Abstract
Myelodysplastic syndromes (MDS) are characterized by abnormal and dysplastic maturation of all blood lineages. Even though epigenetic alterations have been seen in MDS marrow progenitors, very little is known about the molecular alterations in dysplastic peripheral blood cells. We analyzed the methylome of MDS leukocytes by the HELP assay and determined that it was globally distinct from age-matched controls and was characterized by numerous novel, aberrant hypermethylated marks that were located mainly outside of CpG islands and preferentially affected GTPase regulators and other cancer-related pathways. Additionally, array comparative genomic hybridization revealed that novel as well as previously characterized deletions and amplifications could also be visualized in peripheral blood leukocytes, thus potentially reducing the need for bone marrow samples for future studies. Using integrative analysis, potentially pathogenic genes silenced by genetic deletions and aberrant hypermethylation in different patients were identified. DOCK4, a GTPase regulator located in the commonly deleted 7q31 region, was identified by this unbiased approach. Significant hypermethylation and reduced expression of DOCK4 in MDS bone marrow stem cells was observed in two large independent datasets, providing further validation of our findings. Finally, DOCK4 knockdown in primary marrow CD34+ stem cells led to decreased erythroid colony formation and increased apoptosis, thus recapitulating the bone marrow failure seen in MDS. These findings reveal widespread novel epigenetic alterations in myelodysplastic leukocytes and implicate DOCK4 as a pathogenic gene located on the 7q chromosomal region.
Keywords: Epigenetics, Gene Expression, Gene Silencing, Microarray, Stem Cell
Introduction
The myelodysplastic syndromes (MDS)3 are collections of heterogeneous hematological diseases characterized by refractory cytopenias due to ineffective hematopoiesis. Recent evidence suggests that stem cells in MDS are characterized by aberrant transcriptional profiles and that deregulation of gene expression may account for abnormal growth and differentiation of these progenitors (1, 2). One of the ways that gene expression may be dysregulated is through aberrant DNA methylation. Methylation of cytosine has been implicated as a way to silence genes epigenetically and indicates an attractive target for potential therapeutics (3). Aberrant methylation of promoters of genes such as p15,DAPK, and others has been reported in MDS (4, 5). Even though these are important cell cycle and apoptosis genes, methylation of their promoter CpGs has not correlated very well with clinical responses after treatment with DNA methyltransferase inhibitors in most studies (7, 8). It is possible that global studies of the DNA methylome in MDS may yield an epigenetic signature that is better as a diagnostic and prognostic tool than single locus studies. Early attempts at global methylation analysis of MDS using a microarray covering 1,505 CpG islands have shown aberrant hypermethylation of selected genes in MDS and their involvement in progression to AML (9). Their study opened up the possibility that assays with better resolution and coverage not restricted to CpG islands alone may yield more informative insights into the MDS methylome.
Several experimental approaches are available to determine genome-wide DNA methylation levels. Most of these techniques are based on restriction enzyme digestion or DNA immunoprecipitation with antibodies that bind to methylated CpGs (10). Among the restriction enzyme-based methods, some involve comparing the profiles from digestion of DNA with methylation-sensitive and -insensitive restriction enzymes (11, 12). The HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay is based on this principle and relies on differential digestion by a pair of enzymes, HpaII and MspI, that differ on the basis of their methylation sensitivity. These enzymes cut at the same CpG-containing sites (CCGG), but HpaII is unable to cleave the sites that are methylated. Thus, the DNA segments generated by these two digestions will vary in composition based on the amount of methylation. The HpaII and MspI genomic representations can be cohybridized to a custom microarray and their ratio used to indicate the methylation of particular CCGG sites at these loci. The HELP assay has been shown to be a robust discovery tool for flagging loci for subsequent quantitative and nucleotide resolution bisulfite analyses (MassArray and Pyrosequencing) that represent the gold standard tests for cytosine methylation (13–15).
