Abstract
The membrane glycoprotein CD36 binds nanomolar concentrations of long chain fatty acids (LCFA) and is highly expressed on the luminal surface of enterocytes. CD36 deficiency reduces chylomicron production through unknown mechanisms. In this report, we provide novel insights into some of the underlying mechanisms. Our in vivo data demonstrate that CD36 gene deletion in mice does not affect LCFA uptake and subsequent esterification into triglycerides by the intestinal mucosa exposed to the micellar LCFA concentrations prevailing in the intestine. In rodents, the CD36 protein disappears early from the luminal side of intestinal villi during the postprandial period, but only when the diet contains lipids. This drop is significant 1 h after a lipid supply and associates with ubiquitination of CD36. Using CHO cells expressing CD36, it is shown that the digestion products LCFA and diglycerides trigger CD36 ubiquitination. In vivo treatment with the proteasome inhibitor MG132 prevents the lipid-mediated degradation of CD36. In vivo and ex vivo, CD36 is shown to be required for lipid activation of ERK1/2, which associates with an increase of the key chylomicron synthesis proteins, apolipoprotein B48 and microsomal triglyceride transfer protein. Therefore, intestinal CD36, possibly through ERK1/2-mediated signaling, is involved in the adaptation of enterocyte metabolism to the postprandial lipid challenge by promoting the production of large triglyceride-rich lipoproteins that are rapidly cleared in the blood. This suggests that CD36 may be a therapeutic target for reducing the postprandial hypertriglyceridemia and associated cardiovascular risks.
Keywords: ERK, Intestine, Lipid Absorption, Lipid-binding Protein, Lipoprotein, ApoB48, CD36, MTP, Chylomicron
Introduction
CD36 (also known as fatty acid translocase) is a transmembrane glycoprotein expressed in many tissues. It is a multifunctional protein homologous to scavenger receptor class B, type I. CD36 facilitates uptake of long chain fatty acids (LCFA)2 in cardiomyocytes (1) and adipocytes (2, 3) and that of oxidized LDL by macrophages (4). CD36 is involved in platelet aggregation by binding thrombospondin and collagen (5), phagocytosis of apoptotic cells by macrophages (6), and the cytoadhesion of erythrocytes infected with Plasmodium falciparum (7). In addition, CD36 has recently been shown to play a role in taste perception of dietary fatty acid on the tongue by triggering a cell signaling cascade (8–10).
Deletion of CD36 in mice highlighted the importance of this protein for optimal utilization of dietary lipids. Significant impairment in the uptake of LCFA by skeletal muscle, heart, and adipose tissues was shown (2). Insulin- and exercise-dependent translocation of CD36 from an intracellular pool to the sarcolemna was documented and postulated to increase the muscle efficiency by allowing adaptive changes in LCFA uptake and utilization (11, 12). Finally, CD36-deficient mice exhibit a loss of the spontaneous preference for lipid-rich foods and a decrease of orosensory-mediated rise in digestive secretions (8, 9). In humans, variants in the CD36 gene have been associated with abnormalities of lipid and glucose metabolism (13) and with altered susceptibility to the metabolic syndrome (14) and diabetes-associated coronary disease (15).
In the small intestine, expression of CD36 is especially high in the brush border membrane (BBM) of duodeno-jejunal enterocytes both in rodents and humans (16, 17). Although this location correlates with the main site of LCFA absorption, the physiological role played by CD36 in the gut remains incompletely understood. In isolated enterocytes from the proximal intestine, CD36 deletion impairs LCFA uptake (18). However, in vivo, the lack of impairment in intestinal net lipid absorption in CD36−/− mice (18, 19), as well as in subjects with type I CD36 deficiency (20), seems to preclude a quantitative role of intestinal CD36 in LCFA uptake similar to that demonstrated in skeletal muscle, heart, and adipose tissues. By contrast, lipoprotein synthesis and/or secretion are deeply impaired in enterocytes from CD36−/− mice, where smaller chylomicrons are produced. This event might explain the postprandial hypertriglyceridemia found in CD36-deficient mice and humans (20–22). Indeed, the efficiency of blood triglyceride (TG) clearance is positively linked to chylomicron size (23). Therefore, it can be speculated that CD36 plays a role in regulating the metabolic fate of lipids rather than LCFA uptake in the small intestine. However, the molecular mechanisms by which this effect would occur remain unknown. One possibility could be a CD36-dependent modulation of ERK, because it was previously shown to regulate lipoprotein formation (24–26), and it is activated by ligand binding to CD36 (27).
To explore this concept, experiments were conducted in vivo in rats and in wild-type or CD36−/− mice, ex vivo in intestinal segments, and in vitro in stably transfected CHO cells. The present data show that CD36 undergoes rapid ubiquitination and degradation triggered by digestion products during intestinal lipid absorption. This process is associated with CD36-dependent activation of ERK and with increased expression of the key proteins of lipoprotein synthesis apoB48 and MTP.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against rat CD36 were generated in rabbits and used as described previously (16). Anti-mouse CD36 (R & D System or Santa Cruz Biotechnology), mouse anti-ubiquitin (Zymed Laboratories Inc., Invitrogen), anti-phospho-ERK1/2 (Thr-202/Tyr-204) and anti-ERK1/2 (Cell Signaling Technology), anti-apoB, anti-HSC70 (Santa Cruz Biotechnology), anti-MTP (BD Sciences), secondary antibody peroxidase conjugate goat anti-rabbit (Chemicon International), anti-mouse IgG (Sigma-Aldrich), and donkey anti-goat (Santa Cruz Biotechnology) were from commercial sources. Protein A/G Plus-agarose immunoprecipitation reagent was from Santa Cruz; MG132 (Z-Leu-Leu-Leu-al) was from Sigma-Aldrich; the ECL kit was from PerkinElmer Life Sciences; triolein was from Sigma; and oleic acid, 2-monolein, and miolein were from Larodan Fine Chemicals.
Animals and Experimental Procedures
The care and use of laboratory animals followed guidelines of the animal ethics committee of Burgundy University, which approved all protocols. The rats or mice were housed in a controlled environment (constant temperature and humidity, darkness from 6 p.m. to 6 a.m.). They were fed a standard laboratory chow containing 3% (w/w) lipids (UAR A-04 for rats and mice; Usine d'Alimentation Rationnelle). The alipidic diet was prepared by Safe.
Rats
Male Wistar rats weighting 180–200 g (Elevage Dépré) were fasted for 48 h. In a first protocol, the animals were refed either a standard laboratory chow containing 3% lipids (w/w, sunflower oil) or a fat-free diet. After sacrifice (by isoflurane anesthesia), jejunal samples (1 cm) taken 24 cm after the pylorus were prepared for immunohistochemistry using antibody against CD36 raised in rabbits (16).
