Abstract
Cerebral cavernous malformations (CCMs) are alterations in brain capillary architecture that can result in neurological deficits, seizures, or stroke. We recently demonstrated that CCM3, a protein mutated in familial CCMs, resides predominantly within the STRIPAK complex (striatin interacting phosphatase and kinase). Along with CCM3, STRIPAK contains the Ser/Thr phosphatase PP2A. The PP2A holoenzyme consists of a core catalytic subunit along with variable scaffolding and regulatory subunits. Within STRIPAK, striatin family members act as PP2A regulatory subunits. STRIPAK also contains all three members of a subfamily of Sterile 20 kinases called the GCKIII proteins (MST4, STK24, and STK25). Here, we report that striatins and CCM3 bridge the phosphatase and kinase components of STRIPAK and map the interacting regions on each protein. We show that striatins and CCM3 regulate the Golgi localization of MST4 in an opposite manner. Consistent with a previously described function for MST4 and CCM3 in Golgi positioning, depletion of CCM3 or striatins affects Golgi polarization, also in an opposite manner. We propose that STRIPAK regulates the balance between MST4 localization at the Golgi and in the cytosol to control Golgi positioning.
Keywords: Golgi, Mass Spectrometry (MS), PP2A, Protein-Protein Interactions, Serine Threonine Protein Kinase, Serine Threonine Protein Phosphatase, Signal Transduction, Cerebral Cavernous Malformations, MST4, Striatin
Introduction
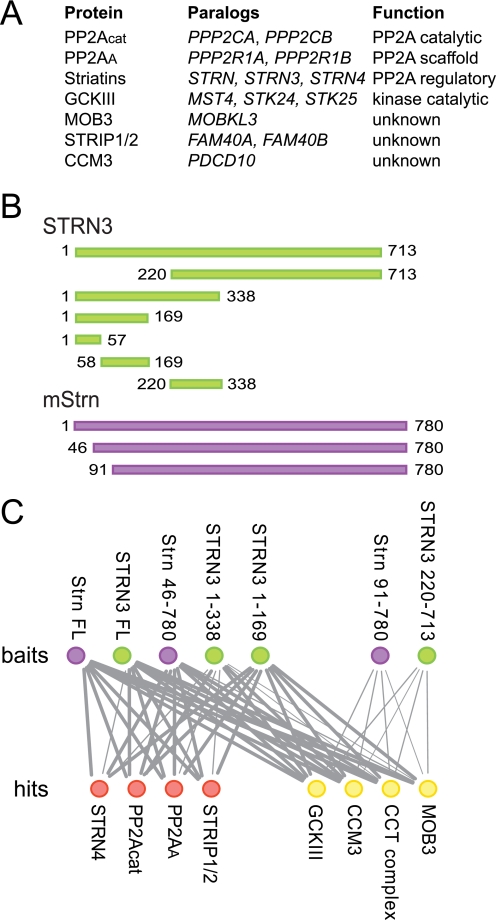
PP2A4 is an essential serine threonine phosphatase involved in many aspects of cell function (1, 2). PP2A acquires substrate and subcellular localization specificity via association with various scaffolding and regulatory subunits to form a number of different holoenzymes, most of which are trimers. In previous studies using affinity purification coupled to mass spectrometry, a portion of PP2A was also found in a higher order complex that we termed STRIPAK (striatin interacting phosphatase and kinase) (3, 4). In addition to the catalytic subunit PP2Acat, its scaffolding subunit PP2AA and members of the striatin family of regulatory subunits (5), the core STRIPAK complex contains the striatin interactor Mob3 (6), the uncharacterized protein STRIP1, members of the germinal center kinase III (GCKIII) group (STK24, STK25, and MST4; Ref. 7), and the small molecular weight protein CCM3 (Fig. 1A). Additional proteins can associate with this core STRIPAK complex in a mutually exclusive manner (4).
FIGURE 1.
Striatin is a scaffolding subunit within STRIPAK. A, composition of core STRIPAK. Proteins, paralogous genes (human nomenclature), and function of proteins are listed. B, schematic of the constructs used in this study. Human STRN3 constructs are in green, mouse Strn constructs are in purple. The first and last amino acid in each construct are indicated. C, summary of AP-MS results for the association of core STRIPAK components with the STRN3 (green) and Strn (purple) deletion mutants (top row). Identified proteins (hits; bottom row) in red do not interact with Strn(91–780) or STRN3(220–713), whereas hits in yellow do. The thickness of each line is proportional to the number of spectral counts (total number of peptides) recovered for each of the proteins in the analysis of the striatin mutants, relative to the spectral counts for the same protein in the AP-MS of full-length Strn. Note that each node (and its associated edges) represents paralogous families, as defined in A. The complete mass spectrometry data used to make this figure are presented in supplemental Table 1.
CCM3 is encoded by one of the three genes mutated in familial cerebral cavernous malformations (CCMs; Ref. 8) and was identified previously as an interactor for the GCKIII proteins (9, 10). CCMs are vascular lesions of the brain characterized by enlarged capillaries that lack structural integrity and that form caverns that tend to bleed, leading to symptoms ranging from headaches and dizziness to severe strokes and death (reviewed in Ref. 11). Recent studies have implicated defective Rho signaling as one of the consequences of depletion (or overexpression) of the CCM1, CCM2, and CCM3 proteins (12–14). Further links between CCM3 and its kinase partners and cytoskeletal dynamics via the Golgi were also uncovered. The Ser/Thr kinases STK25 and MST4 were found to localize to the Golgi apparatus via an association with the Golgi resident protein GM130 (15). Mislocalization of these kinases results in defects in Golgi positioning and cell migration (15). Recently, CCM3 was shown to participate in this effect by stabilizing the GCKIII proteins to promote Golgi orientation and assembly and proper cell orientation (16).
Here, we define the structural organization of the STRIPAK complex, identifying direct interactions and interacting regions within the complex. Specifically, we demonstrate that the striatins and CCM3 act as adapter molecules to bridge the kinase and phosphatase catalytic activities (an accompanying publication by Ceccarelli et al. characterizes interactions between the GCKIII proteins and CCM3; 49). We also report the surprising finding that CCM3 and striatins exhibit opposing functions on the targeting of MST4 to the Golgi and Golgi positioning.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA5-FRT-FLAG was engineered to inducibly express fusion proteins with a single N-terminal FLAG epitope and was constructed from the parent vector pcDNA5-FRT-TO (Invitrogen) and the vector pcDNA3-FLAG (17) as follows. A HindIII/XhoI cassette from pcDNA3-FLAG (containing the FLAG and the multiple cloning site) was subcloned into the pcDNA5-FRT-TO vector also digested with HindIII/XhoI. An internal EcoRI site was subsequently destroyed by mutagenesis. pcDNA5-FRT-eGFP was constructed by subcloning the HindIII/AscI cassette from pcDNA3-eGFP into pcDNA5-FRT-TO. The complete sequences of the cloning vectors are available at the Gingras Laboratory website (Samuel Lunenfeld Research Institute). FLAG-tagged mammalian expression constructs for full-length STRIPAK proteins are described in Ref. 4. Truncations of STRN3 (amino acids 1–169, 1–338, and 220–338) and mouse Strn (amino acids 46–781 and 91–781) were cloned into pcDNA3-FLAG for mammalian expression (supplemental Fig. 1). All point mutations were generated by overlap extension PCR; CCM3 point mutants were subcloned into the pcDNA5-FRT-GFP vector. N-mut is L44D,A47D,I66D,L67D; C-mut (4A) is K132A,K139A,K172A,K179A, and the N-mut/C-mut construct contains both sets of mutations. Inserts were fully sequenced. The full-length and several truncations in human STRN3 (amino acids 1–57, 1–169, 220–713, 58–713, 58–169, and 220–338; supplemental Fig. 1) as well as full-length mouse Strn, mouse Mst4 D162A, and MOB3 were cloned into the GST-tagged expression vector pGEX-2T-TEV HTa for bacterial expression and purification. pGEX-2T-TEV HTa (which expresses a tobacco etch virus-cleavable GST protein was described previously (18). Wild-type CCM3, CCM3 C-mut (4A) (generated by overlap extension PCR), and PP2AA were inserted into the His-tagged expression vector pProEx-HTa for bacterial expression and purification. The coding sequence of GOLGA2 (encoding protein GM130) was amplified by PCR from the cDNA clone from the mammalian gene collection BC069268. The full-length and minimal kinase interaction region at amino acids 72–271 (15) were cloned into pcDNA5-FRT-FLAG.