In addition to epigenetic alterations, MDS is also characterized by many cytogenetic abnormalities that may contribute to its pathogenesis. Recent studies have shown that higher resolution microarray-based technologies such as comparative genomic hybridization (aCGH) and single nucleotide polymorphism microarrays can reveal cytogenetic abnormalities not seen by conventional methods (16–18). In this study, we tested whether it is important to study the effect of genetic and epigenetic abnormalities together to obtain a comprehensive insight into MDS pathogenesis. We have developed an integrated genomics and epigenomics platform based on the combination of the HELP assay and aCGH and have used it on MDS samples. We have used MDS peripheral blood cells as very little is known about the molecular and epigenetic makeup of these dysplastic cells. We wanted to determine whether aberrant epigenetic marks can be observed in MDS peripheral blood cells and whether these cells could be used for these studies instead of hard to obtain marrow samples. Our studies showed that methylation changes could be seen in peripheral blood leukocytes and were of sufficient magnitude to discriminate MDS leukocytes from age-matched controls. Similarly, both novel and well characterized genomic copy number changes were also found in these peripheral blood cells. Using integrative analysis, common sets of genes were identified that were affected in different patients by genetic deletion events and the epigenetic events of aberrant methylation. One of the genes identified by this unbiased approach was DOCK4, which is located in the commonly deleted chromosome 7q31 region. DOCK4 was found to be epigenetically silenced in both peripheral leukocytes and marrow stem cells in MDS. We determined that Dock4 knockdown leads to ineffective hematopoiesis, thus implicating it as a potential candidate gene in MDS and underscoring the power of genome-wide integrative analysis in gene discovery in MDS.
MATERIALS AND METHODS
Patient Samples and Nucleic Acid Extraction
Specimens were obtained from 21 patients diagnosed with MDS and from controls after signed informed consent was approved by the Albert Einstein College of Medicine Institutional Review Board. MDS subtypes included refractory cytopenias with multilineage dysplasia, refractory anemia, refractory anemia with excess blasts, and chronic myelomonocytic leukemia. Peripheral blood leukocytes were isolated after red cell lysis and used for DNA and RNA extraction. Genomic DNA was extracted by a standard phenol-chloroform protocol followed by an ethanol precipitation and resuspension in 10 mm Tris-HCl, pH 8.0. Total RNA was extracted using an RNeasy mini kit from Qiagen (Valencia, CA) and subjected to amplification using the MessageAmp II aRNA kit from Ambion (Foster City, CA).
DNA Methylation Analysis by HELP
The HELP assay was carried out as published previously (14). Intact DNA of high molecular weight was corroborated by electrophoresis on 1% agarose gel in all cases. One microgram of genomic DNA was digested overnight with either HpaII or MspI (New England Biolabs, Ipswich, MA). The following day, the reactions were extracted once with phenol-chloroform and resuspended in 11 μl of 10 mm Tris-HCl, pH 8.0, and the digested DNA was used to set up an overnight ligation of the JHpaII adapter using T4 DNA ligase. The adapter-ligated DNA was used to carry out the PCR amplification of the HpaII- and MspI-digested DNA as described previously (14). Both amplified fractions were submitted to Roche-NimbleGen, Inc. (Madison, WI), for labeling and hybridization onto a human hg17 custom-designed oligonucleotide array (50-mers) covering 25,626 HpaII-amplifiable fragments located at gene promoters. HpaII-amplifiable fragments are defined as genomic sequences contained between two flanking HpaII sites found within 200–2,000 bp from each other. Each fragment on the array is represented by 15 individual probes distributed randomly and spatially across the microarray slide. Thus, the microarray covers 50,000 CpGs corresponding to 14,000 gene promoters.
