Abstract
Niemann-Pick C1-like 1 (NPC1L1) is a multitransmembrane protein playing a crucial role in dietary and biliary cholesterol absorption. Cholesterol promotes the formation and endocytosis of NPC1L1-flotillin-cholesterol membrane microdomains, which is an early step in cholesterol uptake. How cholesterol is sensed in this step is unknown. Here, we find that the N-terminal domain (NTD) of NPC1L1 binds cholesterol. Mutation of residue Leu-216 in NPC1L1-NTD eliminates cholesterol binding, decreases the formation of NPC1L1-flotillin-cholesterol membrane microdomains, and prevents NPC1L1-mediated cholesterol uptake in culture cells and mice livers. NPC1L1-NTD specifically binds cholesterol but not plant sterols, which may account for the selective cholesterol absorption in intestine. Furthermore, 25- or 27-hydroxycholesterol competes with cholesterol to bind NPC1L1-NTD and inhibits the cholesterol induced endocytosis of NPC1L1. Together, these results demonstrate that plasma membrane-localized NPC1L1 binds exogenous cholesterol via its NTD, and facilitates the formation of NPC1L1-flotillin-cholesterol membrane microdomains that are then internalized into cells through the clathrin-AP2 pathway. Our study uncovers the mechanism of cholesterol sensing by NPC1L1 and proposes a mechanism for selective cholesterol absorption.
Keywords: Cholesterol, Cholesterol Metabolism, Membrane Proteins, Protein Domains, Sterol, Cholesterol Absorbtion, N-terminal Domain, NPC1L1, Oxysterol, Plant Sterol
Introduction
NPC1L1 is a multiple transmembrane protein that is essential for dietary cholesterol absorption and biliary cholesterol reabsorption (1–3). Our previous studies have revealed that NPC1L1 can move between the endocytic recycling compartment (ERC)4 and plasma membrane (PM) (4). Cholesterol depletion induces the export of NPC1L1 from ERC to PM in a myosin Vb-rab11a-rab11-FIP2-dependent manner (5, 6). Reversely, cholesterol replenishment promotes the formation of cholesterol-enriched membrane microdomains containing NPC1L1 and flotillins, named NPC1L1-flotillin-cholesterol membrane microdomains (7). These microdomains are in turn internalized through clathrin-AP2-mediated endocytosis and, therefore, lead to the internalization of NPC1L1 and ensure the efficient cholesterol uptake (7). It's still a mystery how cholesterol is sensed to initiate the formation and endocytosis of NPC1L1-flotillin-cholesterol membrane microdomains.
Niemann-Pick C (NPC) 1 is a homologue of NPC1L1 (8, 9). NPC1 and another protein, NPC2, are required for transporting the LDL-derived cholesterol out of late endosomes/lysosomes (10). Mutation of the npc1 or npc2 genes leads to the Niemann-Pick Type C disease, a fatal hereditary disorder characterized by accumulation of cholesterol in late endosomes/lysosomes (9, 11). NPC1 and NPC1L1 show the similar protein topology. Both of them have 13 transmembrane helices containing a sterol-sensing domain, a large luminal N-terminal domain (NTD), and two large luminal loops (12, 13). Recently, it has been shown that the NTD of NPC1 directly binds cholesterol (14). The crystal structure of the cholesterol-bound NPC1-NTD reveals that the cholesterol molecule is embedded in the binding pocket with an orientation of the tetracyclic ring in the interior, whereas the isoocytl chain exposed the entrance of the pocket (15), which is contrary to that in the binding pocket of NPC2 (16, 17). Together with results from cholesterol transfer studies, these lines of evidence favor a model of cholesterol handing over, whereby NPC1-NTD receives cholesterol from NPC2 and transfers it to the membrane of late endosomes/lysosomes (18, 19).
In the current study, we report that the NPC1L1-NTD binds cholesterol. This cholesterol-binding activity is crucial for NPC1L1-mediated cholesterol uptake, biliary cholesterol reabsorption, and the formation of NPC1L1-flotillin-cholesterol membrane microdomains. Moreover, plant sterols, including β-sitosterol and stigmasterol, cannot bind to NPC1L1-NTD, providing a mechanism for the preferential absorption of cholesterol in intestine over plant sterols. In addition, 25- and 27-hydroxycholesterol compete with cholesterol to bind NPC1L1-NTD and inhibit the endocytosis of NPC1L1 by preventing the formation of cholesterol-enriched membrane microdomains, suggesting a possible role of oxysterols in the regulation of cholesterol absorption.
EXPERIMENTAL PROCEDURES
Materials and Plasmids
We obtained sterols from Steraloids, Inc., [1,2-3H]cholesterol (45 Ci/mmol) was from PerkinElmer Life Sciences, and other reagents were from previously described sources (4, 7, 20).
The plasmid expressing EGFP-tagged full-length human NPC1L1 was described previously (4). The plasmids encoding the mutated version of NPC1L1-EGFP and the plasmid of NPC1L1ΔNTD (Δaa 18–260)-EGFP were generated by site-directed mutagenesis (QuikChange II XL, Stratagene). The NTD (aa 1–280; aa 1–17 comprise the signal peptide sequence) of human NPC1L1 was constructed by standard PCR amplified from the plasmid encoding the full-length NPC1L1-EGFP. Eight histidine residues were linked to the C terminus of NPC1L1-NTD during the PCR. The DNA fragments encoding NPC1L1-NTD-His8 were ligated into the pCMV-3×FLAG-14 vector (Sigma). The plasmid encodes the NPC1L1-NTD followed by eight histidines and three FLAGs at the C terminus. Mutants of the NTD were produced by site-directed mutagenesis from NPC1L1-NTD-His8-FLAG. For generation of the GST-NTD-His6-encoding plasmids, pGEX-4T-3 vector (Amersham Biosciences) was modified by inserting the DNA sequence encoding six histidines. Then the DNA fragments encoding the wild-type human NPC1L1 (20–280 aa) were amplified and inserted between the GST and the six histidines of the modified pGEX-4T-3 vector; so the expressed protein has a GST tag at the N terminus and six histidines at the C terminus. All the constructs were verified by DNA sequencing.