Antibodies
Commercial antibodies were as follows (catalogue numbers are in parentheses): anti-PP2Acat (610555), anti-striatin (610838), anti-GM130 (610822), and anti-MST4 (612684) were from BD Transduction Laboratories; anti-PP2AA was from Upstate Biotechnology (07-250); anti-STRN3 was from Cell Signaling Technology (S68); anti-giantin was as described (19); anti-FLAG was from Sigma (F3165); and anti-HA was from Covance Research Products (MMS-101R). Anti-CCM3 antibody was raised in rabbit (Covance) using GST-CCM3 (amino acids 2–212) as an antigen. This antibody was tested for specificity in immunoblots against the GST-CCM3 antigen, FLAG-CCM3 transiently transfected in HEK293T cells and endogenous CCM3 (after silencing by RNAi) (data not shown). Secondary antibodies for immunoblotting were sheep anti-mouse IgG and donkey anti-rabbit IgG, both conjugated to horseradish peroxidase from GE Healthcare (NA931 and NA934). Alexa Fluor-labeled secondary antibodies for immunofluorescence were from Invitrogen Molecular Probes: goat anti-mouse 594 (A11005), goat anti-rabbit 594 (A11012), goat anti-mouse 488 (A11001), and goat anti-rabbit 488 (A11034).
Recombinant Protein Purification, Gel Filtration, and in Vitro Binding Assays
His- or GST-tagged fusion proteins were purified as described (20), using lysis buffer with 20 mm Hepes, pH 7.5, 500 mm NaCl, and 5 mm β-mercaptoethanol at 4 °C. Gel filtration was performed to purify proteins and complexes based on size, in buffer containing 100 or 150 mm NaCl. For the PP2AA:STRN3(58–169) complex, a 1:2 ratio of proteins (purified by gel filtration) was mixed before loading onto a Superdex 200 column. For the PP2AA:STRN3(1–338):CCM3 complex, a 1:2:2 ratio of proteins was mixed before loading onto a Superdex 200 column. Fractions encompassing the elution of these protein complexes (as detected by UV) were run on a SDS-PAGE gel and Coomassie stained. In vitro binding assays (GST pulldowns) were performed essentially as described (18), with the following modifications: GST-tagged proteins were purified as described above, without cleavage of proteins from GST resin. Untagged or His-tagged proteins were incubated with GST-tagged proteins on resin in 150 μl of binding buffer (20 mm Hepes, pH 7.5, 100 mm NaCl, and 5 mm β-mercaptoethanol). Glutathione resin was washed rapidly three times in 500 μl of binding buffer, and elution was performed by boiling in Laemmli sample buffer.
Peptide Overlay Assay
Peptide libraries were produced by automatic SPOT synthesis and probed as described previously (21). They were synthesized on continuous cellulose membrane supports on Whatman 50 cellulose membranes using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry with the AutoSpot-Robot ASS 222 (Intavis Bioanalytical Instruments AG, Köln, Germany). The interaction of spotted peptides with purified, recombinant GST and GST-CCM3 fusion proteins was determined by overlaying the cellulose membranes with 10 μg/ml recombinant protein. Bound recombinant proteins were then detected following wash steps with rabbit anti-GST, and detection was performed with a secondary anti-rabbit horseradish peroxidase-coupled antibody.
Fluorescence Polarization Peptide Binding Assay
A 25-mer peptide (286EGLAADLTDDPDTEEALKEFDFLVT310) from human STRN3 was synthesized by Biomatik Corporation (Wilmington, DE) and used for fluorescence polarization binding studies with purified CCM3 proteins as described (22). Equilibrium binding constant determination was carried out on a Beacon fluorescence polarization system (Pan Vera, WI) and data were analyzed using the GraphPad Prism software (GraphPad Software, Inc.)
Mammalian Cell Culture, Immunoprecipitation, and Mass Spectrometry
Transient transfection and immunoprecipitation followed by immunoblotting and/or mass spectrometry were performed essentially as described (4), using either pools of stable cells (FLAG-Strn, FLAG-STRN3, FLAG-Mst4, and their deletion constructs) or a stable inducible clone (FLAG-GM130). Samples were analyzed on a ThermoFinnigan LTQ or an AB-SCIEX 5600 TripleTOF instrument, as described below.
esiRNA and siRNA-mediated Knockdown
All siRNAs were purchased from Dharmacon: CCM3 (L-004436-00), STRN (D-019572-02, -04, -19, -20), STRN3 (D-019145-01, -03, 04, -17) and STRN4 (D-020389-01, -02, -03, -04), as well as the siGENOME non-targeting RNA (D-001210-0X). For endoribonuclease-prepared siRNA (esiRNA), primers were as follows: CCM3 (TCACTATAGGGAGAGTCTTCGTATGGCAGCTGATG; TCACTATAGGGAGACTAGTCGGTTGGCACTTACGA); STRN (TCACTATAGGGAGAGCTTTCGATCAGCATCACTGC; TCACTATAGGGAGACTTCTCTGTGCTCCTTCAGCA); STRN3 (TCACTATAGGGAGAGAAGTCATCCCACACTTCCTGTT; TCACTATAGGGAGACCTTTCTGATGGCAGTGATGC); and STRN4 (TCACTATAGGGAGAGCAGATCTCACCGTCACCAAC; TCACTATAGGGAGACTAGGGATCCATGCTGAGGTC), as well as esiRNA directed against luciferase as described (23).
esiRNAs were prepared as described (23) and diluted to 100 ng/μl; siRNAs (Dharmacon) were diluted to 20 μm. HeLa or HEK293 cells were transfected with esiRNAs using RNAiMAX (Invitrogen; 100 ng per 1 well in a 24-well dish). For striatins, esiRNAs against all three paralogs were pooled and used at 70 ng each. HeLa cells were transfected with siRNAs using RNAiMAX (Invitrogen; 20 pmol per 1 well in a 24-well dish). (siRNAs for all striatin paralogs were pooled, and 40 pmol total was used.) RNA silencing experiments were performed for 72 h before harvesting or imaging cells. Knockdown was assessed using immunoblotting and/or RT-PCR.