In a second protocol, 48-h fasted rats were force-fed 3 ml of sunflower oil or sucrose (isocaloric, 5.27 m). Mucosa from jejunal segments (25 cm taken 10 cm after pylorus) was obtained at different times after and used to prepare homogenates and BBM. Mucosa samples were rapidly frozen and stored at −80 °C until RNA purification.
Mice
Male C57Bl6 (CD36+/+ and CD36−/− mice (provided by Dr. Jan Glatz) (12 weeks of age) weighing 25–30 g were fasted for 12 h prior to oil gavage (0.5 ml of sunflower oil) and sacrificed by isoflurane anesthesia. The small intestine was divided into three equal parts. The first centimeter was taken from the middle part of the small intestine, considered as the jejunum and treated for immunohistochemistry using antibody anti-CD36 from R & D System. The mucosa from jejunal segments was scraped at 1, 2, and 4 h after the gavage for homogenate preparation and mRNA analysis. The same protocol was performed in mice subjected 30 min before the gavage to an intraperitoneal injection of 100 μl of Me2SO containing 14 mg/kg of body weight of the proteasome inhibitor MG132, whereas the control group received the same volume of Me2SO. The animals were sacrificed 4 h after the oil gavage.
Jejunal Homogenate and Brush Border Membrane
Scraped mucosa obtained from rats were weighed and homogenized (with buffer A, which contained 1 g/16.7 ml of 100 mm mannitol, and 10 mm Tris-HCl, pH 7.1) using a Dounce potter homogenizer. Purified BBM were prepared according to Shirazi-Beechey et al. (28). Briefly, MgCl2 was slowly added to homogenates to reach a final concentration of 10 mm. The samples were stored on ice for 30 min followed by centrifugation (3,000 × g, 30 min, 4 °C). Supernatants were centrifuged (27,000 × g, 30 min, 4 °C) (Beckman LE-70, SW41Ti rotor). The supernatant was removed, and the pellet was resuspended in 3.8 mol of buffer B (300 mm mannitol, 10 mm Tris-HCl, 0.1 mm MgSO4, pH 7.4) and centrifuged (27,000 × g, 30 min, 4 °C) (Beckman centrifuge, 70.1Ti rotor). The supernatant was removed, and the BBM pellet was resuspended in 150 μl of buffer B and stored at −20 °C. The purity of BBM preparation and membrane yield were evaluated by assaying the BBM-specific enzyme sucrase-isomaltase according to the Dahlqvist method (29), and enzyme content was related to 1 mg of starting mucosa. The proportion of the sucrase activity retrieved in the BBM fraction corresponds to the efficiency of BBM extraction. Enrichment was obtained by dividing specific activity (UI/mg of protein) of the sucrase in BBM fraction by that of the homogenate. Yield and enrichment were consistent with previous reports (28). To prepare microsomal and cytosolic fractions, the remaining homogenate from the same animals was centrifuged (18,000 × g, 10 min, 4 °C), and the supernatant was transferred and centrifuged (105,000 × g, 60 min, 4 °C) to yield a microsomal (pellet) and a cytosolic (supernatant) fraction. The pellet was resuspended in 200 μl of buffer B. The fractions were stored at −20 °C. When mice were used, a jejunal homogenate was obtained by adding 50 mg of jejunal mucosa to 0.5 ml of buffer A with 1% Triton X-100.
Jejunal triglyceride content was evaluated after lipid extraction of mucosa according to the Delsal method (30). Dried extracts dissolved in 1% Triton X-100 (in chloroform) were dried then redissolved in water for triglyceride determination using CHOD-PAP (Roche Applied Science).
Intestinal Fatty Acid Uptake: In Situ Isolated Jejunal Loop
Isofluran-anesthetized, 16-h fasted mice were laparatomized, and a 10-cm jejunal loop was isolated in situ and infused with 0.5 ml of a radiolabeled solution of linoleic acid in 2180 μm solution containing 10% of [1-14C]linoleic acid (50 mCi/mmol) emulsified with 10 mm sodium taurocholate to form micelles. Radioactivity incorporation into the different lipid classes in the mucosa was determined (31).
Isolated Intestinal Segment Culture
The proximal small intestine from 16-h fasted wild-type and CD36−/− mice was cut into small segments (3-cm length) taken 3 cm after the pylorus, split as described by Pardo et al. (32). After a 15-min stabilization period, intestinal segments were treated with 240 μm linoleic acid or vehicle (ethanol 0.01%) for 10–20 min. The treatment was ended by submerging the intestinal segment into liquid nitrogen. The mucosa was collected by scraping into ice-cold radioimmune precipitation assay buffer containing anti-protease and anti-phosphatase mixture.
Evaluation of CD36 Ubiquitination in Mice Jejunum and in CHO Cells
CD36 was immunoprecipitated overnight at 4 °C from 500 μg of jejunal homogenate using 1 μg of anti-CD36 (R & D System). Then protein A/G-agarose plus beads were added, and the incubation continued for another 2–3 h. The final pellet was washed three times in radioimmune precipitation assay buffer and boiled for 10 min in SDS sample buffer. After centrifugation, the supernatant was analyzed by SDS-PAGE, and the proteins were electroblotted onto PVDF membranes. The membranes were treated with a denaturating buffer (6 m guanidine HCl, 20 mm Tris-HCl, pH 7.5, 5 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride) for 30 min at 4 °C, followed by extensive PBS washing and then used for detection of ubiquitin or CD36 proteins.
In CHO cells, the carboxyl-FLAG-tagged WT or carboxyl lysine mutant (K/A) human CD36 were generated as described (33). The cells were transfected (Lipofectamine 2000; Invitrogen) with empty vector (control) or with vector containing WT CD36 or CD36 where the two C-terminal lysines (469 and 472) were substituted with alanine (K/A). The cells were then selected for stable transfections using hygromycin. Stably transfected cells plated in 6-well plates were incubated for 2 h with either BSA or BSA together with 100 μm of oleic acid, triolein (Sigma), 2-monolein, or diolein (Larodan Fine Chemicals). The cells were lysed, and clarified lysates were immunoprecipitated with FLAG affinity gel (Sigma). Immunocomplexes were eluted with FLAG peptide, resolved by SDS-PAGE, and immunoblotted with either anti-ubiquitin antibody (Zymed Laboratories Inc.) or anti-CD36 antibody (Santa Cruz).