Buffers
Buffer A contained PBS plus 5 mm EDTA, 5 mm EGTA, 0.5% (w/v) digitonin. Buffer B contained 0.2 m glycine-HAc (pH2.9). Buffer C contained 1 m Tris-HCl (pH 9.5). Buffer D contained PBS plus 0.001% (w/v) Nonidet P-40. Buffer E contained PBS plus 0.002% (w/v) Nonidet P-40. Buffer F contained PBS plus 0.001% (w/v) Nonidet P-40, and 250 mm imidazole.
Medium
Medium A contained DMEM containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B contained medium A supplemented with 10% fetal bovine serum (FBS). Medium C contained medium A supplemented with 5% lipoprotein-deficient serum, 10 μm compactin, and 50 μm mevalonate. Cholesterol-depleting medium was medium C supplemented with 1.5% (w/v) methyl-β-cyclodextrin. Cholesterol-replenishing medium contained medium C supplemented with 37.5 μm cholesterol/methyl-β-cyclodextrin. The cholesterol/methyl-β-cyclodextrin inclusion complexes were prepared as described previously (4).
Cell Culture
HEK293T (human embryonic kidney cell), CRL1601 (McArdle RH7777 rat hepatoma cell), CRL1601-NPC1L1-EGFP, which expresses NPC1L1-EGFP, and CRL1601-NPC1L1ΔNTD-EGFP, which expresses the truncated version of NPC1L1-EGFP, were grown in monolayer at 37 °C in 5% CO2. HEK293T and CRL1601 were maintained in medium B, and CRL1601-NPC1L1-EGFP and CRL1601-NPC1L1ΔNTD-EGFP were maintained in medium B supplemented with 200 μg/ml G418. Transfection of cells with FuGENE HD was performed according to the manufacturer's manual.
Animals
All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Shanghai Institutes for Biological Sciences. Animal protocols and adenovirus-mediated expression of EGFP, NPC1L1-EGFP, NPC1L1ΔNTD-EGFP, and NPC1L1(L216A)-EGFP in mice liver as well as measurement of cholesterol, phospholipid, or triglyceride in the bile, liver, or plasma were described previously (7, 21).
[3H]Cholesterol Binding to Recombinant NPC1L1-NTD from E. coli
The proteins of GST-NPC1L1-NTD-His6, GST-NPC1L1-NTD(L216A)-His6, or GST-His6 were produced in Escherichia coli (BL21, Invitrogen) and purified under non-denaturing conditions, using glutathione-Sepharose 4 Fast Flow affinity chromatography (GE Healthcare), as recommended by the manufacturer (22).
For examination of [3H]cholesterol binding to recombinant NPC1L1-NTDs (WT or L216A), a final volume of 100 μl of buffer D containing 4 pmol of purified proteins, 1 μg of bovine serum albumin (BSA) and indicated concentrations of [3H]cholesterol delivered in ethanol were incubated for 24 h at 4 °C. Then the protein was precipitated by Ni-NTA spin column (Qiagen), and washed with 6 ml of Buffer D. The bound [3H]cholesterol was eluted with buffer F, and then quantified by scintillation counting. Binding background was determined by incubating the proteins with the indicated concentrations [3H]cholesterol in the presence of a large excess of unlabeled cholesterol. In each experiment, all tubes received the same amount of ethanol. For determination of 25-hydroxycholesterol binding, similar procedures were carried out.
[3H]Cholesterol Binding to NPC1L1-NTD in Culture Medium from Transfected HEK293T Cells
The procedure of purification of the secreted NPC1L1-NTD variants was similar to previous report (15, 18, 19). Briefly, HEK293T cells were grown in a monolayer at 37 °C in 5% CO2. On day 0, the cells were plated at a density of 6 × 106 cells per 100-mm dish in medium B. On day 2, each dish was transfected with 5 μg of plasmids encoding NPC1L1-NTDs (WT or mutant versions) with the N-terminal signal peptide and His8-FLAG tags at the C terminus. On day 3, each dish was washed with PBS twice, and then switched to 7 ml of medium A containing 1% (v/v) Cellgro ITS (Fisher Scientific) followed by medium collection after 24 h. The procedure was then repeated twice. Then, the media collected from 3 consecutive days were subjected to centrifugation at 2,500 rpm for 5 min at 4 °C. The supernatants were concentrated to 1 ml using a 10-kDa Amicon Ultracel filter module (Millipore). The concentrated media was stored at 4 °C. The amount of secreted NPC1L1-NTDs was estimated by SDS-PAGE and immunoblot analysis with monoclonal anti-FLAG antibody.
A cholesterol binding assay was performed according to previous reports (15, 18, 19). Briefly, 80 μl of Buffer E and 1.6 μg of BSA were mixed with 80 μl of concentrated culture medium from transfected cells and the indicated concentrations of [3H]cholesterol delivered in ethanol. After incubation for 24 h at 4 °C, the bound [3H]cholesterol was quantified the same as that for binding to the GST fusions.
Sucrose Gradient Ultracentrifugation
Sucrose gradient ultracentrifugation was performed as previously described (7). Briefly, the cells were homogenized in 0.5 m Na2CO3 (pH 11.0) and sonicated. The 2-ml post-nuclear fractions were collected and mixed 1:1 (v/v) with 90% sucrose (90% sucrose, 25 mm Mes, pH 6.5, and 0.15 m NaCl) and then layered with 2 ml of 35% sucrose, 3 ml of 22% sucrose, and 3 ml of 5% sucrose in 25 mm Mes, 0.15 m NaCl, and 0.25 m Na2CO3 (pH 11.0). After centrifugation at 200,000 × g for 20 h, 24 fractions containing 0.5 ml per sample were collected from the top. The gradient fractions were subjected to immunoblot analysis and cholesterol assay. A cholesterol assay was carried out using the Amplex Red Cholesterol Assay Kit (Invitrogen).
Immunofluorescence, Filipin Staining, and Fluorescence Quantification
The immunofluorescence or filipin staining were performed as previously described (4). Images were obtained with a Leica TCS SP5 laser confocal scanning microscope. Filipin signals were obtained from the same confocal microscope equipped with a two-photon laser using an excitatory wavelength of 720 nm. Fluorescence quantification was calculated using Image-Pro Plus 5.02, and the detailed procedure was as described previously (4).