RT-PCR Procedure and Primers
RT-PCR was performed as follows: RNA was purified from cells using the RNeasy kit (Qiagen 74104). Cells were lysed in 600 μl using a 20-gauge needle for homogenization. The final product was eluted in 30 μl of water. 200 ng of RNA was run on an agarose gel to check quality. Reverse transcription of RNA into cDNA was performed following the instructions from the Invitrogen SuperScript III reverse transcriptase guide (Invitrogen 18080-093). cDNA was amplified by PCR and analyzed on an agarose gel. Primers were GGATGACAATGGAAGAGATGAAG and GACAGATTTACTCGTTCTAGCTC for PDCD10 (encoding CCM3) and TGAATGACACGAGACTTTACC and TGAAGAGGGAAGGTGGAAC for TIPRL (encoding an unrelated protein used as a loading control).
Immunofluorescence
Immunofluorescence was performed on HeLa cells as described previously (24) with the following modifications: cells were permeabilized with 0.1% Triton X-100, incubated with primary antibodies for 2 h at room temperature, and mounted with ProLong Gold (Invitrogen, P36930). Images were acquired on a DeltaVision at 60× magnification (with a 2× digital zoom for Fig. 5A, essentially as described (25)). Fig. 5E and supplemental Figs. 5, 6 (C and D), and 7 were acquired on an Olympus epifluorescence microscope at 40× magnification. Fig. 6A was acquired on an Olympus epifluorescence microscope at 20× magnification.
FIGURE 5.
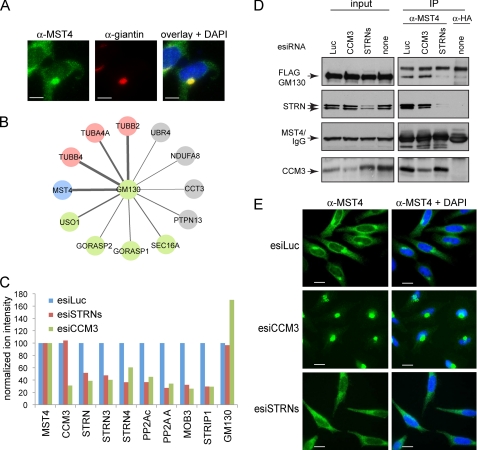
CCM3 and striatins exert opposite effects on MST4 localization. A, co-localization of endogenous MST4 (green) with the Golgi protein giantin (red). In the overlay (right), co-localization to the Golgi is shown in yellow; note that a fraction of MST4 does not localize to the Golgi but is instead detected as green punctae in the cytosol. Scale bar, 7.5 μm. B, GM130 interactors identified by mass spectrometry. AP-MS was performed as described under “Experimental Procedures.” Statistical analysis of the interactions using SAINT was performed; see supplemental Tables 3 and 4 for complete mass spectrometric data. The thickness of the edges is proportional to spectral counts (total number of peptides) for the prey, whereas the color indicates MST4 (blue), known Golgi proteins (green), tubulins (pink), or proteins other than MST4, Golgi proteins, or tubulins (gray). Note that MST4 is a major interaction partner for GM130. C, depletion of CCM3 decreases association of MST4 with STRIPAK but not the interaction with GM130. Stable HEK293 cells expressing FLAG-Mst4 were transfected with the indicated esiRNAs (see Fig. 4C for details). FLAG-Mst4 was immunoprecipitated using anti-FLAG antibodies, and the sample was processed for quantitative mass spectrometry. Relative quantification by mass spectrometry was performed using a TripleTOF 5600 with cells depleted of STRN proteins or CCM3; normalization to Mst4 (bait) levels and to the expression levels in the luciferase samples is shown. See supplemental Table 5 for mass spectrometric results. As shown in Fig. 4C, depletion of CCM3 affects recovery of all STRIPAK components with Mst4; depletion of the striatins affects recovery of all STRIPAK components with the exception of CCM3. By contrast, depletion of CCM3 appeared to increase the GM130 interaction with FLAG-Mst4, indicating that this interaction is not mediated via STRIPAK. D, GM130 interaction with endogenous MST4 is reduced by depletion of striatins in HEK293 cells stably expressing FLAG-GM130. Transfection of esiRNAs was followed by immunoprecipitation of endogenous MST4 and immunoblotting for FLAG-GM130 and STRIPAK proteins. To control for the amount of FLAG-GM130 non-specifically binding to the beads, we performed immunoprecipitation in parallel with an isotype-matched antibody (anti-HA). (There is no HA protein transfected in these cells.) E, esiRNA-mediated depletion of CCM3 in HeLa cells induces near complete localization of MST4 to the Golgi, whereas depletion of striatins prevents Golgi localization. Transfection of indicated esiRNAs was followed by immunofluorescence staining of MST4 and DAPI. Scale bar, 10 μm.
FIGURE 6.
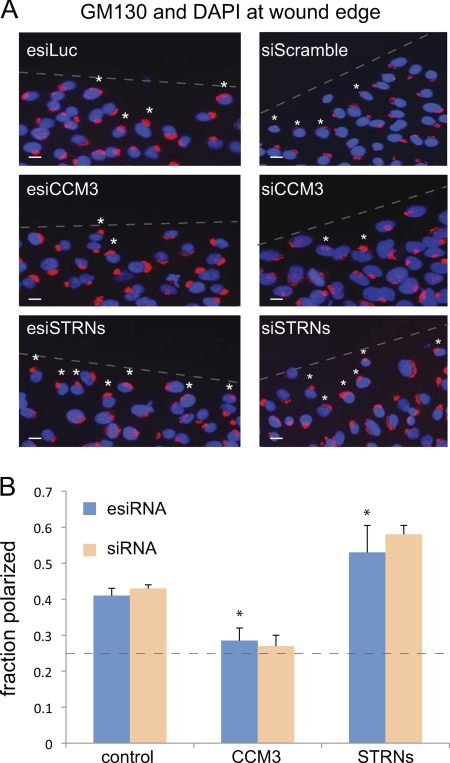
CCM3 and striatins exert opposite effects on Golgi polarization. A, GM130 and DAPI staining at the wound edge in cells depleted of indicated proteins by esiRNA (left) and chemical siRNA (right). esiLuc and siScramble are negative controls. The position of the wound is indicated by a dashed line. A 90° quadrant scoring of the Golgi positioning relative to the wound was performed (see “Experimental Procedures”). Small white asterisks indicate Golgi that are polarized toward the wound within the first cell layer. Scale bar, 10 μm. B, quantification of Golgi polarization from A indicated that ∼40% of control cells had Golgi polarized toward the wound. In cells depleted of CCM3, this value was decreased to ∼25–30%, whereas in cells depleted of striatins, this value increased to ∼54–60% (esiRNA, n = 4, p < 0.05; siRNA, n = 2). The dashed line indicates the number of cells expected to randomly orient their Golgi toward the wound in our scoring system (25%).
Wound Healing and Quantification of Golgi Polarization
Assays were performed as described previously (16) with the following modifications: Cells were plated to confluency on fibronectin-coated coverslips. After 6–8 h, cells were serum-starved in DMEM with 0.1% FBS overnight, and the monolayer was wounded. Cells were incubated in DMEM with 10% FBS for 90 min and fixed with ice-cold 4% paraformaldehyde. Golgi staining was performed using anti-GM130 or anti-giantin, and the first row of cells was counted. A minimum of 150 cells were counted per treatment per experiment; experiments were performed in quadruplicates. Treatments were labeled in code, and polarization was assessed independently by two people. The Golgi of cells on the wound edge were counted as polarized when the majority of the stained Golgi was located within a 90° angle facing the wound (26).