Real Time PCR, Western Blotting, and ELISA Analyses
mRNA levels were measured by real time PCR (9, 31). CD36 protein levels were determined using Western blotting (16) and sandwich ELISA (34). Quantification of immune signals was by densitometry and with Quantity One software (Bio-Rad).
Statistics
All of the data are presented as the means ± S.E. for the indicated number of animals. Significance of differences between groups of observations was tested with a paired Student's t test. p ≤ 0.05 was considered significant.
RESULTS
CD36 Gene Deletion Does Not Affect Micellar LCFA Uptake and TG Re-esterification in Mouse Jejunum
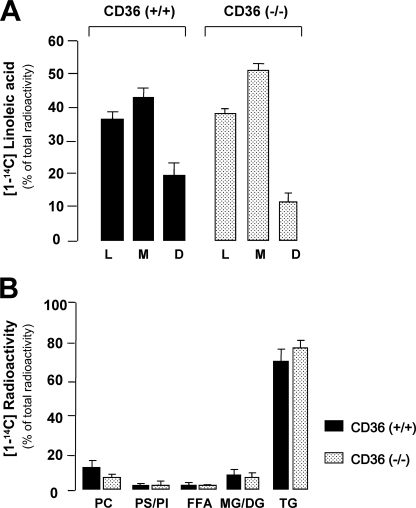
To determine whether CD36 plays a critical role in intestinal LCFA uptake under close to physiological FA delivery conditions, emulsified lipid solution containing [1-14C]linoleic acid as tracer was infused into the lumen of in situ jejunal loops isolated in fasted wild-type and CD36−/− mice. This in vivo technique maintains both intestinal microclimate (i.e. unstirred water layer and cell polarization) and lymph/blood circulations (31). A large lipid load (2180 μm) was used to mimic the postprandial period. Five minutes after infusion, the distribution of radioactivity in lumen and mucosa was similar in CD36+/+ and CD36−/− mice, confirming that CD36 does not contribute to net lipid absorption in the mouse small intestine (Fig. 1A). Similar results were obtained using a lower concentration of linoleic acid (400 μm) (data not shown). Similarly, analysis of radioactivity incorporation into the different lipid classes in the mucosa did not reveal a difference in TG synthesis between CD36−/− mice and controls (Fig. 1B). Cellular radioactivity was recovered mostly in TG 5 min after infusion, supporting physiological relevance of the experimental model used.
FIGURE 1.
CD36 deficiency does not alter jejunal uptake and esterification of micellar fatty acids in the mouse. Linoleic acid (0.5 ml, 2180 μm) in solution containing [1-14C]linoleic acid (51 mCi/mmol) emulsified with 10 mm taurocholic acid was infused into in situ isolated jejunal loops of fasted mice. L and M correspond to the percentage of [1-14C]linoleic acid remaining respectively in the lumen (L) and the mucosa (M). The percentage of [1-14C]linoleic disappeared (D) (mainly absorbed or oxidized) was determined by subtraction of the radioactivity remaining into M and L from the radioactivity initially infused into the lumen. The values shown are the means ± S.E. (n = 3). B, relative distribution of [1-14C] radioactivity in various lipid classes in the mucosa. PC, phosphatidylcholine; PS/PI, phosphatidylserine/phosphatidylinositol; DG/MG, di- and monoglyceride; FFA, free fatty acid.
Lipid-dependent Disappearance of CD36 from the Luminal Side of Jejunal Epithelium
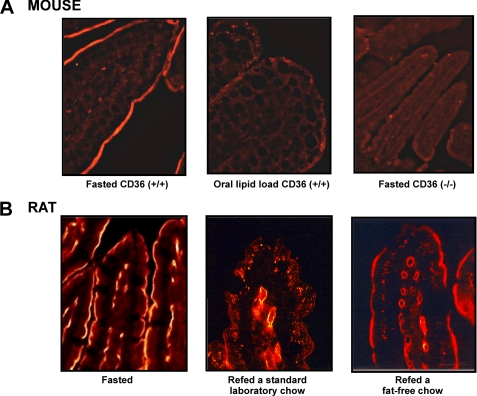
To gain insight into how intestinal CD36 can influence the metabolic fate of LCFA in enterocytes, the effect of nutritional status on CD36 localization in the absorptive epithelium was explored by immunohistochemistry. In fasted mice, prominent staining was detected in the epithelial lining of villi (Fig. 2A). Surprisingly, disappearance of this immunostaining was observed 4 h after an oral lipid load. The fact that no staining was observed in CD36−/− mice confirms signal specificity. Similar results were observed in fasted rats refed a standard laboratory chow containing 3% lipids (w/w). Interestingly, immunostaining persisted when fasted rats were fed a fat-free diet, demonstrating that CD36 removal from the apical side of enterocytes is a physiological event related to dietary lipids.
FIGURE 2.
Lipid-dependent disappearance of CD36 protein from the luminal side of mice and rat jejunum. A, immunolocalization of CD36 in jejunal epithelium in mice. Fasted controls were compared with experimental mice 4 h after force-feeding with 0.5 ml oil. Immunostaining of CD36 on CD36−/− mice demonstrates specificity of the antibody (R & D System). B, immunolocalization of CD36 in jejunal epithelium in the rat. Fasted controls were compared with rats refed for 6 h a standard laboratory chow containing 3% lipids (w/w) or a lipid-free diet. Specific CD36 immunostaining was performed using an antiserum raised in rabbit against purified rat CD36 and a FITC-conjugated anti-rabbit antibody raised in goat. In negative control micrographs, the primary antibody was omitted.
CD36 Removal from the BBM Is Not Associated with CD36 Recovery in Intracellular Fractions
To explore whether the disappearance of staining resulted from a translocation of CD36 from the apical plasma membrane to an intracellular pool as reported for scavenger receptor class B, type I (35), CD36 content in jejunal mucosa homogenates and in purified BBM was assayed by sandwich ELISA. Fasted rats were subjected to an oral lipid load (3 ml of sunflower oil) to optimize the detection of changes in CD36 localization. To confirm lipid dependence, a set of rats was force-fed a sucrose solution providing a similar caloric load to the oil gavage. Activity of sucrase, a BBM enzyme marker, was determined to verify the efficiency of BBM purification. BBM fractions from fasted and force-fed rats exhibited equivalent enrichment and degree of purity and thus can be compared (Table 1).
TABLE 1.