RESULTS
The NTD of Human NPC1L1 Binds Cholesterol
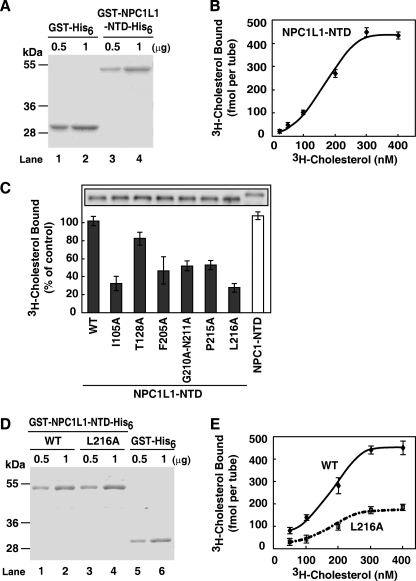
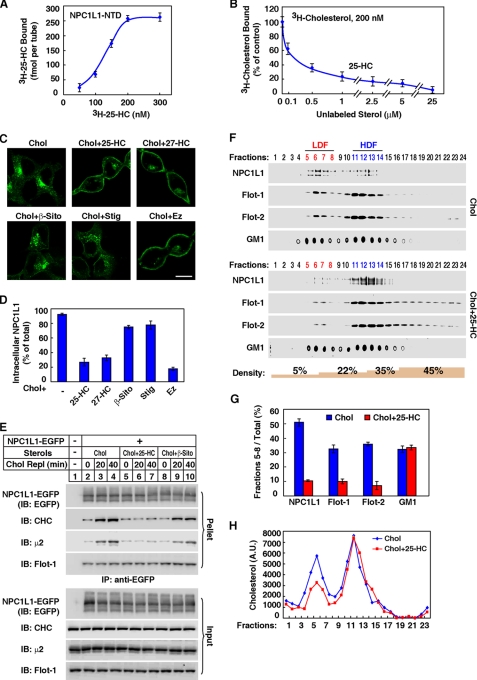
The NTD of NPC1 has been shown to directly bind cholesterol (14, 23). The NPC1L1-NTD (aa 18–287; aa 1–17 comprise the signal peptide sequence) shows 47% homology with NPC1-NTD. To directly assess whether the NPC1L1-NTD binds cholesterol, we prepared recombinant NPC1L1-NTD protein from E. coli (Fig. 1A) and performed in vitro [3H]cholesterol binding assay. The binding of [3H]cholesterol to NPC1L1-NTD was saturable with a Kd of ∼170 nm (Fig. 1B), which is comparable to the Kd of NPC1-NTD-cholesterol binding (14). Interestingly, the binding curve is sigmoidal with a calculated Hill coefficient of 1.8, suggesting a positive cooperative association between NPC1L1-NTD and cholesterol. We further performed cross-linking experiments, and the results showed that NPC1L1-NTD forms a species with the molecular mass of ∼130 kDa, which is 4-fold of the NTD itself (32 kDa) (supplemental Fig. S1). Thus, these data indicate that, in our experimental conditions, the NPC1L1-NTD predominantly forms a tetramer and cholesterol binding to the tetramer shows a positive cooperative effect.
FIGURE 1.
Cholesterol binding to the N-terminal domain of NPC1L1. A, Coomassie blue staining of recombinant GST-NPC1L1-NTD-His6 and GST-His6 proteins purified from E. coli. B, the saturation curve for [3H]cholesterol binding to recombinant NPC1L1-NTD. In each reaction, the indicated concentrations of [3H]cholesterol (25–400 nm) were incubated with 4 pmol of the purified GST-NPC1L1-NTD-His6 or GST-His6 at 4 °C for 24 h in a final volume of 100 μl of buffer D containing 1 μg of BSA. Then the protein was precipitated by using a nickel-nitrilotriacetic acid spin column. After washing with buffer D, the bound [3H]cholesterol was measured by scintillation counting. The specific binding was shown after subtraction of the binding value obtained from parallel experiments with 50 μm unlabeled cholesterol. C, cholesterol-binding properties of the NPC1L1-NTD variants with mutations in the predicted cholesterol-binding pocket. HEK293T cells were transfected with plasmids encoding NPC1L1-NTDs (WT or mutant versions) or NPC1-NTD using FuGENE HD. The media were collected, concentrated, and subjected to a [3H]cholesterol binding assay. In each reaction, indicated concentrations of [3H]cholesterol (200 nm) in 80 μl of buffer E containing 1.6 μg of BSA were incubated with 80 μl of the concentrated culture medium from transfected cells (final volume, 160 μl; final concentrations, 0.001% Nonidet P-40 and 100 nm [3H]cholesterol) at 4 °C. After 24 h, the [3H]cholesterol bound was measured the same as in B. Nonspecific binding was determined by similar experiment with the medium from mock transfected cells. Error bars represent standard deviations of three independent experiments. Inset: immunoblot analysis of the indicated NPC1L1-NTD variants or NPC1-NTD from concentrated medium by anti-FLAG antibody. D, Coomassie blue staining of recombinant GST-NPC1L1-NTD-His6, GST-NPC1L1-NTD(L216A)-His6, and GST-His6 proteins purified from E. coli. E, the saturation curves for [3H]cholesterol binding to NPC1L1-NTD and NPC1L1-NTD(L216A). The binding experiments were performed the same as in B. Error bars represent standard deviations of three independent experiments.