Mass Spectrometric Analysis
Acquired RAW files were converted to mgf format, which were searched with the Mascot search engine (Matrix Sciences, London, UK) against the human RefSeq database (release 37) with a precursor ion mass tolerance of 3.0 and a fragment ion mass tolerance of 0.6. Methionine oxidation and asparagine deamidation were allowed as variable modifications, and trypsin specificity (with one missed cleavage allowed) was selected. The data were analyzed in the “Analyst” module of ProHits (27) and exported into Excel files for spectral normalization and manual curation. For the STRIPAK pulldowns, only interaction partners previously reported and confirmed (4) are reported (supplemental Table 1). For GM130 pulldowns, database searches were performed as above, and the results were analyzed using SAINT (version 2.0) (28, 29), using eight negative control runs as part of the modeling. Hits detected with SAINT AvgP ≥ 0.7 and with a minimum of 10 spectra in at least one of the replicates are reported (supplemental Table 3; detailed mass spectrometry data are presented in supplemental Table 4); interactions with the wild-type protein were deposited to the BioGRID database.
For quantitative analysis (Fig. 5C), immunoprecipitation of FLAG-Mst4 was performed after depletion of CCM3 or all striatins by esiRNA from HEK293 cells stably expressing FLAG-Mst4. Data were acquired on an ABSCIEX 5600 TripleTOF instrument using an Eksigent Ultra nanoLC with NanoFlex cHiPLC columns. The samples were loaded onto a C18 Trap chip at 500 nl/min and separated over a C18 column chip at 250 nl/min (120 min gradient). Data acquisition was done with 1 high resolution MS scan followed by 20 high resolution MS/MS scans. Resulting data were searched using ProteinPilot (version 4.0) against human proteins in Uniprot (release 8.8) (spectral counts are presented in supplemental Table 5). PeakView was used to extract peak areas for all peptides identified for target proteins (STRIPAK core components and GM130). The total number of peptides used for the quantification of each protein is shown in supplemental Table 5 (each of them was manually inspected). Total sum areas for proteins were determined and exported to Markerview where values were normalized to Mst4 across all samples. Further normalization for each protein in the esiCCM3 or esiSTRNs sample to the esiLuc control was performed.
Structure Modeling
The crystal structure of human CCM3 (Protein Data Bank 3L8I (30)) was superimposed on the focal adhesion targeting (FAT) domain of focal adhesion kinase in complex with a peptide derived from paxillin (PDB 1OW7 (31)). Residues of CCM3 FAT domain (Lys-132, Lys-139, Lys-172, and Lys-179) analogous to focal adhesion kinase residues lining the paxillin binding groove were mutated to alanines for the purpose of interaction studies (see Fig. 3).
FIGURE 3.
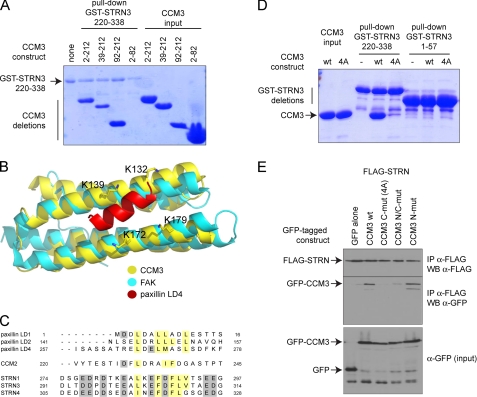
CCM3 interacts with STRN3 via its FAT domain. A. STRN3 interacts with the C-terminal portion of CCM3. A GST pulldown assay was performed with GST-STRN3 220–338 and bacterially expressed and purified CCM3 deletions to map the region on CCM3 responsible for binding to GST-STRN3. Deletion of the first 92 amino acids of CCM3 did not affect the interaction, and a region encompassing amino acids 2–82 was unable to associate to GST-STRN3(220–338). B, structural modeling of the CCM3 FAT domain with a peptide derived from paxillin. The CCM3 crystal structure (30) revealed that the region that we have mapped as interacting with STRN3 also folds as a focal adhesion targeting domain (yellow) similar to focal adhesion kinase (FAK, cyan). Interaction with peptides derived from paxillin (shown in red) are mediated via four lysine residues (highlighted). C, alignment of the peptide derived from STRN3 (and corresponding peptides in the STRN and STRN4 paralogs) with the CCM3 binding regions of CCM2 and paxillin suggests a common mode of association. D, mutation to alanines of the four conserved lysines (Lys-132, Lys-139, Lys-172, and Lys-179) in CCM3 C-mut (4A) abrogates interaction with GST-STRN3(220–338) in vitro. GST pulldown assays were performed with GST-STRN3(220–338) to monitor binding of wild-type CCM3 or CCM3 C-mut (4A). Only wild-type CCM3 is pulled down by GST-STRN3. GST-STRN3(1–57) was used as a negative control. E, mutation to alanines of the four conserved lysines in CCM3 abrogates the interaction with full-length Strn in HEK293T cells. Co-transfection of FLAG-tagged full-length Strn with GFP-tagged CCM3 constructs WT, C-mut (4A), N-mut, and N/C-mut was performed. Immunoprecipitation of FLAG-Strn was followed by immunoblotting with anti-GFP to detect CCM3 association. CCM3 C-mut, 4A is unable to interact with FLAG-Strn, whereas CCM3 N-mut has no effect on the interaction. A combination of both mutations also prevented the interaction, as expected.
RESULTS
Striatins Act as Molecular Scaffolds within STRIPAK
The STRIPAK complex contains members of the evolutionarily conserved striatin family of PP2A regulatory subunits. To investigate the binding topology of the STRIPAK complex with respect to striatin, full-length and truncation mutants of striatin molecules (Fig. 1B) were stably expressed in HEK293 cells and subjected to affinity purification coupled to mass spectrometry (as described in Ref. 4). As expected (Fig. 1C; supplemental Table 1), full-length striatins recovered all core STRIPAK components, as well as members of the Chaperone containing TCP complex. Although deletion of the first 45 amino acids of Strn did not affect any of the interactions, truncation of the amino-terminal 90 amino acids completely abolished the interactions with most STRIPAK components, including PP2Acat and PP2AA (shown as red circles in Fig. 1C). (Note that only “core” STRIPAK components as defined in Fig. 1A are shown on this figure; alternative STRIPAK components, including SLMAP, SIKE, and CTTNBP2NL are also unable to associate with this truncated striatin molecule, see supplemental Table 1). Importantly, however, interactions with components of the Chaperone containing TCP complex, Mob3, CCM3, and the GCKIII proteins were not abrogated by this truncation. Further mapping with STRN3 deletion mutants extended these observations (Fig. 1C), indicating that the N-terminal portion of the molecule is essential for mediating interactions with PP2A and various STRIPAK components, whereas the C-terminal region (amino acids 220–713 in STRN3) is sufficient for interactions with Mob3, CCM3, GCKIII, and the Chaperone containing TCP complex. Interestingly, STRN3 fragments encompassing amino acids 1–169 and 1–338 interacted with all of the STRIPAK proteins (Fig. 1C). We attributed these observations to the fact that a coiled-coil element (amino acids 85–130; Ref. 32) is located within this region and likely mediates homo- and hetero-oligomerization (33) of these deletants with endogenous full-length striatin molecules.