Quantitative DNA Methylation Analysis by Mass Array Epityping
Validation of HELP microarray findings was carried out by MALDI-TOF mass spectrometry using EpiTYPERTM by MassArray (Sequenom) on bisulfite-converted DNA as described previously (19, 20). MassArray primers were designed to cover the flanking HpaII sites for a given HpaII-amplifiable fragment, as well as any other HpaII sites found up to 2,000 bp upstream of the downstream site and up to 2,000 bp downstream of the upstream site, to cover all possible alternative sites of digestion. The primers used were as follows: KLF3_1, forward 5′-AGGAAGAGAGTATTTTAAAGATGAAGTTTATGGGATAGT-3′ and reverse 5′-CAGTAATACGACTCACTATAGGGAGAAGGCTAAACCCTTTAAATTAACCCATCTC-3′; KLF3_2, forward 5′-AGGAAGAGAGTTGAAGGTTATTGAGTTTAGGG-3′ and reverse 5′-CAGTAATACGACTCACTATAGGGAGAAGGCTCTCAACTCACTACAAAAAAAAAA-3′; Dock4-294-593, number 1, forward AGGAAGAGAGGGAGAAAATGTTATGGAATGGTTTTT and reverse CAGTAATACGACTCACTATAGGGAGAAGGCTTCACCTCAACCACAAACTAAACAAA; and Dock4–1299-1597, number 2, forward AGGAAGAGAGGGGTTATTAGTTTAAGATTTAAATTGGTG and reverse CAGTAATACGACTCACTATAGGGAGAAGGCTAAATCATAACTCACCACAACCTCC.
Quantitative Real Time PCR
The expression values of DOCK4 were validated by quantitative RT-PCR. cDNA was synthesized from DNase I-treated total RNA extracted from patient samples using the Superscript III first strand kit from Invitrogen (Superscript III) following the manufacturer's protocol. Real time PCR was performed using SYBR Green PCR master mix from Applied Biosystems (Foster City, CA) with primers specific for DOCK4 and a DNA Engine Opticon 2 real time thermocycler from Bio-Rad. GAPDH was simultaneously amplified with specific primers as housekeeping genes to normalize the DOCK4 expression. The primer sequences are as follows: DOCK4, forward 5′-GGATACCTACGGAGCACGAG-3′ and reverse 5′-AGCCATCACACTTCTCCAGG-3′; glyceraldehyde-3-phosphate dehydrogenase, forward 5′-CGACCACTTTGTCAAGCTCA-3′ and reverse 5′-CCCTGTTGCTGTAGCCAAAT-3′.
Microarray Quality Control
All microarray hybridizations were subjected to extensive quality control using the following strategies. First, uniformity of hybridization was evaluated using a modified version of a previously published algorithm (15) adapted for the NimbleGen platform, and any hybridization with strong regional artifacts was discarded and repeated. Second, normalized signal intensities from each array were compared against a 20% trimmed mean of signal intensities across all arrays in that experiment, and any arrays displaying a significant intensity bias that could not be explained by the biology of the sample were excluded.
HELP Data Processing and Analysis
Signal intensities at each HpaII-amplifiable fragment were calculated as a robust (25% trimmed) mean of their component probe-level signal intensities. Any fragments found within the level of background MspI signal intensity, measured as 2.5 mean-absolute-differences above the median of random probe signals, were categorized as “failed.” These failed loci therefore represent the population of fragments that did not amplify by PCR, whatever the biological (e.g. genomic deletions and other sequence errors) or experimental cause. However, “methylated” loci were so designated when the level of HpaII signal intensity was similarly indistinguishable from background. PCR-amplifying fragments (those not flagged as either methylated or failed) were normalized using an intra-array quantile approach wherein HpaII/MspI ratios are aligned across density-dependent sliding windows of fragment size-sorted data. The log2(HpaII/MspI) was used as a representative for methylation and analyzed as a continuous variable. For most loci, each fragment was categorized as either methylated, if the centered log HpaII/MspI ratio was less than zero, or hypomethylated, if the log ratio was greater than zero.
Microarray Data Analysis
Unsupervised clustering of HELP data by hierarchical clustering was performed using the statistical software R version 2.6.2. A two-sample t test was used for each gene to summarize methylation differences between groups. Genes were ranked on the basis of this test statistic and a set of top differentially methylated genes with an observed log fold change of >1 between group means was identified. Genes were further grouped according to the direction of the methylation change (hypomethylated versus hypermethylated in MDS), and the relative frequencies of these changes were computed among the top candidates to explore global methylation patterns. Extensive validations (shown for KLF3 promoter regions) with MassArray showed good correlation with the data generated by the HELP assay. MassArray analysis validated significant quantitative differences in methylation for differentially methylated genes selected by our approach.