Characteristics of BBM fractions isolated from rat jejunal mucosa
Homogenates and BBM fractions were prepared as indicated under “Experimental Procedures.” Sucrase activity was evaluated in each fraction. Five rats were used in each group. NS, not significant.
| Total sucrase activity |
Enrichment factor (BBM/homogenate) | Efficiency of BBM extraction | ||
|---|---|---|---|---|
| Homogenate | BBM | |||
| units/min/mg of protein | % | |||
| Fasted controls | 0.79 ± 0.09 | 11.53 ± 1.52 | 14.82 ± 1.43 | 13.44 ± 0.96 |
| Oil load | 0.50 ± 0.06 (NS) | 9.72 ± 0.85 (NS) | 20.17 ± 2.38 (NS) | 10.51 ± 1.72 (NS) |
| Sucrose load | 0.66 ± 0.11 (NS) | 11.00 ± 0.42 (NS) | 16.67 ± 0.56 (NS) | 15.27 ± 0.53 (NS) |
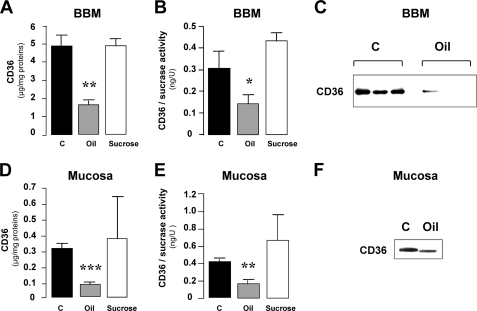
Consistent with earlier data (Fig. 2B), a lipid load led to a 3-fold decrease of CD36 in jejunal BBM from rats 6 h after gavage (Fig. 3, A and C). Similar results were found when the data were expressed as a ratio of CD36 level to sucrase activity, demonstrating that it was BBM-related (Fig. 3B). In agreement with our earlier immunohistochemistry results, this effect was lipid-mediated because no change was observed in sucrose force-fed animals (Fig. 3, A and B). To explore whether lipid-mediated disappearance of CD36 from BBM is associated with intracellular transfer of the protein, CD36 was assayed in jejunal homogenates by both ELISA (Fig. 3, D and E) and Western blotting (Fig. 3F). Total CD36 content in mucosa of lipid force-fed rats remained low as compared with fasted controls.
FIGURE 3.
CD36 removal from BBM does not quantitatively match its intracellular transfer. CD36 content in purified BBM and jejunal mucosa from fasted control rats (C) and from rats 6 h after force-feeding with 3 ml of oil were assayed by both ELISA (A, B, D, and E) and Western blotting (C and F). To determine whether the oil effect on CD36 levels in BBM and jejunal mucosa is specific, a set of animals were also force-fed with a sucrose solution providing a similar caloric load as the oil gavage. The activity of sucrase, a BBM enzyme marker, was determined to verify the efficiency of BBM purification (B and E). The values are the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To further analyze this result, CD36 content in cytosolic and microsomal/endosomal fractions (105,000 × g supernatants and pellets, respectively) isolated from jejunal homogenates was assayed by ELISA. Cytosol CD36 content was 8-fold higher in lipid-fed than in fasted rats (2.47 ± 0.34% of total CD36 versus 0.29 ± 0.09%; p < 0.05, n = 3). However, it remained dramatically low and could not account for the 65% decrease in protein in BBM fractions and homogenates. Because no CD36 was found in microsomal/endosomal fractions (data not shown), these results strongly suggest that a major part of internalized intestinal CD36 is degraded 6 h after the lipid load.
Dietary Lipids Specifically Induce a Rapid Drop in Jejunal CD36 Content
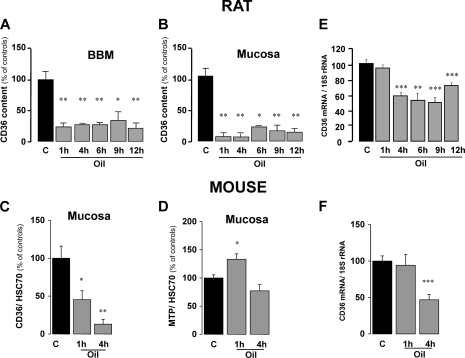
To study the kinetics of the lipid-mediated disappearance of CD36 protein, CD36 content in whole mucosa and in purified BBM was explored at various times following the oral lipid load in rats. As shown in Fig. 4A, the amount of CD36 in BBM dramatically decreased 1 h after force-feeding. This effect remained significant 12 h after the gavage, likely because of the high lipid load used in this experiment. The same pattern was observed when CD36 was assayed in the whole mucosa, which excludes quantitative CD36 accumulation in the intracellular compartment (Fig. 4B). Similar data were also obtained in the mouse (Fig. 4C). The lipid-mediated decrease in CD36 protein in jejunal mucosa is specific to this scavenger receptor because two other lipid-binding proteins, coexpressed with CD36 in differentiated enterocytes, were either unchanged (intestinal fatty acid-binding protein; data not shown) or increased (MTP) (Fig. 4D). The lipid-mediated drop in intestinal CD36 levels did not associate with a change in CD36 mRNA 1 h after the lipid load (Fig. 4, E and F). In contrast, a significant decrease in CD36 mRNA levels occurred 4 h after the load in rats (Fig. 4E) and mice (Fig. 4F). Thus, two regulatory mechanisms seem to be involved in the decrease of intestinal CD36 content triggered by dietary lipids: an early post-translational mechanism followed by a more delayed transcriptional regulation.
FIGURE 4.
Time course of lipid-induced drop in jejunal CD36 protein content. A and B, CD36 content in purified BBM (A) and jejunal mucosa (B) from fasted and oil gavaged rats was assayed by ELISA at 1, 4, 6, 9, and 12 h after the gavage. C, expression of CD36 in mice mucosa was analyzed by Western blotting (15 μg) at 1 and 4 h after force-feeding with oil and compared with fasted controls. D, to explore the specificity of the oil effect on CD36, expression of MTP was also examined. These data were normalized to HSC70 expression, which remained unchanged in these experimental conditions and served as an internal control for protein loading. The values are the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01. E and F, CD36 mRNA levels were assayed in jejunal mucosa from fasted control rats and experimental animals at 1, 4, 6, 12, and 30 h after force-feeding with 3 ml of sunflower oil (E) and in jejunal mucosa from fasted and experimental mice at 1 or 4 h after force-feeding with 0.5 ml of oil (F). The data were normalized to 18 S rRNA for differences in RNA loading. The values are the means ± S.E. (n = 5). **, p < 0.01; ***, p < 0.001.