STRN3 Binds Directly to PP2AA and CCM3
Yeast two-hybrid interactions between PP2AA and striatins were detected previously in a high-throughput experiment, suggesting a direct association between these molecules (AfCS yeast two-hybrid screen). To demonstrate that PP2AA and striatin did interact directly in the absence of bridging proteins, an in vitro binding assay was performed using bacterially expressed and purified proteins. Soluble His-PP2AA was incubated with immobilized GST-STRN3 in a pulldown experiment followed by an SDS-PAGE gel and Coomassie staining. His-PP2AA was efficiently captured by full-length GST-STRN3 (Fig. 2A, top panel, compare lanes 1 and 2). To delimit the STRN3 region responsible for interacting with PP2AA, a series of STRN3 truncation mutants was assayed. This analysis revealed that amino acids 1–57 and 169–713 were dispensable for PP2AA binding activity, whereas further truncations inside this region prevented interaction (Fig. 2A). To test whether residues 58–169 were sufficient to mediate the interaction, GST-STRN3(58–169) and a negative control, GST-STRN3(1–57), were incubated with His-PP2AA. Only the GST-STRN3(58–169) protein efficiently pulled down PP2AA (Fig. 2B, left panel).
FIGURE 2.
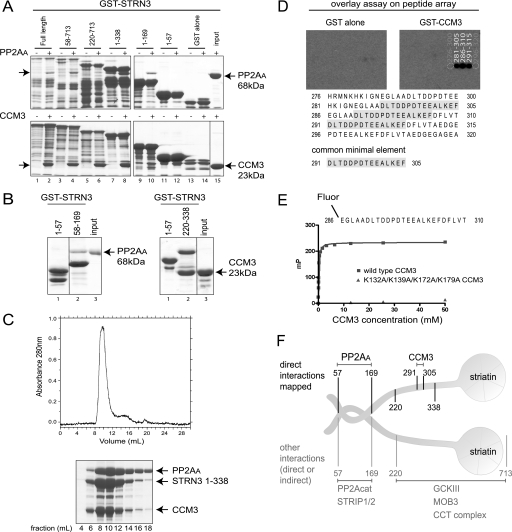
Striatin binds directly to PP2AA and CCM3. A, mapping of the direct in vitro association between GST-STRN3 truncation mutants and PP2AA (top) or CCM3 (bottom). Bacterially expressed and purified GST-STRN3 deletion proteins were used for GST pulldown assays with soluble PP2AA or CCM3; GST alone was used as a negative control. Proteins were visualized by SDS-PAGE and Coomassie staining. The soluble proteins were added to even numbered lanes only. The position of the soluble proteins are indicated by arrows. B, amino acids 58–169 of GST-STRN3 are sufficient to mediate an interaction with PP2AA (left) and amino acids 220–338 are sufficient to mediate the interaction with CCM3 (right) in a GST pulldown assay. GST-STRN3(1–57) was used as a negative control. C, PP2AA, STRN3(1–338), and CCM3 form a complex that is stable throughout the course of gel filtration. Bacterially expressed recombinant proteins were purified and loaded onto a Superdex 200 gel filtration column. The proteins elute as one major peak, ∼8–12 ml, as detected by A280 nm (top) and SDS-PAGE followed by Coomassie staining (bottom). D, peptide array identifies the core STRN3 residues (amino acids 291–305) responsible for binding to GST-CCM3 in an overlay assay. 25-mer peptides derived from STRN3(220–338) were spotted on a membrane (see supplemental Fig. 3 for Coomassie staining of the membrane) and subjected to an overlay assay with GST-CCM3 (or GST alone) followed by detection with anti-GST and horseradish peroxidase-coupled secondary antibodies. The sequence of the common minimal element from the peptides that display association is highlighted. E, fluorescence polarization indicates that a fluorescent 25-mer STRN3 peptide (amino acids 286–310) interacts with wild-type CCM3. The minimal sequence of STRN3 determined in D, plus five flanking residues on either side, was synthesized as a fluorescent peptide and used in a fluorescence polarization assay. This peptide readily interacts with wild-type CCM3 (blue curve). However, substitution of Lys-132, Lys-139, Lys-172, and Lys-179 in CCM3 for alanines completely abrogated association with the STRN3 peptide (see Fig. 3 for description of this mutant). F, summary of the binding surfaces mapped on striatin (results from Figs. 1 and 2).
Our mass spectrometry results indicated that Mob3, CCM3, and the GCKIII proteins interact with a portion of the C terminus of both Strn and STRN3 (from amino acids 220–713 in STRN3). GST pulldown assays were conducted to uncover possible direct interactions. Full-length GST-STRN3 efficiently pulled down CCM3 in vitro (Fig. 2A, bottom panel). To map the interaction region(s), truncation mutants of STRN3 were tested for CCM3 binding, as above. Deletion of the first 219 amino acids or residues 338–713 of STRN3 did not prevent association of CCM3 (Fig. 2A). We next demonstrated that STRN3(220–338) was sufficient to mediate the interaction with CCM3 (Fig. 2B, right panel). To further examine the interactions between PP2AA, STRN3, and CCM3, the recombinant proteins were mixed and analyzed by gel filtration chromatography followed by Coomassie staining. PP2AA and STRN3(58–169) form a complex that is stable throughout chromatographic fractionation (supplemental Fig. 4). Similarly, a trimeric complex formed of PP2AA, STRN3(1–338), and CCM3(2–212) also eluted as a stable complex from gel filtration experiments (Fig. 2C).
To further refine the location of the CCM3 binding site on STRN3, overlay assays of peptides derived from STRN3(220–338), using full-length GST-CCM3 as a probe, were performed. STRN3 peptides containing amino acids 291–305 were able to interact with CCM3 on a membrane (Fig. 2D). Furthermore, a peptide encompassing amino acids 286–310 was sufficient for interaction in solution (as detected by fluorescence polarization) and had an apparent Kd of 132 ± 0.003 nm when modeled as one site binding (Fig. 2E and supplemental Table 2). Taken together, our mapping studies identified striatin as a scaffolding molecule within the STRIPAK complex and revealed direct interactions with both PP2AA (amino acids 58–169 of STRN3) and CCM3 (amino acids 291–305 of STRN3; Fig. 2F).
CCM3 Associates with Striatin via Its FAT Domain
Deletion mutants of CCM3 were next analyzed for their ability to bind to GST-STRN3(220–338) in vitro. Although deletion of CCM3 amino acids 82–212 precluded interaction with STRN3, a construct expressing only amino acids 92–212 was sufficient to bind to STRN3 (Fig. 3A). This region of CCM3 forms a globular domain consisting of four α-helices exhibiting structural resemblance to the FAT domain, which mediates the interaction between focal adhesion kinase and paxillin. On this basis, an interaction of CCM3 with paxillin was validated previously (30) and shown to require four lysine residues that establish interactions with paxillin (Fig. 3B); the same residues were also implicated in mediating the interaction between CCM3 and CCM2, as CCM2 shares a stretch of homology to paxillin (30). The striatin peptide responsible for association with CCM3 exhibits an amino acid composition similar to the paxillin and CCM2-derived peptide (Fig. 3C), suggesting that the same mode of binding may be employed for striatin-CCM3 interactions. These four surface lysine residues at the interface of the paxillin-CCM3 model (Lys-132, Lys-139, Lys-172, Lys-179) were mutated to alanines (called CCM3 C-mut 4A). As measured by fluorescence polarization, these mutations abrogated the interaction with the STRN3 peptide (Fig. 2E). These mutations also abrogated the interaction with a STRN3(220–338) protein in a GST pulldown assay (Fig. 3D). (WT CCM3 is pulled down but not CCM3 C-mut 4A.) Lastly, we tested the recovery with endogenous striatin of transiently transfected GFP-tagged versions of CCM3 WT, C-mut 4A, N-mut (which should not abrogate the interaction) as well as N/C-mut (a combination of both the N and C mutations). Only those constructs with the C-mut 4A lost the interaction (Fig. 3E). Taken together, these data indicate a similar mode of association for CCM3-paxillin, CCM3-CCM2, and CCM3-striatin, suggesting mutually exclusive interactions between these proteins. (The CCM3-striatin interaction is more readily detected by AP-MS in our cells.) Consistent with this, an interaction between CCM2 and striatin was never detected by AP-MS (data not shown).