Array-based Comparative Genomic Hybridization (aCGH)
Gene copy number changes were analyzed by high resolution (6 kb) microarray-based comparative genomic hybridization (aCGH) performed on Roche-NimbleGen 385K whole genome tiling arrays (2006–11-01_HG17_WG_CGH). Pooled DNA from healthy cases was used as controls during hybridization. These arrays contain 50–75-mer probes at average spacing of 6270 bp (6 kb). This probe-level aCGH data were analyzed by DNA copy algorithm (Nimblescan software package, Roche-Nimblegen) using five adjacent oligonucleotides and confirmed by circular binary segmentation algorithm (22). Significant DNA copy number changes were cross-referenced from the HapMap data base from NCBI to remove normal variants.
Pathway Analysis and Transcription Factor-binding Site Analysis
Using the Ingenuity Pathway Analysis software (Redwood City, CA), we carried out an analysis of the biological information retrieved by each of the individual platforms alone, and we compared it with the information obtained by the integrated analysis of all three platforms. Enrichment of genes associated with specific canonical pathways was determined relative to the ingenuity knowledge data base for each of the individual platforms and the integrated analysis at a significance level of p < 0.01. Biological networks captured by the different microarray platforms were generated using Ingenuity Pathway Analysis software and scored based on the relationship between the total number of genes in the specific network and the total number of genes identified by the microarray analysis. The list of hypermethylated genes was examined for enrichment of conserved gene-associated transcription factor-binding sites using the Molecular Signatures Database (MSigDB) (23). Their functional gene sets were obtained from Gene Ontology (GO) (24).
This analysis was performed by Gene Set Enrichment Analysis (GSEA) (23), a computational method that determines whether an a priori defined set of genes (commonly hypermethylated genes in MDS) shows statistically significant, concordant differences between two biological states. GSEA calculates an enrichment score (ES) for a given gene set using a rank of genes and infers statistical significance of each ES against ES background distribution calculated by permutation of the original data set. The ES is the maximum deviation from zero of the cumulative sum and can be interpreted as a weighted Kolmogorov-Smirnov statistic. When an entire data base of gene sets is scored, an adjustment was made to the resulting p values to account for multiple hypotheses testing. In this study, the javaGSEA implementation was used for GSEA analysis. The list of differentially methylated HpaII fragments was analyzed using GSEA “pre-ranked” algorithm, which is used when a pre-ordered ranked list is to be analyzed with GSEA. 1,000 permutations were applied to sample labels to test if genes from each a priori defined positional gene sets were randomly distributed along the gene list.
The same method was applied to determine whether transcription-binding sites are randomly distributed in the differentially methylated genes. The a priori defined gene sets used in this analysis is transcription factor target, which contains genes that share a transcription factor-binding site defined in the TRANSFAC (version 7.4) database (25). Using GSEA pre-ranked algorithm, 1000 permutations were applied to sample labels to test if genes from each transcription factor target gene sets were randomly distributed along the differentially methylated gene list. The result shows significant over-representation of binding sites for SP1, AHR, FOXO4, LEF1, NF1, and SOX9 and other transcription factors.
Meta-analysis of MDS and Normal CD34+ Gene Expression Studies
A human bone marrow gene expression dataset, including profiles of 89 cases of MDS CD34+ cells and 61 normal CD34+ profiles was constructed. Individual datasets were obtained from seven independent studies (2, 26–31) from NCBI Gene Expression Omnibus database, an on-line repository of all gene expression profiles reported in the literature (26). Methods to find and extract data have been described previously (32, 33). The datasets were integrated based on UniGene identifications and were quantile-normalized to ensure cross-study comparability, based on our previous approach (32, 33). Analyses were performed using SAS (SAS Institute, Cary, NC) and the R language.
