similar binding affinity (Kd) to dUTP and TTP. In addition, HIV-1 RT also has a very similar isomerization/catalysis capability with dUTP and TTP, giving minimal selectivity change. Thus, both steady and pre-steady state kinetic analysis of HIV-1 RT with TTP and dUTP demonstrate that HIV-1 RT has a very poor capability to distinguish TTP from dUTP (less than 1-fold difference). These data further imply that HIV-1 may incorporate dUTP more frequently per infection in macrophages than in activated T cells due to not only the unusual large disparity of dUTP/TTP pools in macrophages but also the poor enzymatic dUTP/TTP selectivity of HIV-1 RT.
TABLE 3.
Pre-steady state kinetic parameters for TTP/dUTP for HIV-1
| Kd | Kpol | kpol/kd | Selectivity | |
|---|---|---|---|---|
| μm | s−1 | μm−1s−1 | ||
| TTP | 3.4 ± 0.62 | 85.72 ± 7.3 | 25 | 0.66 |
| dUTP | 5.1 ± 1.93 | 194.39 ± 30.0 | 38 |
Primate Lentiviral RTs Readily Incorporate dUTP, whereas Other Retroviral RTs Are Selective for TTP
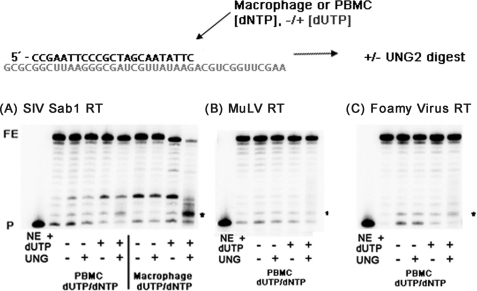
Next, we investigated whether the lack of enzymatic dUTP and TTP selectivity demonstrated by HIV-1 RT is common. We measured and compared the steady state kinetics parameters for three lentiviral RTs: HIV-1 RT, simian immunodeficiency virus (SIV) RT (SIVagm Sab1), and feline immunodeficiency virus (FIV) RT. As shown in Table 2, SIVagm RT has also very similar Km and kcat values with TTP and dUTP, as was observed with HIV-1 RT (Table 1), suggesting that SIV RT has a poor capability of differentiating dUTP from TTP. However, FIV RT displayed a higher Km and lower kcat values with dUTP than TTP, indicating that FIV RT was able to differentiate dUTP from TTP ∼3–4 times better than HIV-1 and SIV RT. Next, we also simulated RT-mediated DNA synthesis using macrophage and PBMC dNTP/dUTP pools as used in Fig. 2 for HIV-1 RT. As shown in Fig. 4A, SIVagm Sab-1 RT also showed significant degradation of the full-length product and accumulation of the small cleavage product in the reaction with the macrophage dNTP/dUTP mixture pool, suggesting that, like HIV-1 RT, SIV RT incorporates dUTP efficiently in the macrophage nucleotide pools but not in the PBMC nucleotide pools (Fig. 2A).
FIGURE 4.
dUTP incorporation by other lentiviral and non-lentiviral RT proteins. UNG-2 digestion assays were carried out as described in Fig. 2 for SIVagm, MuLV, and FV RTs. Uracil digestion products . are marked with an asterisk. FE, fully extended product; P, primer.