Dietary Lipids Trigger Degradation of Intestinal CD36 via the Ubiquitin-Proteasome Pathway
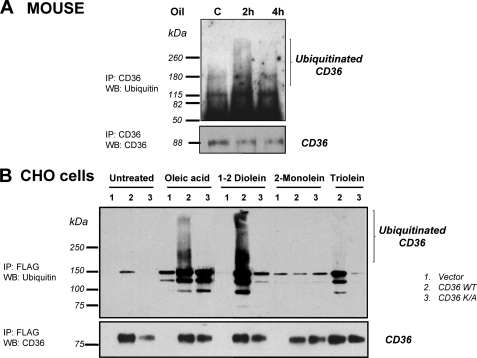
Polyubiquitination tags proteins targeted to proteasomes or lysosomes for subsequent degradation (36). To determine whether the lipid-mediated drop in CD36 levels measured in intestinal mucosa involves the ubiquitin-proteasome pathway, jejunal CD36 from mice subjected or not to a lipid load was immunoprecipitated before immunoblotting with an antibody against ubiquitin. The CD36 signal obtained at 2 h was smeared, which is typical of polyubiquitination. This smearing was diminished at 4 h after the lipid load, likely reflecting degradation of the ubiquitinated protein (Fig. 5A). Therefore, dietary fat induces in the small intestine rapid ubiquitination of CD36.
FIGURE 5.
Lipids induce polyubiquitination of jejunal CD36 in vivo and in CD36-transfected CHO cells in vitro. A, fasted control mice were force-fed with 0.5 ml of oil and sacrificed 2 or 4 h later. The jejunal mucosa homogenates were immunoprecipitated (IP) with anti-CD36 antibody. Immunocomplexes were analyzed by Western blotting (WB) with anti-ubiquitin or anti-CD36 antibodies. The data are representative of three different experiments. B, CHO cells transfected with an empty vector (lane 1), FLAG-CD36 (lane 2), or double mutated CD36 (K469A and K472A) (lane 3) were treated 2 h with (100 μm) of the indicated lipid; oleic acid, 1–2 diolein, and 2-monolein were added in presence of BSA. The cells were lysed, and clarified lysates were immunoprecipitated (IP) with FLAG affinity gel. The immunocomplexes were then analyzed by Western blotting (WB) using an anti-ubiquitin antibody or an anti-CD36 antibody.
Fat digestion produces 1,2-diglycerides, 2-monoglycerides, and free LCFA. To explore which among these lipids might be responsible for down-regulation of CD36 levels, ubiquitination of FLAG-tagged human CD36 (FLAG-CD36) stably expressed in CHO cells was examined next. To determine whether Lys-469 and Lys-472 in the carboxyl-cytoplasmic tail of CD36 are the putative ubiquitination sites, CHO cells stably expressing a FLAG-CD36 double mutant where the Lys were substituted by Ala were used. Incubation of CHO cells for 2 h in presence of 100 μm of the various lipids showed that oleic acid and 1,2-diolein led to CD36 ubiquitination (Fig. 5B, lanes 2). This effect was dependent on presence of Lys-469 and Lys-472 because it was not observed with mutated CD36 (Fig. 5B, lanes 3). In contrast, 2-monoolein and triolein were inefficient in inducing CD36 ubiquitination. To assess whether ubiquitination induces proteosomal degradation of CD36, fasted mice were treated with the proteasome inhibitor MG132 before gavage with oil. Consistent with our earlier data (Fig. 4C), a dramatic drop in CD36 content in the intestinal mucosa took place 4 h after the gavage. MG132 treatment prevented this lipid-mediated decrease, demonstrating involvement of the proteasome in CD36 degradation (Fig. 6, A and B).
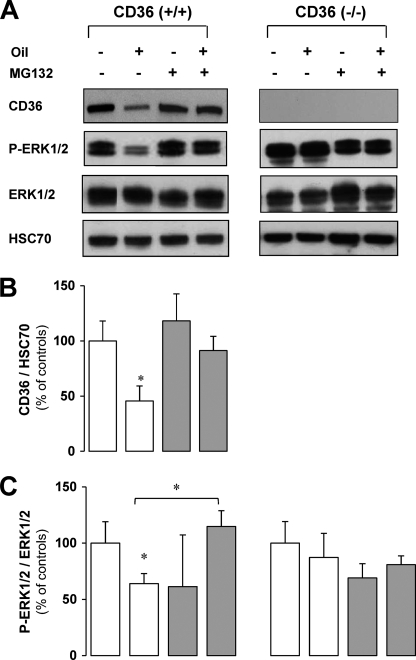
FIGURE 6.
Degradation of CD36 by the proteasome (A and B) leads to a decrease in the ERK1/2 (A and C) activation in CD36+/+ mice but not in CD36−/− mice. Control fasted CD36+/+ and CD36−/− mice were injected intraperitoneally with MG132 (14 mg/kg) 30 min before gavage (0.5 ml oil) and sacrificed 4 h later. CD36, HSC70, phosphorylated ERK1/2 (P-ERK1/2), and ERK1/2 levels in jejunal mucosa were analyzed by Western blotting. A representative signal is shown from four independent experiments, quantified by densitometry, and standardized to HSC70 signal or total ERK1/2 as the loading control. The data are expressed as percentages of controls not treated with MG132. The values are the means ± S.E. (n = 4). *, p < 0.05.
The Lipid-mediated Degradation of Intestinal CD36 Affects the Activation Levels of ERK1/2, ApoB48, and MTP Protein Expression
There is evidence from the literature indicating that ligand binding to CD36 mediates cell-specific responses especially via activation of the MAPKs family pathway (37) including ERK1/2 (27, 38). Because the modulation of ERK1/2 activation has been demonstrated to be involved in the assembly of apoB containing lipoproteins (24, 25), we hypothesized that a defect in CD36 signaling could be involved in the reduction of intestinal lipoprotein production in CD36−/− mice (21).