shRNA Gene Knockdown
The human GIPZ lentiviral shRNAmir individual clones targeting DOCK4 (catalog nos. RHS4430-98480907, RHS4430-99166546, and RHS4430-98521322) and the nonsilencing control (catalog no. RHS4346) were obtained from OpenBiosystems (Huntsville, AL). Nucleofection of CD34+ cells was performed according to the manufacturer's instruction using the Nucleofector machine (Amaxa, Cologne, Germany). 106 CD34+ cells (AllCells) were thawed, cultured for 2 h, resuspended in 100 μl of human CD34+ Nucleofection solution (Amaxa), then transferred into cuvettes, and electroporated using program U008. CD34+ cells were collected and cultured in 24-well plates containing 1 ml of prewarmed Stemspan (Stemcell Technologies, Vancouver, British Columbia, Canada), supplemented with 100 ng/ml human Flt-3, stem cell factor, and Tpo for another 24 h before the analysis.
Hematopoietic Colony Assays
24 h after gene knockdown, CD34+ cells were collected, and the shRNA-transfected cells were sorted according to the GFP intensity using Moflow (BD Biosciences). The same numbers of GFP-positive cells were culture in MethoCult GF4434 (StemCell Technologies) containing recombinant human stem cell factor, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-3, and erythropoietin. Granulocyte/macrophage colony-forming units and erythroid burst-forming units were scored on day 14 of culture.
Apoptosis Assay
To detect apoptotic cells, annexin V-APC staining was performed 24 h after the lentiviral transfection using the annexin V-APC apoptosis detection kit (eBioscience) according to the manufacturer's instructions. 7-Aminoactinomycin D was used for the viability staining. Apoptotic cells were analyzed using a FACScan (BD Biosciences).
Immunohistochemistry on Bone Marrow Tissue Microarray
Tissue microarrays were constructed from formalin-fixed, paraffin-embedded bone marrow core biopsies from patients with MDS and control patients with anemia whose bone marrow showed no evidence of neoplasia. The tissue blocks were procured from Jacobi Hospital (Bronx, NY) after approval by the Internal Review Board. For each patient, three 0.5-mm cores were placed in a tissue array using a manual arrayer (Chemicon International, Temecula, CA). Sections of the tissue microarrays were cut to a 5-μm thickness, placed on positively charged slides, and heated to 60 °C for 1 h. They were then deparaffinized in xylene and rehydrated with graded alcohols. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide. Antigen retrieval was accomplished by microwaving the slides in Dako Target Retrieval Solution, pH 6.0 (Dako Cytomation, Dako, Carpinteria, CA), and subsequently steaming them in a vegetable steamer for 30 min. The slides were stained using a rabbit polyclonal anti-DOCK4 antibody, provided by Yajnik and co-workers (34), at 1:200 dilution, followed by Dako EnVision labeled polymer-HRP anti-rabbit antibody. Antibody binding was detected using 3,3-diaminobenzidine chromogen (Cell Marque, Rocklin, CA). The slides were lightly counterstained with hematoxylin, dehydrated with graded alcohols, cleared with xylene, and coverslipped using Cytoseal 60 (Thermo Scientific, Waltham, MA). The tissue cores were then scored for weak versus strong staining for DOCK4 by a hematopathologist who was blinded to the patient identities. Tissue cores that did not contain at least 10% evaluable marrow were excluded from the analysis.