Next, we tested if RTs of retroviruses such as MuLV and foamy virus (FV), which do not replicate in non-dividing cells, can incorporate dUTPs in the dividing PBMC dNTP/dUTP pool. We previously reported that RTs from this type of the retroviruses do not synthesize DNA at the dNTP concentrations found in macrophages and stay active only with the high dNTP concentrations found in dividing cells (24, 33, 34). When we simulated MuLV RT-mediated DNA synthesis with the PBMC dNTP/dUTP pools, DNA products did not produce the small cleavage products upon UNG/heat treatment (Fig. 4B). This indicates that MuLV RT does not incorporate dUTPs during the DNA synthesis with the PBMC dNTP/dUTP pools. When we measured the steady state kinetic parameter of MuLV RT with TTP and dUTP (Table 3), we found that MuLV RT has better discrimination capability for dUTP and TTP than HIV-1 and SIV RTs. Next, we simulated the FV-mediated DNA synthesis with the dividing PBMC dNTP/dUTP pools. As observed with MuLV RT, no significant dUTP incorporation was observed for FV RT (Fig. 4C). Therefore, the data from the biochemical simulations (Fig. 4) suggest that RTs of all of the retroviruses tested in this study infrequently incorporate dUTP during DNA synthesis with the dividing PBMC dNTP/dUTP pools. However, as shown in Table 3, the steady state kinetic experiments demonstrate that RTs of FIV and MuLV preferentially incorporate TTP over dUTP, whereas RTs of HIV-1 and SIV have poor selectivity of TTP over dUTP. Possible links of these biochemical findings with their virological consequences are addressed under “Discussion.”
ddU Inhibits HIV-1 Reverse Transcription Exclusively in Primary Human Macrophage
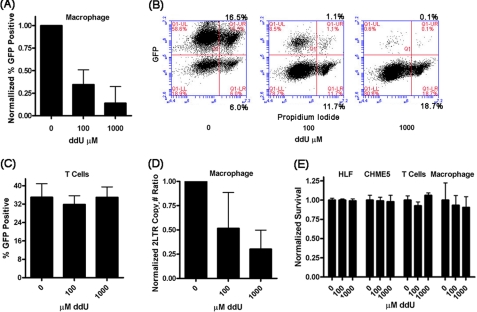
In this study, we observed 1) a high abundance of dUTP in macrophages, 2) a poor capability of HIV-1 RT to discriminate between dNTP and dUTP, and 3) the frequent incorporation of dUTP by HIV-1 RT in the macrophage dNTP/dUTP pools. These observations led us to postulate that if dUTP is readily incorporated during reverse transcription, specifically in macrophages, a dU analog, ddU, would be inhibitory against HIV-1 replication in macrophages but not in activated CD4+ T cells. To test this, we transduced primary human macrophages and activated CD4+ T cells isolated from three donors with an HIV-1 vector and determined the transduction efficiency with or without different concentrations of ddU. The HIV-1 vector used in this test was pseudotyped with the vesicular stomatitis virus glycoprotein envelope, which encodes the entire HIV-1 genome except the env gene and the nef gene, which was replaced with enhanced GFP. When the transduced (GFP-positive) macrophages were measured by flow cytometry, we observed a ddU dose-dependent inhibition of transduction efficiency (Fig. 5, A and B). However, no significant ddU inhibition was observed in activated CD4+ T cells (Fig. 5C). Finally, to verify that the inhibition of HIV-1 vector transduction results from the inhibition of the proviral DNA synthesis step, we performed a quantitative 2LTR quantitative RT-PCR. As shown in Fig. 5D, fewer 2LTR circles accumulated in the cells treated with higher concentrations of ddU. Importantly, the ddU did not display detectable cytotoxicity in both cell types during this treatment (Fig. 5E). Thus, the data presented in Fig. 5 indicate that ddU inhibits HIV-1 reverse transcription and that this inhibition specifically occurs in macrophages but not in T cells, which we previously reported (35).
FIGURE 5.
Effect of ddU treatment on HIV-1 replication in primary human macrophages and activated CD4+ T cells. Primary human macrophage were pretreated with ddU and then transduced with a single round vesicular stomatitis virus glycoprotein pseudotyped HIV-1 vector (D3HIV). A, the normalized vector transduction efficiency in macrophages at the indicated ddU concentrations from n = 5 independent human donors. B, FACS analysis for the macrophages in A for GFP-expressing cells. C, the normalized vector transduction efficiency in human primary activated CD4+ T cells at the indicated ddU concentrations from three independent human donors. D, 2LTR circle quantitative PCR plotted by ddU concentration from four independent human donors. E, ddU toxicity assessed by the live and dead cell assay for primary human lung fibroblasts (MRC5), human microglial cell line (CHME5), human primary activated CD4+ T cells, and primary human macrophage.