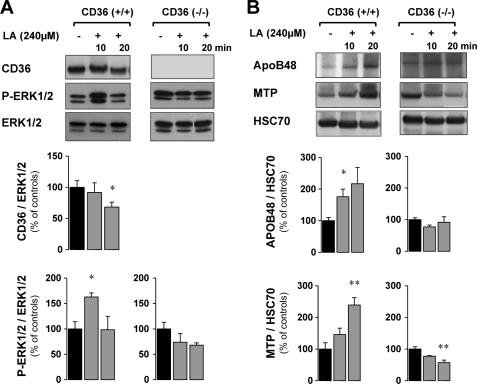
To examine whether the lipid-mediated degradation of CD36 observed in the postprandial period is linked to ERK activation in vivo, the phosphorylation of this MAPK was measured in mouse intestines after the lipid load when CD36 was degraded or not by MG132 treatment (Fig. 6A). As shown in Fig. 6B, there was basal activation of ERK1/2 in the fasted state (control), which was similar in CD36+/+ and CD36−/− mice. However, in CD36+/+ mice, 4 h after the lipid ingestion as CD36 protein levels significantly decreased, an associated decrease in ERK1/2 activation was observed (Fig. 6B). Moreover, this decrease was blunted when degradation of CD36 was prevented by MG132. In contrast, in CD36−/− mice neither the lipid load nor MG132 treatment significantly affected ERK1/2 activation (Fig. 6, A and C). Together, these in vivo data suggested that dietary LCFA mediated ERK1/2 activation during intestinal absorption is dependent on CD36 protein level. This interpretation would imply that LCFA are able to activate ERK1/2 via CD36 before its degradation. The direct effect of LCFA on CD36 protein level and ERK activation was determined ex vivo, in isolated intestinal segments from CD36+/+ and CD36−/− mice incubated in presence of linoleic acid (240 μm, complexed with BSA) for a short time (10 to 20 min) to avoid CD36 degradation. As expected, LCFA treatment led to activation of ERK1/2 after 5 min (data not shown) and 10 min (+ 60%), when the level of CD36 had not decreased yet. After 20 min of LCFA treatment, ERK1/2 activation was reduced, but at that time CD36 protein levels were significantly decreased (−30%) (Fig. 7A). In agreement with our in vivo data, LCFA treatment did not affect ERK1/2 activation in intestinal segments derived from CD36−/− mice (Fig. 7A).
FIGURE 7.
The LCFA-mediated ERK1/2 activation requires CD36 and triggers an up-regulation of apoB48 and MTP protein expression. Isolated intestinal segments from CD36+/+ and CD36−/− mice were cultured in the presence of linoleic acid (240 μm, complexed with BSA) for 10–20 min. A, CD36, phosphorylated ERK1/2 (P-ERK1/2), and ERK1/2 levels in jejunal mucosa were analyzed by Western blotting. A representative signal is shown from four independent experiments, quantified by densitometry, and standardized to total ERK1/2 as the loading control. B, apoB48, MTP, and HSC70 levels in jejunal mucosa were analyzed by Western blotting. A representative signal is shown from four independent experiments, quantified by densitometry, and standardized to HSC70 as the loading control. The data are expressed as percentages of controls not treated with LCFA. The values are the means ± S.E. (n = 4). *, p < 0.05; **, p < 0.01.
These data demonstrated that LCFA induce CD36-dependent ERK1/2 activation and subsequently CD36 degradation, which correlates with a reduction of ERK1/2 activation. Finally, to determine whether the CD36-associated modulation of ERK1/2 affects proteins involved in formation of chylomicrons, apoB48 and MTP protein content was evaluated in the mucosa of intestinal segment isolated from CD36+/+ and CD36−/− mice and cultured with LCFA. Our data demonstrated that LCFA treatment increased apoB48 after 10 min when lipid-mediated activation of ERK1/2 was highest and MTP after 20 min when ERK1/2 activation was down-regulated in CD36+/+ mouse segments. These effects were not observed in CD36−/− mouse segments (Fig. 7B).
DISCUSSION
CD36 deficiency has been documented to impair intestinal chylomicron production, but how this occurs remains unclear. In this report, we provide novel insight into the potential underlying mechanisms. We present the first demonstration that CD36 disappears from the luminal side of intestinal villi early during the postprandial period. This phenomenon is lipid-dependent and is triggered by LCFA and/or diglycerides derived from gastric and pancreatic digestion of dietary TG. In addition it is highly lipid-sensitive and observed in both rats and mice fed a standard chow containing only 3% lipids. We also show that lipid-induced down-regulation of CD36 in the intestinal mucosa reflects targeted CD36 proteolysis by the ubiquitin-proteasome pathway. Moreover, we document in vivo and ex vivo that CD36 is required for dietary lipid activation of ERK1/2 and that the activation level parallels that of CD36 protein. Finally, we demonstrate, using ex vivo experiments, that CD36/lipid-dependent modulation of ERK1/2 associates a sequential up-regulation of apoB48 and MTP, two proteins required for the assembly of large chylomicrons.
Indirect evidence suggesting lipid induced down-regulation of CD36 was previously reported in rat red quadriceps where CD36 level decreased 2 h after intravenous infusion of a TG emulsion (39). A potential mechanism was provided by the finding in C2C12 myotubes that LCFA can induce CD36 ubiquitination and its subsequent degradation by the proteasome (33). Our findings indicate that during lipid absorption, digestive products, in particular LCFA and diglycerides, induce down-regulation of luminal CD36 content in the intestinal mucosa via its internalization and targeting to degradation.
This rapid degradation of CD36 in the intestine argues against its efficient participation in LCFA uptake during the postprandial period. Consistent with this, CD36 deletion does not affect lipid uptake (Fig. 1) using in situ isolated intestinal segments and a micellar FA delivery system. This model keeps intact the unstirred water layer lining the BBM, a property of enterocytes that is not applicable to other lipid-utilizing cells (i.e. adipocytes, myocytes, and hepatocytes). Our data are in agreement with the fact that fecal lipid content was not altered in the CD36−/− mice (22). Enterocytes are subjected to very high levels of lipid during postprandial periods, which is not the case for other cells. Luminal lipid levels would greatly exceed CD36 capacity because the protein functions at low (nanomolar) concentrations of LCFA. In addition, the unstirred water layer, which has a low pH gradient, induces LCFA protonation, which facilitates their membrane permeation (for review see Ref. 40). The findings in this study together with previously published work (19) suggest that CD36 likely functions in the early stages of absorption to initiate events that regulate intracellular FA processing.
There is a large body of evidence to support the function of membrane CD36 in transducing intracellular signals after binding a number of ligands such as thrombospondin-1 in endothelial cells (41, 42), oxidized LDL in macrophages (43), and LCFA in taste bud cells (8, 9). CD36 localizes to plasma membrane detergent-resistant microdomains implicated in cell signaling (44). CD36-dependent signal transduction is generally associated with activation of Src and MAP family kinases (for review see Ref. 37). The cytoplasmic C-terminal tail of CD36 mediates these interactions in various tissues (10, 42, 43, 45), including mouse taste buds (9, 10). Moreover, in macrophages it has been demonstrated that the C-terminal domain of CD36 is associated with MEKK2 (a MAP kinase pathway component) (43), and CD36-dependent activation of the ERK1/2 has been demonstrated in at least two studies (27, 38). Our data support the involvement of ERK1/2 in CD36-mediated regulation of chylomicron formation. Indeed, in vivo as well as ex vivo LCFA activation of ERK1/2 is tightly correlated with CD36 protein levels and is reduced in parallel with CD36 during lipid absorption. Furthermore CD36 is required for LCFA activation of ERK1/2 as shown by our in vivo and ex vivo studies using CD36−/− mice. As with numerous surface receptors (36) exposed to persistent and high supply of ligand, intestinal CD36 is down-regulated by its LCFA ligands via ubiquitination and degradation. This would serve as a feedback regulatory and potentially protective mechanism (36, 46).