RESULTS
Methylation Profiling on Peripheral Blood Leukocytes Separates Distinct Subsets of MDS from Normal Controls
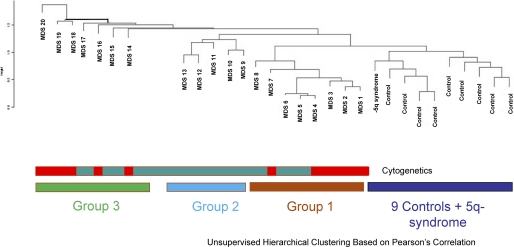
Even though the hallmark of myelodysplastic syndromes is dysplastic appearance of peripheral blood cells, epigenetic and other molecular alterations in these cells have not been examined in detail. We wanted to determine the methylome of these cells by the HELP assay, which is an unbiased high resolution-based assay that has led to the discovery of novel epigenetic alterations in leukemias and other cancers (13, 35, 37). DNA methylation profiles were generated from 21 MDS patient peripheral leukocyte samples and 9 age-matched controls. The MDS samples included all subtypes of this disease (Table 1). The controls included six elderly healthy cases and three patients with anemia of chronic disease. Unsupervised hierarchal clustering showed that the controls formed a cluster that was distinct from MDS samples, demonstrating epigenetic dissimilarity between these groups. Interestingly, a sample from a patient with a 5q syndrome clustered with normals (Fig. 1). The MDS samples formed two clusters with epigenomic similarity to each other (groups 1 and 2), in addition to the rest of samples that demonstrated greater epigenetic heterogeneity (group 3). Because we used peripheral blood leukocytes for these analyses, we wanted to determine whether these epigenetic clusters were due the differing myeloid and lymphoid cell percentages in these samples. We observed that most of the MDS samples had lymphoid and neutrophil percentages that were in the normal range, and clustering was not found be dependent on their relative ratios (Table 1 showing sample characteristics, no significant differences between myeloid and lymphoid percentages between the cases p > 0.05, Proportions Test). Furthermore, epigenetic similarity between clusters of samples was neither dependent on the histological subtypes of MDS nor cytogenetic alterations within these samples. These data demonstrate that significant changes in DNA methylation are seen in MDS leukocytes and are sufficient to clearly distinguish these cases from controls (Fig. 1).
TABLE 1.
MDS patient characteristics
The following abbreviations are used: HGB, GM/DL, NML, normal; RAEB, refractory anemia with excess blasts; RCMD, refractory cytopenias with multilineage dysplasia; RA, refractory anemia; CMML, chronic myelomonocytic leukemia;.
| Sample ID | Subtype | HGB (GM/DL) | WBC (×103) | Platelets (×103) | Cytogenetics | Lymphocytes | Neutrophils |
|---|---|---|---|---|---|---|---|
| % | % | ||||||
| MDS 1 | RAEB | 7.7 | 1 | 24 | −7, −5 | 33 | 44 |
| MDS 2 | RCMD | 11.7 | 1.7 | 32 | −20q | 40 | 45 |
| MDS 3 | RAEB | 9.6 | 3.8 | 1 | Complex | 39 | 29 |
| MDS 4 | RAEB | 9 | 1.4 | 19 | +8 | 47 | 51 |
| MDS 5 | RCMD | 14 | 2.1 | 126 | NML | 26 | 59 |
| MDS 6 | RCMD | 12.6 | 2.1 | 85 | NML | 27 | 56 |
| MDS 7 | RAEB | 9.9 | 2.1 | 5 | Complex | 57 | 32 |
| MDS 8 | RCMD | 8.2 | 4.5 | 101 | NML | 36 | 54 |
| MDS 9 | RCMD | 11.2 | 5.1 | 170 | NML | 15 | 76 |
| MDS 10 | RA | 9 | 6.7 | 335 | NML | 38 | 45 |
| MDS 11 | RCMD | 8.1 | 2.9 | 131 | NML | 37 | 49 |
| MDS 12 | RAEB | 8.9 | 2.2 | 26 | NML | 32 | 49 |
| MDS 13 | RA | 9.8 | 5.8 | 165 | NML | 42 | 41 |
| MDS 14 | RCMD | 9.3 | 4.1 | 67 | −7, +1 | 38 | 8 |
| MDS 15 | CMML | 12.8 | 60 | 468 | NML | 5 | 81 |
| MDS 16 | RCMD | 7.3 | 11.6 | 123 | −5q, −1 | 30 | 56 |
| MDS 17 | RAEB | 4.6 | 9.2 | 169 | NML | 42 | 39 |
| MDS 18 | RAEB | 8.4 | 44 | 89 | +8 | 10 | 89 |
| MDS 19 | RA | 8.9 | 6.3 | 181 | +8 | 45 | 39 |
| MDS 20 | RAEB | 8.1 | 0.8 | 51 | Complex | 73 | 25 |
| 5q Syndrome | RA | 6.6 | 11.6 | 562 | −5q | 16 | 74 |
FIGURE 1.