To gain more insight into the NPC1L1-NTD-cholesterol interaction, homology modeling of the human NPC1L1-NTD based on the established crystal structure of human NPC1-NTD (PDB: 3GKI) was performed (supplemental Fig. S2, A–C) (15). The predicted structure of NPC1L1-NTD with cholesterol revealed the enclosure of the cholesterol tetracyclic ring in the binding pocket analogous to that of NPC1-NTD. In the modeling structure, there are 15 residues surrounding the cholesterol binding pocket, which may contribute to the cholesterol binding (supplemental Fig. S2A, blue asterisks, and supplemental Fig. S2D). To investigate the importance of these residues, 13 single-point mutations and one double mutation of Gly-210 to Asn-211 to alanine were introduced individually into the full-length NPC1L1. Eight of these mutations (supplemental Fig. S2, gray dots) showed endoplasmic reticulum co-localization with calnexin (an endoplasmic reticulum marker) and could not be targeted to the ERC (not co-localized with ERC marker rab11a) (24), suggesting that they are misfolded (supplemental Figs. S3 and S4). However, the other six mutants (I105A, T128A, F205A, G210A-N211A, P215A, and L216A) could correctly fold and be transported to the ERC, the compartment where mature NPC1L1 arrives (25) (supplemental Figs. S3 and S4). So these six mutants of NPC1L1-NTD were individually generated, and their cholesterol-binding activities were examined. Five of the six NPC1L1-NTD mutants, including I105A, F205A, G210A-N211A, P215A, and L216A showed dramatically decreased cholesterol binding to <50% of that of WT (Fig. 1C). Only the NPC1L1-NTD (T128A) bound [3H]cholesterol similarly to NPC1L1-NTD (WT) (Fig. 1C). Because L216A showed the lowest cholesterol-binding activity, we used it as a representative mutation in the following experiments. A detailed binding assay with increasing concentrations of [3H]cholesterol confirmed that Leu-216 is critical for cholesterol binding (Fig. 1E).
Cholesterol Binding by NTD Is Essential for NPC1L1-mediated Cellular Cholesterol Uptake
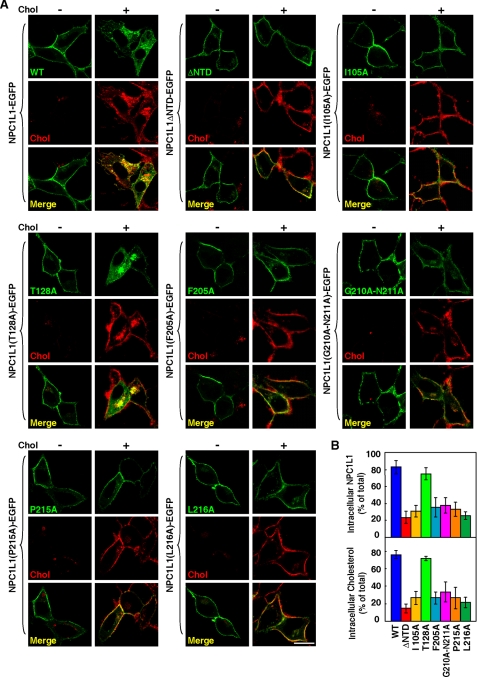
To examine the role of cholesterol binding by NTD in NPC1L1-mediated cholesterol uptake, EGFP-fused NPC1L1, NPC1L1ΔNTD, and the six NPC1L1 mutants were expressed in CRL1601 cells. After cholesterol depletion, they all localized to the PM, suggesting the transport of NPC1L1 from ERC to PM is not affected by deletion of NTD or point mutations (Fig. 2A). Interestingly, after cholesterol replenishment, NPC1L1ΔNTD and the five cholesterol binding-deficient mutants (I105A, F205A, G210A-N211A, P215A, and L216A) were unable to be internalized or promote cholesterol uptake comparing with wild-type NPC1L1 (Fig. 2). On the contrary, the T128A mutant that retains cholesterol-binding ability showed normal endocytosis and promotion of cholesterol uptake (Fig. 2). Thus, these data suggest that the cholesterol binding by NTD is required for NPC1L1 endocytosis and it mediated cholesterol uptake.
FIGURE 2.
The cholesterol uptake mediated by NPC1L1 variants. A, cellular cholesterol uptake mediated by NPC1L1 variants. CRL1601 cells were transfected with indicated NPC1L1-EGFP variants. 48 h after transfection, the cells were incubated in cholesterol-depleting medium for 60 min (Chol−) followed by cholesterol replenishment with 37.5 μm cyclodextrin-coated cholesterol for 60 min (Chol+). The cells were fixed, stained with filipin, and examined by two-photon confocal microscopy. Bar: 10 μm. B, quantification of intracellular localized NPC1L1 and cholesterol in A. Error bars represent standard deviations (n ≥ 100).
Cholesterol Binding by NTD Is Required for NPC1L1-mediated Biliary Cholesterol Reabsorption in Mice
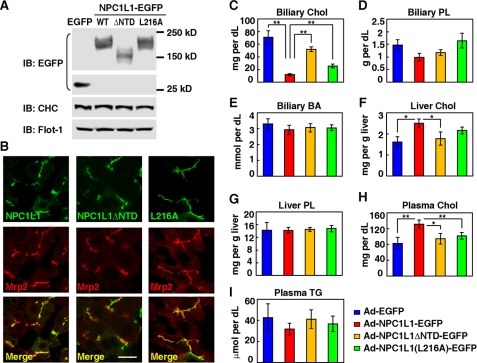
In mice, NPC1L1 is mainly expressed in the small intestine, whereas it is highly expressed in both liver and intestine in humans (3, 26). Previous studies have shown that the exogenously expressed NPC1L1-EGFP in mice liver facilitates biliary cholesterol reabsorption (2, 7). To determine the role of NTD in NPC1L1-mediated cholesterol uptake in vivo, we expressed EGFP, NPC1L1(WT)-EGFP, NPC1L1ΔNTD-EGFP, and NPC1L1(L216A)-EGFP in mice liver using an adenovirus system (Fig. 3A). The NPC1L1-EGFP co-localized with Mrp2 (Fig. 3B), a liver apical membrane marker, indicating it localizes on the canalicular membrane of hepatocytes in liver (27). Deletion of NTD (NPC1L1ΔNTD) or L216A mutation did not affect the canalicular membrane targeting of NPC1L1 (Fig. 3B). Comparing with EGFP overexpression mice, the biliary cholesterol from NPC1L1-EGFP overexpression mice was greatly decreased, whereas liver and plasma cholesterol was increased. For mice expressing NPC1L1ΔNTD-EGFP or NPC1L1(L216A)-EGFP, their biliary cholesterol was higher and their liver and plasma cholesterol was lower than those of NPC1L1(WT)-EGFP expression mice (Fig. 3, C, F, and H). As controls, the biliary phospholipids, bile acids, liver phospholipids, or plasma triglycerides (Fig. 3, D, E, G, and I) of different groups were not changed. These data indicate that deletion or mutation of NTD hampers the cholesterol binding and impairs NPC1L1-mediated cholesterol reabsorption in mice liver.