DISCUSSION
Most DNA polymerases must function in a cellular milieu with their choice substrate being dNTPs. However, dUTP, a key metabolic intermediate of TTP biosynthesis, is also present in most living organisms. It is also clear that most DNA polymerases are minimally selective for TTP over dUTP (36, 37), and thus, the cellular levels of dUTP and TTP generally determine the probability of their incorporation. It is exceedingly difficult to enzymatically select TTP over dUTP (which differ only by a 5′-methyl group present on thymidine) during DNA synthesis. Both cellular and viral polymerases responsible for DNA replication are not highly selective for dUTP over TTP (36, 37) when compared with the incorrect base and ribonucleotides (38–40). In actively dividing cells undergoing DNA synthesis, dUTP is actively degraded by cellular dUTPases (41). However, in the event of aberrant dUTP incorporation or the well studied deamination of dCMP in viral DNA by APOBEC3G, dUMP incorporated into DNA is quickly removed by a series of highly developed and conserved cellular repair mechanisms, including various enzymes, such as UNGs, apurinic/apyrimidinic endonucleases, DNA repair polymerases, and ligases (42, 43). Although many cell types are equipped with these repair mechanisms to remove the incorporated dUMP, dUTPase is still essential in dividing Saccharomyces cerevisiae, E. coli, and human cells (13, 44–46), and knock-outs result in excessive dUMP incorporation beyond the capability of the cellular repair systems. This argues that there is a threshold dUTP/TTP ratio capable of repair below which host cellular DNA replication occurs. However, the status of dUTP availability and the cellular impact of dUTP in terminally differentiated/non-dividing cell types, such as macrophage, where chromosomal DNA replication machinery is permanently dormant are still relatively unknown.
dUTP is an effective cellular antiviral, and it is clear from numerous examples in the virosphere infecting both non-dividing and dividing cells that many of the parasitic viruses typically encode or require either a dUTPase or uracil gylcosylase to infect dividing cells (reviewed in Ref. 4). Specifically, lentiviruses, such as HIV-1, uniquely replicate in non-dividing cells and rely on cellular nucleotides, which freely diffuse into the retroviral capsid after entry, for reverse transcription. Similar to the cell, lentiviruses must prevent uracilation of their genomes in order to minimize the lethal mutagenic effect of cellular dUTP. Lentiviruses employ two distinct mechanisms to prevent or remove uracil prior to and after reverse transcription. All non-primate lentiviruses encode dUTPases to hydrolyze dUTP prior to reverse transcription (47), as do many dsDNA viruses that replicate in non-dividing cells (48, 49). Furthermore, it has been postulated that HIV-1 may have encoded a dUTPase, which is believed to be vestigial (50, 51) and has acquired the ability to package host UNG2 from cells, possibly replacing this dUTPase activity during evolution (19, 52). However, other retroviruses, which do not replicate in non-dividing cells, carry neither viral dUTPases nor host UNG repair enzymes. This supports the idea that uracilation of viral genomes may occur more readily in non-dividing cells, and only lentiviruses that infect non-dividing cells are equipped with mechanisms to counteract the cellular antiviral dUTP.