CCM3 Bridges GCKIII Proteins to STRIPAK via Striatin
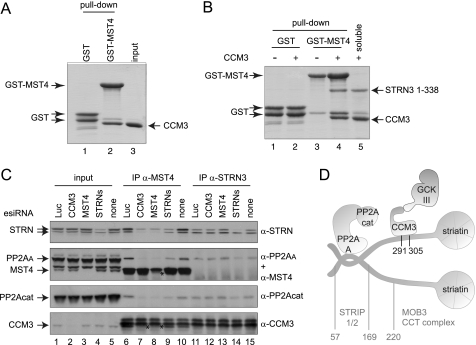
It was reported previously that CCM3 interacts with members of the GCKIII protein family (9, 10), and direct associations between this kinase family and CCM3 were mapped to a CCM3 region (amino acids 2–82) different from that implicated in striatin binding (49). Motivated by the finding that CCM3 binds directly to striatins, we sought to determine whether CCM3 could bridge GCKIII proteins to striatin. A direct interaction between the GCKIII protein Mst4 and CCM3 was first recapitulated in vitro (Fig. 4A). To test whether CCM3 acts as a bridge between the kinases and striatins, pulldown assays using GST-Mst4 and untagged STRN3(1–338) were performed in the presence or absence of untagged CCM3. STRN3 alone did not associate with GST-Mst4 (Fig. 4B, lane 3). However, in the presence of CCM3, GST-Mst4 efficiently pulls down STRN3 (in addition to CCM3; lane 4). CCM3 is therefore able to bridge interactions between the GCKIII protein Mst4 and STRN3.
FIGURE 4.
Striatin and CCM3 bridge the kinase and phosphatase components of STRIPAK. A, direct in vitro interaction between GST-Mst4 and CCM3. GST-Mst4 kinase or GST alone were incubated with soluble CCM3, and a GST pulldown followed by SDS-PAGE and Coomassie staining was performed. B, CCM3 bridges Mst4 to STRN3 in vitro. GST-Mst4 was incubated with soluble STRN3(1–338) in the absence or presence of soluble CCM3. GST pulldown was followed by SDS-PAGE and Coomassie staining. STRN3(1–338) could be precipitated with GST-Mst4 only when incubated with free CCM3. The soluble lane shows amount of CCM3 and/or STRN3(1–338) in the binding assay. C, depletion of CCM3 or striatins by esiRNA prevents association of the PP2A phosphatase and MST4 kinase. HeLa cells were transfected with the indicated esiRNAs. (Note that the esiRNA mixture for striatins targets all three paralogs.) Luc indicates that a non-targeting esiRNA directed against luciferase was employed; none indicates a mock transfection. After cell lysis (total cell lysate input shown on the left), immunoprecipitation of MST4 (center) or STRN3 (right) was performed using antibodies against the endogenous proteins. SDS-PAGE was followed by immunoblotting using antibodies against the indicated endogenous proteins. Arrows indicate the position of each protein; stars indicate the decrease in intensities of the MST4 and CCM3 bands in the immunoprecipitates, as these proteins migrate close to cross-reacting species in the immunoprecipitates. Depletion of striatins, CCM3, or MST4 all prevent recruitment of PP2Acat and PP2AA to MST4, indicating that CCM3 and striatins are responsible for the interaction between the kinase and the phosphatase components of STRIPAK. STRN association with MST4 is prevented by the depletion of MST4 and CCM3, consistent with the bridge model described in B. Depletion of striatins has no effect on the recruitment of CCM3 to MST4. Although depletion of the striatins alters the recovery of PP2Acat and PP2AA to STRN3, as expected, depletion of CCM3 or MST4 has no effect on these interactions. D, model for the structural organization of core STRIPAK.
To determine whether CCM3 and the striatin proteins were responsible for bridging MST4 to PP2A in vivo, endogenous MST4 or STRN3 were immunoprecipitated from cells in which CCM3, MST4, or the three striatin family members were depleted by esiRNA (Fig. 4C). Recovery of PP2A was monitored using antibodies directed against either PP2Acat or PP2AA. Depletion of CCM3 or striatins largely abrogated the interaction between MST4 and both phosphatase subunits (Fig. 4C, lanes 7–9). Note, however, that depletion of CCM3 or MST4 does not affect the interaction between STRN3 and PP2A (lanes 12 and 13).
On the basis of the data presented above, we propose the following architectural model for the STRIPAK complex (Fig. 4D). Striatin functions as a core scaffold within STRIPAK, mediating homo- and hetero-oligomerization, as well as (minimally) direct interactions with PP2AA and CCM3, via two separate regions. Through direct interactions, CCM3 docks onto striatin and recruits the GCKIII proteins to the phosphatase component of STRIPAK.
Localization of MST4 to Golgi and Interaction with GM130 Is Regulated by CCM3 and Striatin in Opposite Manner
The kinase MST4 had been shown previously to localize to the Golgi and had been implicated in Golgi positioning and integrity (15, 16). To begin to understand the functional consequences of the interaction between the kinase and phosphatase components of STRIPAK, the localization of MST4 to the Golgi was monitored in HeLa cells. Consistent with previous studies, we observed strong co-localization between MST4 and the Golgi protein giantin, although localization of MST4 in punctate structures in the cytoplasm (that are not stained with giantin) was also readily apparent (Fig. 5A; supplemental Fig. 4). MST4 was reported to be targeted to the Golgi at least partially due to its interaction with the protein GM130 (15). In agreement with these data, when we conducted an AP-MS analysis on FLAG-Mst4, GM130 was also recovered (data not shown), and the reciprocal AP-MS analysis of FLAG-GM130 recovered MST4 (but not the additional components of STRIPAK) as a major interactor (Fig. 5B; supplemental Tables 3 and 4). These results suggested that MST4 can be found in at least two separate complexes, one with STRIPAK and one with GM130.
To test the effect of CCM3 depletion on the interactions established by MST4, AP-MS was performed in HEK293 cells expressing FLAG-Mst4 after CCM3 knockdown, using a quantitative mass spectrometric approach. CCM3 knockdown resulted in decreased interactions between FLAG-Mst4 and the remaining STRIPAK components (green bars), but not between FLAG-Mst4 and GM130. (In fact, the interaction with GM130 appeared to increase in some experiments (Fig. 5C and supplemental Table 5).) Similarly, depletion of CCM3 in FLAG-GM130 expressing HEK293 cells did not disrupt the interaction between immunoprecipitated endogenous MST4 and FLAG-GM130, when evaluated by immunoblot. This confirms that MST4 is not recruited to GM130 via CCM3.