FIGURE 3.
Biliary cholesterol reabsorption mediated by NPC1L1, NPC1L1ΔNTD, and NPC1L1(L216A) in mice livers. A, immunoblot analysis of the adenovirus-mediated expression of EGFP, NPC1L1-EGFP, NPC1L1ΔNTD-EGFP, and NPC1L1(L216A)-EGFP in mice liver. Flot-1, flotillin-1; CHC, clathrin heavy chain. B, the localization of NPC1L1-EGFP, NPC1L1ΔNTD-EGFP, and NPC1L1(L216A)-EGFP in mice liver. Frozen sections of the liver were stained with polyclonal anti-Mrp2 antibody. Bar: 10 μm. C–I, measurement of biliary cholesterol (Chol) (C), biliary phospholipids (PL) (D), biliary bile acid (BA) (E), liver cholesterol (F), liver PL (G), plasma cholesterol (H), and triglyceride (TG) (I) in the mice administrated with adenovirus expressing EGFP, NPC1L1-EGFP, NPC1L1ΔNTD-EGFP, or NPC1L1(L216A)-EGFP as shown in A. Error bars represent standard deviations (n = 5). **, p < 0.02; *, p < 0.05 (two-tailed Student's t test).
Cholesterol Binding by NTD Is Required for the Formation of NPC1L1-flotillin-cholesterol Membrane Microdomains
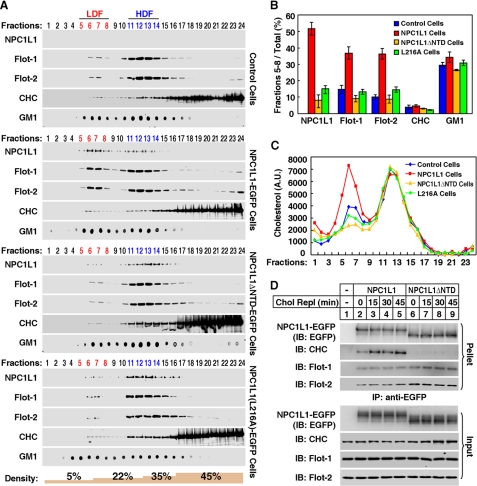
Our previous work has shown that NPC1L1 associates with flotillins to form cholesterol-enriched membrane microdomains (NPC1L1-flotillin-cholesterol membrane microdomains), and they are essential for efficient cholesterol uptake. These membrane microdomains are characterized by their distribution in low density fractions (LDFs) in the sucrose gradient ultracentrifugation assay. Formation of these microdomains occurs upstream of clathrin recruitment and is required for the endocytosis of NPC1L1 (7). To understand the role of cholesterol binding in this process, we investigated whether cholesterol binding of NTD affects the formation of NPC1L1-flotillin-cholesterol membrane microdomains by sucrose gradient ultracentrifugation. The NPC1L1(WT) mainly distributed in LDF, and its expression led to the increased LDF distribution of both flotillins and cholesterol compared with control cells, indicating the formation of NPC1L1-flotillin-cholesterol membrane microdomains (Fig. 4, A–C). In contrast, NPC1L1ΔNTD and NPC1L1(L216A) were mainly present in high density fractions (HDFs) and failed to increase the distribution of flotillins or cholesterol in LDF (Fig. 4, A–C). Co-immunoprecipitation experiments showed that NPC1L1ΔNTD barely associated with clathrin heavy chain, although it still bound flotillins similarly to NPC1L1 (Fig. 4D), suggesting NTD is not involved in the association between NPC1L1 and flotillins but is required for the formation of cholesterol-enriched membrane microdomain.
FIGURE 4.
The cholesterol binding by NPC1L1-NTD is required for the formation of NPC1L1-flotillin-cholesterol membrane microdomains. A, sucrose gradient fractionation showing the distribution of indicated proteins and monosialotetrahexosylganglioside (GM1). CRL1601 (control) cells, CRL1601-NPC1L1-EGFP cells, CRL1601-NPC1L1ΔNTD-EGFP cells, and CRL1601-NPC1L1(L216A)-EGFP cells were depleted of cholesterol for 60 min, and then replenished with 37.5 μm cyclodextrin-coated cholesterol for 60 min. The cells were subjected to sucrose gradient ultracentrifugation analysis. Low density fractions (LDF, fractions 5–8) are highlighted in red, and high density fractions (HDF, fractions 11–14) are highlighted in blue. Flot-1/2, flotillin-1/2; GM1, ganglioside GM1; CHC, clathrin heavy chain. B, quantification of the indicated proteins and GM1 in LDF shown in A. Error bars represent standard deviations of three independent experiments. C, analysis of cholesterol distribution in different fractions. A.U., arbitrary units. D, co-immunoprecipitation of NPC1L1-EGFP or NPC1L1ΔNTD-EGFP with clathrin. CRL1601-NPC1L1-EGFP and CRL1601-NPC1L1ΔNTD-EGFP cells were depleted of cholesterol for 60 min, and then replenished with 37.5 μm cyclodextrin-coated cholesterol for the indicated time durations (Chol Repl 0–45 min). Then co-immunoprecipitation was performed with anti-EGFP antibody-coupled agarose followed by immunoblotting with indicated antibodies. CRL1601 cells were used as a negative control.
These data indicate that loss of the cholesterol-binding ability of NTD impairs the formation of NPC1L1-flotillin-cholesterol membrane microdomains and, therefore, impedes the NPC1L1 endocytosis and cholesterol uptake.