However, the dUTP level in human primary macrophages has not been investigated to date with LC-MS/MS. Because our previous studies reported that the dNTP concentrations in macrophages are extremely low, we assumed that dUTP, which is a metabolic byproduct of TTP, should also be scarce in macrophages. Surprisingly, our measurements of dUTP by LC-MS/MS technology, which accurately differentiates dUTP from TTP and UTP, revealed that dUTP is unusually abundant in macrophages compared with TTP, leading to a significant disparity between dUTP and TTP. This is surprising, considering that the ribnucleotide reductase R2 subunit, which reduces UDP to dUDP, should not be expressed in macrophages (53), and thus the origin of these dUTP levels is under investigation. In contrast, activated PBMCs showed almost equal concentrations of TTP and dUTP as expected for dividing cells actively expressing dUTPase. Next, our biochemical simulations demonstrated that dUTP is frequently incorporated during reverse transcription by HIV-1 RT in the macrophage dNTP/dUTP pools but not in the activated PBMC nucleotide pools. In addition, HIV-1 RT has a very poor enzymatic capability of differentiating dUTP from TTP, supporting a possibility that dUTPs may be more frequently incorporated during HIV-1 proviral DNA synthesis in macrophages than in dividing T cells. Indeed, this possibility was further supported by our experiment with the dUTP analog, ddU, which inhibits HIV-1 proviral DNA synthesis in macrophages, but not in activated CD4+ T cells. These biochemical and virological observations also indicate that HIV-1 may be more dependent on the UNG repair mechanism in macrophages than CD4+ T cells per infection, which was previously addressed (18). However, a study has demonstrated that in both dividing and non-dividing cells, co-packaged UNG is not required for HIV-1 replication (54). This is expected for dividing cells when considering our results; however, for non-dividing cells, the infectivity of virus produced from these UNG minus virus-infected macrophage was not measured, and this perhaps is where mutant virus would be observed.
Our biochemical data with multiple RTs for their enzymatic discrimination capability between dUTP and TTP also raise several interesting possible interactions between RT enzymology and viral dUTP repair strategies. Among the RTs of three lentiviruses analyzed in this study, both HIV-1 and SIV RTs have poor selectivity between these two nucleotides, whereas FIV RT has better discrimination capability for TTP over dUTP. FIV is equipped with the viral dUTPase, whereas HIV-1 and SIV have the UNG2-based repair machinery. Possibly, the dUTPase-based system may still require the additional anti-dUTP mechanism exerted by the better dUTP/TTP discrimination capability of FIV RT for viral viability. However, the UNG2-based repair system may be sufficient to deal with the frequent dUTP incorporation in the case of HIV-1 and SIV, and these lentiviruses may not need the help from their RTs, which have poor dUTP/TTP discrimination power. However, this hypothesized interplay requires further data to generalize, which could be obtained by characterization of lentiviral RTs from other dUTPase minus and UNG-based viruses. Interestingly, RTs of MuLV and FV, which do not infect non-dividing cells, also have better dUTP/TTP discrimination capability, supporting the idea that the dUTP repair may not be necessary in these retroviruses because their RTs can minimize dUTP incorporation in dividing cells where repair is available.
In conclusion, the biochemical and virological observations presented in this study reveal how distinct cellular and enzymatic environments impact HIV-1 reverse transcription and thus its cell type-specific infectivity. Non-canonical dUTP may serve as a cellular chemical defense mechanism, which has been proposed previously; however, there has been little evidence to support this claim. This abundant cellular dUTP can be more frequently incorporated by HIV-1 RT in macrophages and may require efficient repair (e.g. UNG2) mechanisms to avoid both lethal mutagenesis, promoter damage, and to ultimately complete the HIV replication cycle.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI049781 (to B. K.), T32 AI007362 (to E. M. K.), 1R01-RR-025996-01A1 (to R. F. S.), and 2P30-AI-050409 (to R. F. S.). This work was also supported by a grant from the Department of Veterans Affairs (to R. F. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- PBMC
- peripheral blood mononuclear cell
- ddU
- 2′,3′-dideoxy uracil
- MuLV
- murine leukemia virus
- 2LTR
- two-long terminal repeat
- SIV
- simian immunodeficiency virus
- FIV
- feline immunodeficiency virus
- FV
- foamy virus
- UNG
- uracil DNA glycosylase.