We next evaluated the effect of CCM3 knockdown on the MST4 localization pattern in HeLa cells. Interestingly, in cells in which CCM3 was silenced, the localization of MST4 is shifted almost completely to the Golgi region (Fig. 5E). Similar results were obtained using independent silencing reagents (supplemental Fig. 7), demonstrating that the observed effects are caused by the depletion of CCM3, and very little of the protein remains localized to the cytosol. These data suggest that CCM3 may favor the cytosolic (and perhaps punctate) localization of MST4 over Golgi localization.
Because CCM3 and striatin bridge MST4 to other STRIPAK components, we expected that silencing striatins would have the same effect on MST4 Golgi localization as silencing CCM3. However, when striatins were depleted, MST4 Golgi localization was strikingly perturbed, leading to a more prominent cytosolic localization (Fig. 5E). This was accompanied by a reduction in the amount of FLAG-GM130 precipitated with endogenous MST4 (Fig. 5D). (Note that this reduction was not significantly detected in the quantitative mass spectrometry experiment with FLAG-Mst4 cells shown in Fig. 5C.) Taken together, these results suggest that CCM3 and striatin exhibit opposing roles on the localization of the kinase MST4 to the Golgi, with striatins favoring a Golgi localization and CCM3 promoting cytosolic location.
CCM3 and Striatin Oppose Each Other in Golgi Positioning
Depletion of MST4, and more recently CCM3, was shown to affect the positioning of the Golgi toward the leading edge of a wound (15, 16). Prompted by the surprising results that CCM3 and striatin knockdowns have opposing effects on MST4 localization (and in some cases, on the interaction with GM130), the effects of depletion of each of these proteins on Golgi orientation was assessed. Golgi orientation was determined by a well established criterion: the Golgi of cells on the wound edge were considered to be oriented toward the leading edge when the majority of the stained Golgi was located within the quadrant facing the wound (26).
In cells transfected with control esiRNA, ∼40% (± 2%, n = 4) of the cells displayed Golgi positioned toward the leading edge 1.5 h after wounding (Fig. 6, A and B). As reported previously, depletion of CCM3, using either esiRNA or siRNA, reduced the percentage of properly positioned Golgi to roughly 25% (the number of cells expected to randomly orient their Golgi toward the wound). By contrast, but consistent with the data presented above, depletion of striatin using esiRNA markedly enhanced Golgi orientation toward the wound, from ∼40% in controls to >54%. Similar results were observed following striatin knockdown with siRNA (n = 2). These data indicate that depletion of CCM3 and striatin not only have opposite effects on MST4 localization but also have opposing effects on Golgi repositioning during wound healing.
DISCUSSION
We have described the molecular organization of the STRIPAK complex and assigned a role to the disease-related CCM3 protein as an adaptor that links the kinase and phosphatase subunits of STRIPAK. We have also described a means by which the functions of striatin and CCM3 oppose each other, through the regulation of MST4 interactions and localization as well as their effect on Golgi positioning after stimulation by wounding the cell monolayer. These data suggest that the interaction between CCM3 and STRIPAK via direct association with striatin may serve as a regulatory mechanism to control the function of the MST4 kinase. Importantly, these results also suggest that Golgi localization of MST4 may be detrimental to polarization. The Golgi apparatus has emerged as a critical hub for intracellular signaling (34), and signaling is essential for Golgi polarization. For example, phosphorylation of the Golgi protein, GORASP1 (also known as GRASP65, a GM130 interaction partner), by the kinase ERK is required for Golgi reorientation (35). Interestingly, ERK activity has been shown to be modulated by CCM3 and MST4 (9); whether or not GORASP1 phosphorylation is modulated in our system remains to be tested.
A large body of evidence suggests important roles for polarized localization of the Golgi (36). Cell migration requires polarized secretion at the leading edge for the regulated transport of vesicles, the delivery of adhesion molecules and cytoskeletal components, as well as the addition of new membranes. The polarized localization of the Golgi has also been intimately linked to the small proteins of the Rho-GTPase family (26, 37). In light of the defects in Rho signaling following modulation of CCM1, CCM2, or CCM3 expression (12–14), it is tempting to postulate that CCM3, MST4, and perhaps STRIPAK may regulate Golgi polarization via regulation of Rho-GTPases. Whether striatins, CCM3, and MST4 play a role in all aspects of Golgi polarization, including cell migration, remains to be answered.
Given that STRIPAK contains both kinase and phosphatase activities, our results suggest the existence of a molecular switch defined by the balance of phosphorylation and dephosphorylation at the Golgi. At this point, the target(s) for the MST4 kinase (or the PP2A phosphatase) in the Golgi polarization process are still unknown. Additionally, whether and how Golgi polarization may contribute to the vascular defects observed in CCM patients remains to be investigated.
New roles for STRIPAK complex components are beginning to emerge, in large part through analysis of STRIPAK paralogs across species. It is noteworthy that a portion of the STRIPAK complex (lacking CCM3 and the GCKIII protein component) has been conserved throughout eukaryotic evolution. Ancestral roles for STRIPAK point to cytoskeletal and membrane dynamics functions. In Saccharomyces cerevisiae, Far8 (striatin), Far11 (STRIP1/2), Vps64/Far10 (orthologous to the alternate STRIPAK component SLMAP), along with Far3 and Far7 (for which no human orthologs are known) form a protein complex implicated in cell cycle arrest following pheromone treatment (38). Orthologs of these ancestral STRIPAK genes are required for proper vegetative membrane fusion in filamentous fungi (39, 40). The function of STRIPAK in mediating membrane fusion appears to have been conserved in mammals, as deregulation of SLMAP prevents myoblast fusion to myotubes (41). More recently, deletion of the orthologs of striatin (FAR8), STRIP1/2 (FAR11), or one of the PP2A catalytic subunits (PPG1) was demonstrated to suppress lethality and actin cytoskeleton disorganization caused by mutations of TORC2 (target of rapamycin complex 2) (42). Interestingly, TORC2 controls actin cytoskeleton assembly across multiple species, in part via regulation of the Rho1 GTPases (43–45). CCM disease, CCM3, and MST4 are intimately linked to Rho signaling in human cells (12–14, 46), suggesting that this function of STRIPAK has been evolutionarily conserved. In addition to these roles in cytoskeleton and membrane dynamics, a surprising recent report implicated the Drosophila STRIPAK complex (including CCM3) in Hippo signaling (47), indicating that STRIPAK may control multiple signaling pathways. The elucidation of the substrates of the kinase and phosphatase components of STRIPAK will be required for a full molecular understanding of STRIPAK function.
Finally, although our data point to the STRIPAK complex as the major interactor for epitope-tagged or endogenous CCM3 protein in HEK293 cells (4), HeLa cells, C2C12 myoblasts, and myotubes and in bovine endothelial aortic cells (data not shown) CCM3 is also capable of interacting with CCM2 (48) and paxillin (30). Because these interactions are apparently mediated via the same surface as the striatin binding site on CCM3, we propose here that they may be mutually exclusive. Further studies on CCM3 function in vascular disease and elsewhere will need to take these alternative protein assemblies into consideration.
Supplementary Material
Acknowledgments
We thank A. Fernandes and members of the Gingras, Pelletier, and Sicheri laboratories for discussion and technical help and J.-P. Lambert for comments on the manuscript.
This work was supported in part by Canadian Institutes of Health Research Grants MOP-36399 (to F. S.) and MOP-84314 (to A.-C. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I–V, Figs. 1–7, and additional references.