Plant Sterols do Not Bind NPC1L1-NTD and Cannot Induce the Endocytosis of NPC1L1
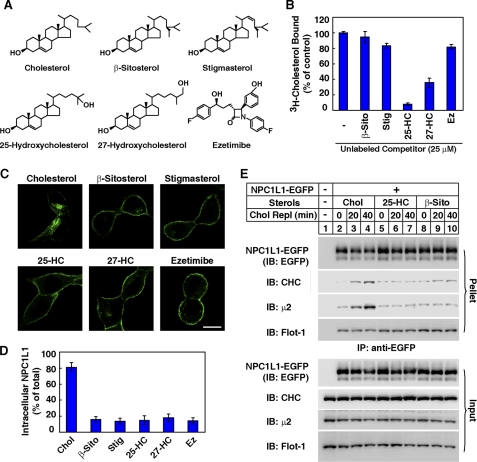
We next performed competition experiments to test the sterol-binding specificity of NPC1L1-NTD (Fig. 5A). In the sterols tested, 25-hydroxycholesterol (25-HC) and 27-hydroxycholesterol (27-HC) but not β-sitosterol or stigmasterol competed with [3H]cholesterol for the binding to NPC1L1-NTD (Fig. 5B), suggesting that 25-HC and 27-HC may bind to NPC1L1-NTD, whereas β-sitosterol and stigmasterol may not. Meanwhile, the cholesterol absorption inhibitor ezetimibe, which has been shown to bind the second large luminal loop of NPC1L1 (28), did not inhibit [3H]cholesterol binding to NPC1L1-NTD (Fig. 5B), which is consistent with the previous report that ezetimibe bound to the second large luminal loop of NPC1L1 (28, 29). β-Sitosterol and stigmasterol lacked the activity to promote the endocytosis of NPC1L1 (Fig. 5C) similar to a previous report (4). Because mammals selectively absorb cholesterol but not plant sterols from diet (30, 31), the cholesterol-binding specificity by NPC1L1-NTD may account for the cholesterol absorption specificity. Although both 25-HC and 27-HC bound to NPC1L1-NTD (Fig. 5B), neither of them could efficiently induce the endocytosis of NPC1L1 (Fig. 5C). This is probably because the hydroxyl group on the isooctyl chain of 25-HC and 27-HC interferes with their insertion into the membrane to form the functional membrane microdomain. In co-immunoprecipitation experiments, cholesterol, but not 25-HC or β-sitosterol, could induce the interaction between NPC1L1 and clathrin-AP2 (Fig. 5E).
FIGURE 5.
Plant sterols do not compete off cholesterol binding to NPC1L1-NTD and cannot promote the endocytosis of NPC1L1. A, structure of different sterols and ezetimibe. B, [3H]cholesterol binding to the NPC1L1-NTD in the presence of excessive indicated sterols or ezetimibe. A cholesterol binding assay was performed as described in Fig. 1B with 100 nm [3H]cholesterol as well as 25 μm of the indicated sterols or ezetimibe. The binding values of [3H]cholesterol to NPC1L1-NTD in the presence of the indicated sterols or ezetimibe were calculated as percentage to control (−). 25-HC, 25-hydroxycholesterol; 27-HC, 27-hydroxycholesterol; β-Sito, β-sitosterol; Stig, stigmasterol; Ez, ezetimibe. C, endocytosis of NPC1L1-EGFP induced by different sterols. CRL1601-NPC1L1-EGFP cells were depleted of cholesterol by incubating in cholesterol-depleting medium for 60 min, and then treated with 37.5 μm cyclodextrin-coated sterols for 60 min. The cells were then examined by confocal microscopy. Bar, 10 μm. D, quantification of the intracellular localized NPC1L1-EGFP shown in C. E, co-immunoprecipitation of NPC1L1-EGFP with clathrin and AP2 subunit μ2. CRL1601-NPC1L1-EGFP cells were depleted of cholesterol for 60 min, and then replenished with 37.5 μm cyclodextrin-coated sterols for the indicated time durations (Chol Repl 0–40 min). Then co-immunoprecipitation was performed with anti-EGFP antibody-coupled agarose followed by immunoblotting with indicated antibodies. CRL1601 cells were used as a negative control.
Oxysterol Competes with Cholesterol to Bind NPC1L1-NTD and Inhibits the Endocytosis of NPC1L1 and the Formation of NPC1L1-flotillin-cholesterol Membrane Microdomains
A detailed [3H]25-HC binding assay and a competition assay confirmed that 25-HC bound NPC1L1-NTD and competed off cholesterol binding to NPC1L1-NTD (Fig. 6, A and B). Similar to that of cholesterol binding, the binding curve of 25-HC is sigmoidal with a Hill coefficient of 2.5, suggesting a positive cooperativity. We next sought to determine whether competing with cholesterol binding to NPC1L1-NTD could interfere with the endocytosis of NPC1L1 induced by cholesterol. As shown in Fig. 6C, cholesterol replenishment promoted NPC1L1 endocytosis, which could be blocked by 25-HC or 27-HC but not β-sitosterol or stigmasterol. As a control, ezetimibe also inhibited the NPC1L1 endocytosis that has been shown before (4). 25-HC but not β-sitosterol abolished cholesterol-induced interaction between NPC1L1 and clathrin-AP2 (Fig. 6E). But none of them had effect on NPC1L1-flotillin interaction (Fig. 6E). Moreover, 25-HC interfered with the formation of NPC1L1-flotillin-cholesterol membrane microdomains (Fig. 6, F–H). In summary, these data suggest that 25-HC or 27-HC competes with cholesterol to bind NPC1L1-NTD and impairs the formation of NPC1L1-flotillin-cholesterol membrane microdomains, thereby inhibiting NPC1L1 internalization.
FIGURE 6.
25-Hydroxycholesterol competes with cholesterol to bind NPC1L1-NTD and inhibits the endocytosis of NPC1L1. A, the saturation curve for [3H]25-HC binding to recombinant NPC1L1-NTD. The binding assay was performed as shown in Fig. 1B except adding [3H]25-HC (50–300 nm) instead of [3H]cholesterol. Nonspecific binding was determined by a similar experiment with 50 μm unlabeled 25-HC. B, competitive binding of [3H]cholesterol to NPC1L1-NTD by 25-HC. The binding assay was performed as shown in Fig. 5B, except with 200 nm of [3H]cholesterol and adding unlabeled sterols with indicated concentrations (0.1–25 μm) for competition. C, 25-HC, 27-HC, and ezetimibe blocked the cholesterol-induced endocytosis of NPC1L1. CRL1601-NPC1L1-EGFP cells were depleted of cholesterol and then incubated with 37.5 μm cyclodextrin-coated cholesterol plus 37.5 μm indicated sterols or ezetimibe for 60 min. The cells were examined by confocal microcopy. Bar: 10 μm. D, quantification of the intracellular localized NPC1L1-EGFP shown in C. E, co-immunoprecipitation of NPC1L1-EGFP with clathrin and AP2 subunit μ2. CRL1601-NPC1L1-EGFP cells were depleted of cholesterol for 60 min, and then replenished with 37.5 μm cyclodextrin-coated cholesterol plus 37.5 μm indicated sterols for the indicated time durations (Chol Repl 0–40 min). Then co-immunoprecipitation was performed with anti-EGFP antibody-coupled agarose followed by immunoblotting with indicated antibodies. CRL1601 cells were used as a negative control. F–H, CRL1601-NPC1L1-EGFP cells were treated as shown in C. Then sucrose gradient ultracentrifugation was performed (F). The indicated proteins or GM1 in LDF was quantified (G). The cholesterol level in each fraction was analyzed (H). A.U., arbitrary units.