REFERENCES
- 1. Brynolf K., Eliasson R., Reichard P. (1978) Cell 13, 573–580 [DOI] [PubMed] [Google Scholar]
- 2. Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nilsen H., Rosewell I., Robins P., Skjelbred C. F., Andersen S., Slupphaug G., Daly G., Krokan H. E., Lindahl T., Barnes D. E. (2000) Mol. Cell 5, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 4. Chen R., Wang H., Mansky L. M. (2002) J. Gen. Virol. 83, 2339–2345 [DOI] [PubMed] [Google Scholar]
- 5. Chugh P., Fan S., Planelles V., Maggirwar S. B., Dewhurst S., Kim B. (2007) J. Mol. Biol. 366, 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelly J., Beddall M. H., Yu D., Iyer S. R., Marsh J. W., Wu Y. (2008) Virology 372, 300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Montaner L. J., Crowe S. M., Aquaro S., Perno C. F., Stevenson M., Collman R. G. (2006) J. Leukoc. Biol. 80, 961–964 [DOI] [PubMed] [Google Scholar]
- 8. Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., Kim B. (2004) J. Biol. Chem. 279, 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiss K. K., Chen R., Skasko M., Reynolds H. M., Lee K., Bambara R. A., Mansky L. M., Kim B. (2004) Biochemistry 43, 4490–4500 [DOI] [PubMed] [Google Scholar]
- 11. Thelander L., Reichard P. (1979) Annu. Rev. Biochem. 48, 133–158 [DOI] [PubMed] [Google Scholar]
- 12. Björnberg O., Neuhard J., Nyman P. O. (2003) J. Biol. Chem. 278, 20667–20672 [DOI] [PubMed] [Google Scholar]
- 13. Gadsden M. H., McIntosh E. M., Game J. C., Wilson P. J., Haynes R. H. (1993) EMBO J. 12, 4425–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aquaro S., Caliò R., Balestra E., Bagnarelli P., Cenci A., Bertoli A., Tavazzi B., Di Pierro D., Francesconi M., Abdelahad D., Perno C. F. (1998) J. Biol. Regul. Homeost. Agents 12, 23–27 [PubMed] [Google Scholar]
- 15. Horowitz R. W., Zhang H., Schwartz E. L., Ladner R. D., Wadler S. (1997) Biochem. Pharmacol. 54, 635–638 [DOI] [PubMed] [Google Scholar]
- 16. Traut T. (1994) Mol. Cell. Biochem. 140, 1573–4919 [DOI] [PubMed] [Google Scholar]
- 17. Sire J., Quérat G., Esnault C., Priet S. (2008) Retrovirology 5, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Priet S., Gros N., Navarro J. M., Boretto J., Canard B., Quérat G., Sire J. (2005) Mol. Cell 17, 479–490 [DOI] [PubMed] [Google Scholar]
- 19. Bouhamdan M., Benichou S., Rey F., Navarro J. M., Agostini I., Spire B., Camonis J., Slupphaug G., Vigne R., Benarous R., Sire J. (1996) J. Virol. 70, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mansky L. M., Preveral S., Selig L., Benarous R., Benichou S. (2000) J. Virol. 74, 7039–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fromentin E., Gavegnano C., Obikhod A., Schinazi R. F. (2010) Anal. Chem. 82, 1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. (1991) EMBO J. 10, 3905–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skasko M., Diamond T. L., Kim B. (2009) Biochemistry 48, 5389–5395 [DOI] [PubMed] [Google Scholar]
- 24. Santos-Velazquez J., Kim B. (2008) J. Virol. 82, 8235–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weiss K. K., Bambara R. A., Kim B. (2002) J. Biol. Chem. 277, 22662–22669 [DOI] [PubMed] [Google Scholar]
- 26. Weiss K. K., Isaacs S. J., Tran N. H., Adman E. T., Kim B. (2000) Biochemistry 39, 10684–10694 [DOI] [PubMed] [Google Scholar]