Methylation profiling on peripheral blood leukocytes separates distinct subsets of MDS from normals. Methylation profiles generated by the HELP assay were used to cluster 21 MDS and 9 control samples by hierarchical clustering. The controls formed a cluster that was distinct from MDS samples. The MDS samples included two clusters (groups 1 and 2) of epigenetically similar samples with a greater amount of resemblance to controls. The remaining seven MDS samples demonstrated greater heterogeneity. No correlation with cytogenetics (normal represented as green and abnormal as red) was seen.
Most Differentially Methylated Genes Are Hypermethylated in MDS Leukocytes
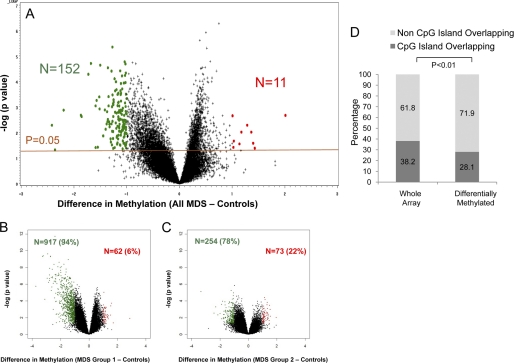
Having demonstrated epigenetic dissimilarity between MDS and control samples, we next determined the qualitative epigenetic differences between these groups by performing a supervised analysis of the respective DNA methylation profiles. A volcano plot comparing the differences between mean methylation of individual loci in MDS versus control samples plotted against the significance (log (p value) based on t test) of the difference is used to represent these data in Fig. 2A. We observed that most significantly differentially methylated loci were hypermethylated in all cases of MDS (n = 152) when compared with controls (p value < 0.05; Fig. 2 and supplemental Tables 1 and 2 listing all genes). This is consistent with previous reports demonstrating hypermethylation of selected loci in MDS bone marrow progenitors (38). The two subgroups of MDS samples based on unsupervised clustering (Fig. 1) also had predominantly hypermethylated genes, although group 2 had a slightly higher proportion of significantly hypomethylated genes when compared with controls (Fig. 2, B and C). Most interestingly, only 28% (43/153) of the commonly differentially hypermethylated CCGG loci (Fig. 2A) were located in the CpG islands (Fig. 2D). This was significant even after the correction for the proportion of non-CpG island probes present in the HELP array and shows that these non-CpG island loci are preferentially dysregulated in this disease (Fig. 2D).
FIGURE 2.
Majority of differentially methylated loci are hypermethylated in MDS leukocytes and reside outside of CpG islands. A volcano plot is shown demonstrating the difference in mean methylation between all MDS samples and controls on the x axis and the log of the p values between the means on the y axis. A two-tailed t test was used to calculate the p values. Significantly methylated loci with a log fold change in mean methylation are labeled in green, and significantly hypomethylated loci are labeled in red (A). Volcano plots for MDS subgroups 1 and 2 also reveal mostly hypermethylated loci with a variable number of hypomethylated loci. B and C, genomic position of every HpaII-amplifiable fragment on the HELP array was compared with the location of known CpG islands, and the fragments on the array were divided into two categories, those overlapping with these genomic elements and those not overlapping. To determine whether the differentially methylated genes between MDS and controls were enriched for either one of these types of elements, a proportions test was used to compare the relative proportion of the two types of HpaII fragments in the signature with the relative proportion on the array. Stacking bars are used to illustrate the finding of a significant enrichment for HpaII-amplifiable fragments not overlapping with CpG islands (D).
A transcription factor, KLF3 (39, 40), that was significantly methylated in MDS was chosen for validation. Promoter regions of the Kruppel-like factor-3 (KLF-3) (supplemental Fig. 1) was examined by MALDI-TOF-based quantitative methods (MassArray, Sequenom). DNA was bisulfite-converted, and primers were designed to amplify regions of interest, and quantitative assessment of methylation was performed by mass spectroscopic analysis. We observed a strong correlation of quantitative methylation obtained from MassArray with the findings of our HELP microarrays, demonstrating the validity of our findings (supplemental Fig. 1). Furthermore, MassArray analysis of CG dinucleotides surrounding the assayed HpaII sites revealed distinct hypermethylation of these cytosines in MDS samples when compared with controls (supplemental Fig. 1), potentially pointing to their role as potential biomarkers in this disease, as shown for the KLF3 gene promoter.