DISCUSSION
In the current study, we found that the NTD of NPC1L1 binds cholesterol. Homology modeling and alanine scanning followed by an in vitro cholesterol binding assay suggested the structural basis for the cholesterol binding of NPC1L1-NTD. We also demonstrated that cholesterol binding by NPC1L1-NTD is essential for NPC1L1-mediated cholesterol uptake in culture cells and mice livers.
It has been known for a long time that mammals absorb cholesterol specifically. About 50% of cholesterol passing the intestine is absorbed, whereas only <5% of plant sterols is taken up (30, 31). Our previous studies showed that NPC1L1 prefers to mediate cholesterol uptake rather than plant sterols (4). The molecular basis on the sterol selectivity is unknown. Here we found that the binding of [3H]cholesterol to NPC1L1-NTD is efficiently competed by oxysterols but not plant sterols (Fig. 5B), indicating that NPC1L1-NTD binds plant sterols poorly. This binding specificity may contribute to the selective cholesterol absorption in mammals.
The efficiency of cholesterol absorption is ensured by formation and endocytosis of NPC1L1-flotillin-cholesterol membrane microdomains (7). How is cholesterol enriched in the local membrane around NPC1L1-flotillin to form the cholesterol-enriched microdomains? Here we showed that NPC1L1-NTD binds cholesterol, which is crucial for the formation of cholesterol-enriched membrane microdomains, although the NTD is dispensable for NPC1L1-flotillin interaction (Fig. 4). Given the recent finding that NPC1 binds cholesterol via its NTD and transfers cholesterol to liposome membrane (14, 15, 18, 19, 23), NPC1L1 may possess the similar abilities. NPC1L1 may bind cholesterol through its own NTD and then transfer cholesterol to the local membrane where NPC1L1 and flotillins reside. Alternatively, the binding of cholesterol to NTD may alter the conformation of NPC1L1 protein to allow the increased cholesterol-loading capacity in the local membrane. In the latter case, other domains of NPC1L1, such as the sterol-sensing domain, may be responsible for holding large amounts of cholesterol together with flotillins. Further studies are necessary to examine these possibilities.
An interesting discovery in our study is that 25- and 27-HC can compete with cholesterol to bind NPC1L1-NTD. Although the competitive binding of 25- and 27-HC against cholesterol to NPC1-NTD has been reported (14, 15, 23), the effect of oxysterols on NPC1-mediated cholesterol transport has not been verified, possibly because most of the assays (i.e. sterol regulatory element-binding protein cleavage and cholesterol esterification) used to analyze the function of NPC1 are indirectly measuring endoplasmic reticulum cholesterol level, and the readouts can be affected by oxysterols themselves (32, 33). On the other hand, here we show that 25- and 27-HC block the endocytosis of NPC1L1 and inhibit the formation of NPC1L1-flotillin-cholesterol membrane microdomains (Fig. 6). These cellular effects are consistent with their in vitro competition with cholesterol in NPC1L1-NTD binding (Fig. 6). In the future, the possibility that 25- and 27-HC may inhibit cholesterol absorption in vivo can be further verified. It is well known that oxysterols mediate the feedback inhibition of cholesterol synthesis by blocking the cleavage of sterol regulatory element-binding protein and promoting the degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase (34). Our current data implicate another regulatory role of oxysterols in cholesterol metabolism, i.e. feedback inhibition of cholesterol absorption by competing with cholesterol to bind NPC1L1-NTD. During the review of this study, a report just came out on the crystal structure of NPC1L1-NTD. The authors also showed that [3H]cholesterol efficiently binds NPC1L1-NTD that could be competed by 25-HC (35). This is consistent with our study.
Although both 25-HC (or 27-HC) and ezetimibe inhibit the formation of NPC1L1-flotillin-cholesterol membrane microdomains and NPC1L1 endocytosis (Fig. 6) (7), they act through different mechanisms. 25-HC (or 27-HC) competes with cholesterol to bind NPC1L1-NTD (the first large luminal loop), thereby preventing the cholesterol-enriched membrane microdomain formation (Fig. 6). However, ezetimibe binds to the second large luminal loop of NPC1L1, dissociates the interaction between NPC1L1 and flotillins, and therefore blocks NPC1L1-flotillin-cholesterol membrane microdomains assembly (7, 28). This finding provides a new way to develop cholesterol absorption inhibitors by searching for compounds binding NPC1L1-NTD, similar to 25-HC and 27-HC.
In summary, our study reveal that NPC1L1 senses cholesterol through direct binding by its NTD, then cholesterol may be transferred to local membrane to form the NPC1L1-flotillin-cholesterol membrane microdomain, which is internalized via the clathrin-AP2 pathway. The cholesterol-binding specificity may serve as a molecular basis for selective cholesterol absorption in intestine. Oxysterols may regulate cholesterol uptake by competing cholesterol binding.
Supplementary Material
Acknowledgments
We thank Yu-Xiu Qu, Su-Zhe Pan, Qin Li, Jie Xu, Chang Xie, and Zhang-Sen Zhou for technical assistance and Na Li for critical reading of the manuscript.
This work was supported by the Ministry of Science and Technology of China (Grants 2009CB919000 and 2011CB910900), the National Natural Science Foundation of China (Grant 30925012), and the Shanghai Science and Technology Committee (Grant 10QH1402900).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, references, and Figs. S1–S4.