- 27. Kennedy E. M., Hergott C., Dewhurst S., Kim B. (2009) Biochemistry 48, 11161–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jamburuthugoda V. K., Chugh P., Kim B. (2006) J. Biol. Chem. 281, 13388–13395 [DOI] [PubMed] [Google Scholar]
- 29. Jamburuthugoda V. K., Santos-Velazquez J. M., Skasko M., Operario D. J., Purohit V., Chugh P., Szymanski E. A., Wedekind J. E., Bambara R. A., Kim B. (2008) J. Biol. Chem. 283, 9206–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skasko M., Kim B. (2008) J. Virol. 82, 7716–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. (1977) J. Biol. Chem. 252, 3286–3294 [PubMed] [Google Scholar]
- 32. Devchand P. R., McGhee J. D., van de Sande J. H. (1993) Nucleic Acids Res. 21, 3437–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diamond T. L., Kimata J., Kim B. (2001) J. Biol. Chem. 276, 23624–23631 [DOI] [PubMed] [Google Scholar]
- 34. Skasko M., Weiss K. K., Reynolds H. M., Jamburuthugoda V., Lee K., Kim B. (2005) J. Biol. Chem. 280, 12190–12200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Graciet J. C., Shi J., Schinazi R. F. (1998) Nucleosides Nucleotides 17, 711–727 [DOI] [PubMed] [Google Scholar]
- 36. Dube D. K., Kunkel T. A., Seal G., Loeb L. A. (1979) Biochim. Biophys. Acta 561, 369–382 [DOI] [PubMed] [Google Scholar]
- 37. Mosbaugh D. W. (1988) Nucleic Acids Res. 16, 5645–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bebenek K., Joyce C. M., Fitzgerald M. P., Kunkel T. A. (1990) J. Biol. Chem. 265, 13878–13887 [PubMed] [Google Scholar]
- 39. Astatke M., Grindley N. D., Joyce C. M. (1998) J. Mol. Biol. 278, 147–165 [DOI] [PubMed] [Google Scholar]
- 40. Yang G., Franklin M., Li J., Lin T. C., Konigsberg W. (2002) Biochemistry 41, 10256–10261 [DOI] [PubMed] [Google Scholar]
- 41. Strahler J. R., Zhu X. X., Hora N., Wang Y. K., Andrews P. C., Roseman N. A., Neel J. V., Turka L., Hanash S. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4991–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindahl T. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krokan H. E., Drabløs F., Slupphaug G. (2002) Oncogene 21, 8935–8948 [DOI] [PubMed] [Google Scholar]
- 44. el-Hajj H. H., Zhang H., Weiss B. (1988) J. Bacteriol. 170, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McIntosh E. M., Ager D. D., Gadsden M. H., Haynes R. H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8020–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tóth J., Varga B., Kovács M., Málnási-Csizmadia A., Vértessy B. G. (2007) J. Biol. Chem. 282, 33572–33582 [DOI] [PubMed] [Google Scholar]
- 47. Elder J. H., Lerner D. L., Hasselkus-Light C. S., Fontenot D. J., Hunter E., Luciw P. A., Montelaro R. C., Phillips T. R. (1992) J. Virol. 66, 1791–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McGeoch D. J. (1990) Nucleic Acids Res. 18, 4105–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Preston V. G., Fisher F. B. (1984) Virology 138, 58–68 [DOI] [PubMed] [Google Scholar]
- 50. Abergel C., Robertson D. L., Claverie J. M. (1999) J. Virol. 73, 751–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Priet S., Navarro J. M., Gros N., Quérat G., Sire J. (2003) Virology 307, 283–289 [DOI] [PubMed] [Google Scholar]
- 53. Björklund S., Skog S., Tribukait B., Thelander L. (1990) Biochemistry 29, 5452–5458 [DOI] [PubMed] [Google Scholar]
- 54. Kaiser S. M., Emerman M. (2006) J. Virol. 80, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.