Interestingly, the LCFA-induced down-regulation may also be important for optimization of chylomicron formation. The lipid CD36-dependent ERK1/2 activation associated with an early increase in intestinal apoB48 versus a more delayed increase in MTP. These two proteins are essential for the assembly and secretion of chylomicrons (40). In the small intestine, MTP is required for transfer of the TG to the apoB-containing prechylomicrons in the lumen of the endoplasmic reticulum, a process that plays a rate-limiting role in lipoprotein synthesis. Mice with conditional intestine-specific MTP deficiency exhibit fat malabsorption, cytoplasmic lipid droplet accumulation in enterocytes, and virtual disappearance of apoB48 lipoproteins (47). ApoB48 is the preferred protein coat for chylomicron lipid and is needed for efficient chylomicron formation (48). Furthermore, our data are in agreement with the lower lymphatic apoB48 secretion observed in CD36−/− mice (22).
MAPK activation has been previously implicated in regulating levels of the apoB48 and MTP proteins. MEK1/2 inhibition (and consequently ERK1/2 inhibition) decreased apoB48 production in hamster enterocytes (25). In contrast, the ERK1/2 cascade exerts an inhibitory effect on MTP gene transcription (26). In the liver, inhibition of the ERK phosphorylation pathway triggers formation of large sized apoB lipoproteins (24). Consistent with these findings, in our studies, MG132 treatment of CD36+/+ mice, which prevents the reduction in ERK1/2 phosphorylation, also prevents up-regulation of MTP mRNA (data not shown). Together, the data suggest that dietary lipid-induced CD36 degradation may play a role in the formation of large chylomicrons via down-regulation of ERK1/2 activation and subsequent up-regulation of MTP.
We thus propose a model in which LCFA binding to the extracellular domains of CD36 at the early stages of intestinal lipid absorption activates ERK1/2, which triggers an increase in mucosal apoB48 protein. LCFA also induce CD36 ubiquitination. In presence of high LCFA, a significant proportion of CD36 is progressively ubiquitinated and degraded, leading to a decrease in CD36 levels. Consequently, CD36-dependent activation of intestinal ERK1/2 also decreases, which would be important to trigger the increase in MTP protein required for apoB48 lipidation (24) and chylomicron formation. These CD36-dependent events may explain why in CD36−/− mice subjected to high fat diet, more intestinal lipid retention is observed and why smaller lipoprotein particles are secreted (21). In conclusion, our data indicate that CD36 degradation, which may be dependent on the LCFA content of the diet, could be the signal leading to the formation of large apoB48-rich TG lipoproteins via reducing ERK1/2 activation and increasing MTP level. Overall, these novel observations suggest that CD36 operates as a lipid sensor, responsible for transducing signals related to the dietary lipid content that optimize the formation of large chylomicrons containing apoB48.
A better understanding of the factors that regulate CD36 and its signaling pathways and how they contribute to lipid-dependent intestinal adaptation (31, 40) is important. It might provide new therapeutic approaches and/or dietary recommendations that optimize chylomicron size and consequently blood chylomicron clearance to decrease the prevalence of postprandial hypertriglyceridemia and the associated risk of cardiovascular diseases.
This work was supported, in whole or in part, by the National Institute of Health and Medical Research (INSERM) and by grants from the Research Program in Human Nutrition SensoFAT of the French National Research Agency (L'Agence Nationale de la Recherche, ANR) (to P. B.) and from the Burgundy Council (to P. B.). This work was also supported by National Institutes of Health Grants R01 DK60022 and DK33301 (to N. A. A.).
- LCFA
- long chain fatty acid(s)
- MTP
- microsomal triglyceride transfer protein
- apoB
- apolipoprotein B
- HSC70
- heat shock cognate protein 70
- TG
- triglycerides
- BBM
- brush border membrane.
REFERENCES
- 1. Habets D. D., Coumans W. A., Voshol P. J., den Boer M. A., Febbraio M., Bonen A., Glatz J. F., Luiken J. J. (2007) Biochem. Biophys. Res. Commun. 355, 204–210 [DOI] [PubMed] [Google Scholar]
- 2. Coburn C. T., Knapp F. F., Jr., Febbraio M., Beets A. L., Silverstein R. L., Abumrad N. A. (2000) J. Biol. Chem. 275, 32523–32529 [DOI] [PubMed] [Google Scholar]
- 3. Ibrahimi A., Sfeir Z., Magharaie H., Amri E. Z., Grimaldi P., Abumrad N. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. (1993) J. Biol. Chem. 268, 11811–11816 [PubMed] [Google Scholar]
- 5. Chen C. H., Cartwright J., Jr., Li Z., Lou S., Nguyen H. H., Gotto A. M., Jr., Henry P. D. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 1303–1312 [DOI] [PubMed] [Google Scholar]
- 6. Ren Y., Silverstein R. L., Allen J., Savill J. (1995) J. Exp. Med. 181, 1857–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oquendo P., Hundt E., Lawler J., Seed B. (1989) Cell 58, 95–101 [DOI] [PubMed] [Google Scholar]
- 8. Laugerette F., Passilly-Degrace P., Patris B., Niot I., Febbraio M., Montmayeur J. P., Besnard P. (2005) J. Clin. Invest. 115, 3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaillard D., Laugerette F., Darcel N., El-Yassimi A., Passilly-Degrace P., Hichami A., Khan N. A., Montmayeur J. P., Besnard P. (2008) FASEB J. 22, 1458–1468 [DOI] [PubMed] [Google Scholar]
- 10. El-Yassimi A., Hichami A., Besnard P., Khan N. A. (2008) J. Biol. Chem. 283, 12949–12959 [DOI] [PubMed] [Google Scholar]
- 11. Luiken J. J., Dyck D. J., Han X. X., Tandon N. N., Arumugam Y., Glatz J. F., Bonen A. (2002) Am. J. Physiol. Endocrinol. Metab. 282, E491–E495 [DOI] [PubMed] [Google Scholar]