- ERC
- endocytic recycling compartment
- NPC1L1
- Niemann-Pick C1-like 1
- PM
- plasma membrane
- NTD
- N-terminal domain
- Chol
- cholesterol
- 25-HC
- 25-hydroxycholesterol
- LDF
- low density fraction
- HDF
- high density fraction
- aa
- amino acid(s)
- Mes
- 4-morpholineethanesulfonic acid
- GM1
- monosialotetrahexosylganglioside.
REFERENCES
- 1. Davis H. R., Jr., Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., Detmers P. A., Graziano M. P., Altmann S. W. (2004) J. Biol. Chem. 279, 33586–33592 [DOI] [PubMed] [Google Scholar]
- 2. Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. (2007) J. Clin. Invest. 117, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M. P. (2004) Science 303, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 4. Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. (2008) Cell Metab. 7, 508–519 [DOI] [PubMed] [Google Scholar]
- 5. Yu L., Bharadwaj S., Brown J. M., Ma Y., Du W., Davis M. A., Michaely P., Liu P., Willingham M. C., Rudel L. L. (2006) J. Biol. Chem. 281, 6616–6624 [DOI] [PubMed] [Google Scholar]
- 6. Chu B. B., Ge L., Xie C., Zhao Y., Miao H. H., Wang J., Li B. L., Song B. L. (2009) J. Biol. Chem. 284, 22481–22490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ge L., Qi W., Wang L. J., Miao H. H., Qu Y. X., Li B. L., Song B. L. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ioannou Y. A. (2000) Mol. Genet. Metab. 71, 175–181 [DOI] [PubMed] [Google Scholar]
- 9. Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B., Nagle J., Polymeropoulos M. H., Sturley S. L., Ioannou Y. A., Higgins M. E., Comly M., Cooney A., Brown A., Kaneski C. R., Blanchette-Mackie E. J., Dwyer N. K., Neufeld E. B., Chang T. Y., Liscum L., Strauss J. F., 3rd, Ohno K., Zeigler M., Carmi R., Sokol J., Markie D., O'Neill R. R., van Diggelen O. P., Elleder M., Patterson M. C., Brady R. O., Vanier M. T., Pentchev P. G., Tagle D. A. (1997) Science 277, 228–2319211849 [Google Scholar]
- 10. Chang T. Y., Reid P. C., Sugii S., Ohgami N., Cruz J. C., Chang C. C. (2005) J. Biol. Chem. 280, 20917–20920 [DOI] [PubMed] [Google Scholar]
- 11. Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., Lobel P. (2000) Science 290, 2298–2301 [DOI] [PubMed] [Google Scholar]
- 12. Davies J. P., Ioannou Y. A. (2000) J. Biol. Chem. 275, 24367–24374 [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Chu B. B., Ge L., Li B. L., Yan Y., Song B. L. (2009) J. Lipid Res. 50, 1653–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Infante R. E., Radhakrishnan A., Abi-Mosleh L., Kinch L. N., Wang M. L., Grishin N. V., Goldstein J. L., Brown M. S. (2008) J. Biol. Chem. 283, 1064–1075 [DOI] [PubMed] [Google Scholar]
- 15. Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., Infante R. E. (2009) Cell 137, 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedland N., Liou H. L., Lobel P., Stock A. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2512–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu S., Benoff B., Liou H. L., Lobel P., Stock A. M. (2007) J. Biol. Chem. 282, 23525–23531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Infante R. E., Wang M. L., Radhakrishnan A., Kwon H. J., Brown M. S., Goldstein J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang M. L., Motamed M., Infante R. E., Abi-Mosleh L., Kwon H. J., Brown M. S., Goldstein J. L. (2010) Cell Metab. 12, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., Bose-Boyd R. A., Tang J. J., Li B. L., Song B. L. (2007) Cell Metab. 6, 115–128 [DOI] [PubMed] [Google Scholar]
- 21. Tang J. J., Li J. G., Qi W., Qiu W. W., Li P. S., Li B. L., Song B. L. (2011) Cell Metab. 13, 44–56 [DOI] [PubMed] [Google Scholar]
- 22. Chen H. Y., Yuan Y. A. (2010) J. Mol. Cell. Biol. 2, 366–374 [DOI] [PubMed] [Google Scholar]
- 23. Infante R. E., Abi-Mosleh L., Radhakrishnan A., Dale J. D., Brown M. S., Goldstein J. L. (2008) J. Biol. Chem. 283, 1052–1063 [DOI] [PubMed] [Google Scholar]
- 24. Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996) J. Cell Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L. J., Wang J., Li N., Ge L., Li B. L., Song B. L. (2011) J. Biol. Chem. 286, 7397–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davies J. P., Scott C., Oishi K., Liapis A., Ioannou Y. A. (2005) J. Biol. Chem. 280, 12710–12720 [DOI] [PubMed] [Google Scholar]
- 27. Nies A. T., Cantz T., Brom M., Leier I., Keppler D. (1998) Hepatology 28, 1332–1340 [DOI] [PubMed] [Google Scholar]
- 28. Weinglass A. B., Kohler M., Schulte U., Liu J., Nketiah E. O., Thomas A., Schmalhofer W., Williams B., Bildl W., McMasters D. R., Dai K., Beers L., McCann M. E., Kaczorowski G. J., Garcia M. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia-Calvo M., Lisnock J., Bull H. G., Hawes B. E., Burnett D. A., Braun M. P., Crona J. H., Davis H. R., Jr., Dean D. C., Detmers P. A., Graziano M. P., Hughes M., Macintyre D. E., Ogawa A., O'neill K. A., Iyer S. P., Shevell D. E., Smith M. M., Tang Y. S., Makarewicz A. M., Ujjainwalla F., Altmann S. W., Chapman K. T., Thornberry N. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8132–8137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoenheimer R. (1931) Science 74, 579–584 [DOI] [PubMed] [Google Scholar]
- 31. Glover J., Morton R. A. (1958) Br. Med. Bull. 14, 226–233 [DOI] [PubMed] [Google Scholar]
- 32. Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 33. Cheng D., Chang C. C., Qu X., Chang T. Y. (1995) J. Biol. Chem. 270, 685–695 [DOI] [PubMed] [Google Scholar]
- 34. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 35. Kwon H. J., Palnitkar M., Deisenhofer J. (2011) PLoS One 6, e18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.