- PP2A
- phosphoprotein phosphatase 2A
- CCM
- cerebral cavernous malformation
- FAT
- focal adhesion targeting
- eGFP
- enhanced GFP
- STRIPAK
- striatin-interacting phosphatase and kinase
- AP-MS
- affinity purification coupled to mass spectrometry
- GCKIII
- germinal center kinase III
- esiRNA
- endoribonuclease-prepared siRNA
- N-mut
- L44D,A47D,I66D,L67D
- C-mut(4A)
- K132A,K139A,K172A,K179A.
REFERENCES
- 1. Virshup D. M., Shenolikar S. (2009) Mol. Cell. 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 2. Shi Y. (2009) Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 3. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) Mol. Syst. Biol. 5, 237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) Mol. Cell Proteomics. 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moreno C. S., Park S., Nelson K., Ashby D., Hubalek F., Lane W. S., Pallas D. C. (2000) J. Biol. Chem. 275, 5257–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moreno C. S., Lane W. S., Pallas D. C. (2001) J. Biol. Chem. 276, 24253–24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dan I., Ong S. E., Watanabe N. M., Blagoev B., Nielsen M. M., Kajikawa E., Kristiansen T. Z., Mann M., Pandey A. (2002) J. Biol. Chem. 277, 5929–5939 [DOI] [PubMed] [Google Scholar]
- 8. Guclu B., Ozturk A. K., Pricola K. L., Bilguvar K., Shin D., O'Roak B. J., Gunel M. (2005) Neurosurgery 57, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 9. Ma X., Zhao H., Shan J., Long F., Chen Y., Chen Y., Zhang Y., Han X., Ma D. (2007) Mol. Biol. Cell. 18, 1965–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 11. Riant F., Bergametti F., Ayrignac X., Boulday G., Tournier-Lasserve E. (2010) FEBS. J. 277, 1070–1075 [DOI] [PubMed] [Google Scholar]
- 12. Borikova A. L., Dibble C. F., Sciaky N., Welch C. M., Abell A. N., Bencharit S., Johnson G. L. (2010) J. Biol. Chem. 285, 11760–11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitehead K. J., Chan A. C., Navankasattusas S., Koh W., London N. R., Ling J., Mayo A. H., Drakos S. G., Jones C. A., Zhu W., Marchuk D. A., Davis G. E., Li D. Y. (2009) Nat. Med. 15, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stockton R. A., Shenkar R., Awad I. A., Ginsberg M. H. (2010) J. Exp. Med. 207, 881–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F. A. (2004) J. Cell Biol. 164, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fidalgo M., Fraile M., Pires A., Force T., Pombo C., Zalvide J. (2010) J. Cell Sci. 123, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 17. Gingras A. C., Caballero M., Zarske M., Sanchez A., Hazbun T. R., Fields S., Sonenberg N., Hafen E., Raught B., Aebersold R. (2005) Mol. Cell Proteomics. 4, 1725–1740 [DOI] [PubMed] [Google Scholar]
- 18. Mao D. Y., Neculai D., Downey M., Orlicky S., Haffani Y. Z., Ceccarelli D. F., Ho J. S., Szilard R. K., Zhang W., Ho C. S., Wan L., Fares C., Rumpel S., Kurinov I., Arrowsmith C. H., Durocher D., Sicheri F. (2008) Mol. Cell. 32, 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pelletier L., Jokitalo E., Warren G. (2000) Nat. Cell. Biol. 2, 840–846 [DOI] [PubMed] [Google Scholar]
- 20. Lee K. P., Dey M., Neculai D., Cao C., Dever T. E., Sicheri F. (2008) Cell 132, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X., Baillie G. S., Houslay M. D. (2009) J. Biol. Chem. 284, 16170–16182 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Smith M. J., Hardy W. R., Murphy J. M., Jones N., Pawson T. (2006) Mol. Cell. Biol. 26, 8461–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kittler R., Pelletier L., Heninger A. K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A. L., Habermann B., Hyman A. A., Buchholz F. (2007) Nat. Cell Biol. 9, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 24. Gonon E. M., Skalski M., Kean M., Coppolino M. G. (2005) FEBS. Lett. 579, 6169–6178 [DOI] [PubMed] [Google Scholar]
- 25. Lawo S., Bashkurov M., Mullin M., Ferreria M. G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J. R., Hegemann B., Pinchev D., Buchholz F., Peters J. M., Hyman A. A., Gingras A. C., Pelletier L. (2009) Curr. Biol. 19, 816–826 [DOI] [PubMed] [Google Scholar]
- 26. Etienne-Manneville S., Hall A. (2001) Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]
- 27. Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z. Y., Breitkreutz B. J., Ding Y., Colwill K., Pasculescu A., Pawson T., Wrana J. L., Nesvizhskii A. I., Raught B., Tyers M., Gingras A. C. (2010) Nat. Biotechnol. 28, 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skarra D. V., Goudreault M., Choi H., Mullin M., Nesvizhskii A. I., Gingras A. C., Honkanen R. E. (2011) Proteomics 11, 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., Nesvizhskii A. I. (2011) Nat. Methods. 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Zhang R., Zhang H., He Y., Ji W., Min W., Boggon T. J. (2010) J. Biol. Chem. 285, 24099–24107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoellerer M. K., Noble M. E., Labesse G., Campbell I. D., Werner J. M., Arold S. T. (2003) Structure 11, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 32. Lupas A., Van Dyke M., Stock J. (1991) Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 33. Gaillard S., Bailly Y., Benoist M., Rakitina T., Kessler J. P., Fronzaroli-Molinières L., Dargent B., Castets F. (2006) Traffic 7, 74–84 [DOI] [PubMed] [Google Scholar]
- 34. Farhan H., Rabouille C. (2011) J. Cell Sci. 124, 171–180 [DOI] [PubMed] [Google Scholar]
- 35. Bisel B., Wang Y., Wei J. H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. (2008) J. Cell Biol. 182, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yadav S., Puri S., Linstedt A. D. (2009) Mol. Biol. Cell 20, 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 38. Kemp H. A., Sprague G. F., Jr. (2003) Mol. Cell. Biol. 23, 1750–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simonin A. R., Rasmussen C. G., Yang M., Glass N. L. (2010) Fungal Genet Biol. 47, 855–868 [DOI] [PubMed] [Google Scholar]
- 40. Xiang Q., Rasmussen C., Glass N. L. (2002) Genetics 160, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guzzo R. M., Wigle J., Salih M., Moore E. D., Tuana B. S. (2004) Biochem. J. 381, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., Toufighi K., Youn J. Y., Ou J., San Luis B. J., Bandyopadhyay S., Hibbs M., Hess D., Gingras A. C., Bader G. D., Troyanskaya O. G., Brown G. W., Andrews B., Boone C., Myers C. L. (2010) Nat. Methods 7, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 44. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell. 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 45. Helliwell S. B., Howald I., Barbet N., Hall M. N. (1998) Genetics 148, 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J., Pack M. A., Sessa W. C., Kahn M. L. (2010) J. Clin. Invest. 120, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010) Mol. Cell. 39, 521–534 [DOI] [PubMed] [Google Scholar]
- 48. Hilder T. L., Malone M. H., Bencharit S., Colicelli J., Haystead T. A., Johnson G. L., Wu C. C. (2007) J. Proteome. Res. 6, 4343–4355 [DOI] [PubMed] [Google Scholar]
- 49. Ceccarelli D. F., Laister R. C., Mulligan V. K., Kean M. J., Goudreault M., Scott I., Derry W. B., Chakrabartty A., Gingras A. C., Sicheri F. (2011) J. Biol. Chem. 286, 25056–25064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.