- 12. Bonen A., Han X. X., Habets D. D., Febbraio M., Glatz J. F., Luiken J. J. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1740–E1749 [DOI] [PubMed] [Google Scholar]
- 13. Yamashita S., Hirano K., Kuwasako T., Janabi M., Toyama Y., Ishigami M., Sakai N. (2007) Mol. Cell Biochem. 299, 19–22 [DOI] [PubMed] [Google Scholar]
- 14. Love-Gregory L., Sherva R., Sun L., Wasson J., Schappe T., Doria A., Rao D. C., Hunt S. C., Klein S., Neuman R. J., Permutt M. A., Abumrad N. A. (2008) Hum. Mol. Genet. 17, 1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma X., Bacci S., Mlynarski W., Gottardo L., Soccio T., Menzaghi C., Iori E., Lager R. A., Shroff A. R., Gervino E. V., Nesto R. W., Johnstone M. T., Abumrad N. A., Avogaro A., Trischitta V., Doria A. (2004) Hum. Mol. Genet. 13, 2197–2205 [DOI] [PubMed] [Google Scholar]
- 16. Poirier H., Degrace P., Niot I., Bernard A., Besnard P. (1996) Eur. J. Biochem. 238, 368–373 [DOI] [PubMed] [Google Scholar]
- 17. Lobo M. V., Huerta L., Ruiz-Velasco N., Teixeiro E., de la Cueva P., Celdrán A., Martín-Hidalgo A., Vega M. A., Bragado R. (2001) J. Histochem. Cytochem. 49, 1253–1260 [DOI] [PubMed] [Google Scholar]
- 18. Nassir F., Wilson B., Han X., Gross R. W., Abumrad N. A. (2007) J. Biol. Chem. 282, 19493–19501 [DOI] [PubMed] [Google Scholar]
- 19. Drover V. A., Nguyen D. V., Bastie C. C., Darlington Y. F., Abumrad N. A., Pessin J. E., London E., Sahoo D., Phillips M. C. (2008) J. Biol. Chem. 283, 13108–13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Masuda D., Hirano K., Oku H., Sandoval J. C., Kawase R., Yuasa-Kawase M., Yamashita Y., Takada M., Tsubakio-Yamamoto K., Tochino Y., Koseki M., Matsuura F., Nishida M., Kawamoto T., Ishigami M., Hori M., Shimomura I., Yamashita S. (2009) J. Lipid Res. 50, 999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drover V. A., Ajmal M., Nassir F., Davidson N. O., Nauli A. M., Sahoo D., Tso P., Abumrad N. A. (2005) J. Clin. Invest. 115, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nauli A. M., Nassir F., Zheng S., Yang Q., Lo C. M., Vonlehmden S. B., Lee D., Jandacek R. J., Abumrad N. A., Tso P. (2006) Gastroenterology 131, 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quarfordt S. H., Goodman D. S. (1966) Biochim. Biophys. Acta 116, 382–385 [DOI] [PubMed] [Google Scholar]
- 24. Tsai J., Qiu W., Kohen-Avramoglu R., Adeli K. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 211–218 [DOI] [PubMed] [Google Scholar]
- 25. Federico L. M., Naples M., Taylor D., Adeli K. (2006) Diabetes 55, 1316–1326 [DOI] [PubMed] [Google Scholar]
- 26. Au W. S., Kung H. F., Lin M. C. (2003) Diabetes 52, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 27. Kuda O., Jenkins C. M., Skinner J. R., Moon S. H., Su X., Gross R. W., Abumrad N. A. (2011) J. Biol. Chem. 286, 17785–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shirazi-Beechey S. P., Davies A. G., Tebbutt K., Dyer J., Ellis A., Taylor C. J., Fairclough P., Beechey R. B. (1990) Gastroenterology 98, 676–685 [DOI] [PubMed] [Google Scholar]
- 29. Dahlqvist A. (1970) Enzymol. Biol. Clin. 11, 52–66 [PubMed] [Google Scholar]
- 30. Delsal J. L. (1954) Bull. Soc. Chim. Biol. 36, 1329–1334 [PubMed] [Google Scholar]
- 31. Petit V., Arnould L., Martin P., Monnot M. C., Pineau T., Besnard P., Niot I. (2007) J. Lipid Res. 48, 278–287 [DOI] [PubMed] [Google Scholar]
- 32. Pardo V. G., Facchinetti M. M., Curino A., Boland R., de Boland A. R. (2007) Biogerontology 8, 13–24 [DOI] [PubMed] [Google Scholar]
- 33. Smith J., Su X., El-Maghrabi R., Stahl P. D., Abumrad N. A. (2008) J. Biol. Chem. 283, 13578–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelsers M. M., Lutgerink J. T., Nieuwenhoven F. A., Tandon N. N., van der Vusse G. J., Arends J. W., Hoogenboom H. R., Glatz J. F. (1999) Biochem. J. 337, 407–414 [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen G. H., Niels-Christiansen L. L., Immerdal L., Danielsen E. M. (2003) Gut 52, 1424–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miranda M., Sorkin A. (2007) Mol. Interv. 7, 157–167 [DOI] [PubMed] [Google Scholar]
- 37. Silverstein R. L., Febbraio M. (2009) Sci. Signal. 2, re3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baranova I. N., Bocharov A. V., Vishnyakova T. G., Kurlander R., Chen Z., Fu D., Arias I. M., Csako G., Patterson A. P., Eggerman T. L. (2010) J. Biol. Chem. 285, 8492–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hevener A. L., Reichart D., Janez A., Olefsky J. (2001) Diabetes 50, 2316–2322 [DOI] [PubMed] [Google Scholar]
- 40. Niot I., Poirier H., Tran T. T., Besnard P. (2009) Prog. Lipid Res. 48, 101–115 [DOI] [PubMed] [Google Scholar]
- 41. Lisanti M. P., Scherer P. E., Vidugiriene J., Tang Z., Hermanowski-Vosatka A., Tu Y. H., Cook R. F., Sargiacomo M. (1994) J. Cell Biol. 126, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiménez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., Bouck N. (2000) Nat. Med. 6, 41–48 [DOI] [PubMed] [Google Scholar]
- 43. Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., Silverstein R. L. (2006) Cell Metab. 4, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ehehalt R., Sparla R., Kulaksiz H., Herrmann T., Füllekrug J., Stremmel W. (2008) BMC Cell Biol. 9, 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang M. M., Bolen J. B., Barnwell J. W., Shattil S. J., Brugge J. S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 47. Xie Y., Newberry E. P., Young S. G., Robine S., Hamilton R. L., Wong J. S., Luo J., Kennedy S., Davidson N. O. (2006) J. Biol. Chem. 281, 4075–4086 [DOI] [PubMed] [Google Scholar]
- 48. Lo C. M., Nordskog B. K., Nauli A. M., Zheng S., Vonlehmden S. B., Yang Q., Lee D., Swift L. L., Davidson N. O., Tso P. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G344–G352 [DOI] [PubMed] [Google Scholar]