Genes Hypermethylated in MDS Display Specific Functional and Genomic Characteristics
A gene ontology analysis of the 152 commonly hypermethylated genes (p <0.05 and methylation change >1 log fold) showed specific enrichment of GTPase regulators with DOCK4, DOCK2, ARHGEF4, CDC42SE1, FARP1, GIT2, IQGAP2, and RALGPS1 as the genes that were hypermethylated in MDS (Table 2). Other gene pathways with significant involvement of hypermethylated genes included those regulating calcium-dependent cell-cell adhesion, spermatid development, small nuclear ribonucleoprotein complex, and nuclear organization. Table 2 shows the genes associated with each of these enriched GO categories, which include many potentially novel relevant candidate genes such as DOCK4 as well as genes already implicated in hematological malignancies such as HOXB3 and RUNX3. Further functional pathway analysis revealed cancer as the top functional pathway affected by hypermethylation in MDS (supplemental Fig. 2). Involvement of these important pathways by genes commonly affected by hypermethylation even in this heterogeneous mix of patients supports the biological validity of our dataset.
TABLE 2.
Functional grouping of hypermethylated genes in MDS (Gene Ontology)
| Biogroup | p value | Genes |
|---|---|---|
| GTPase regulator activity | 6.90E-06 | DOCK4, DOCK2, ARHGEF4, CDC42SE1, FARP1, GIT2, IQGAP2, RALGPS1 |
| Calcium-dependent cell-cell adhesion | 9.10E-05 | PCDH12, PCDHB11 |
| Spermatid development | 9.10E-05 | H1FNT, NME5 |
| Small nuclear ribonucleoprotein complex | 0.0001 | GEMIN4, PRPF8, SNRPF |
| Nuclear organization and biogenesis | 0.0003 | H1FNT, SYNE1 |
| Transcription | 0.0008 | HOXB3, RUNX3, CCRN4L, KLF3, PLAGL2, MED12, PPARGC1A, HMGN3, ZFX, ENG, PRKAR1A |
Aberrant methylation was not distributed randomly across chromosomes. Differentially methylated HpaII fragments showed significant regional differences on chromosomes 11 and 16 compared with the genomic distribution of all HpaII fragments from the HELP array. Furthermore, to determine whether these hypermethylated genes shared any common DNA elements, we performed a search for transcription factor-binding sites enriched in these genes. Significant over-representation of binding sites for SP1, AHR, FOXO4, LEF1, NF1, and SOX9 and other transcription factors was seen in MDS (Table 3).
TABLE 3.
Transcription factor binding sites enriched in hypermethylated genes in MDS
| Transcription factors | No. of genes | p value | Genes | Motif |
|---|---|---|---|---|
| SP1 | 12 | 3.10E-05 | DOCK4, PLAGL2, ZFX, C11ORF30, DDAH2, SYNE1, SV2A, ARRDC4, SEZ6, STRN, NXT2, SLC35D1 | GGGCGGR |
| AHR | 5 | 4.19E-04 | PLAGL2, ZFX, RUNX3, ANGPTL2, VAMP3 | CCYCNRRSTNGCGTGASA |
| E12 | 14 | 3.78E-03 | PLAGL2, C11ORF30, DDAH2, SYNE1, SV2A, DUSP4, PPARGC1A, HOXB3, ADAMTS15, GIT2, ODC1, ITPK1, IQGAP2, LEPREL1 | CAGGTG |
| FOXO4 | 11 | 4.93E-03 | MDS1, HSPG2, KLF3, C11ORF30, DOCK4, DUSP4, PPARGC1A, HOXB3, HMGN3, ABCC1, DNAH11 | TTGTTT |
| NFY | 4 | 5.80E-03 | ARRDC4, ADAMTS15, JMJD2A, IMP4 | GATTGGY |
| CHX10 | 2 | 8.60E-03 | SEZ6, MDS1 | TAATTA |
| AREB6 | 2 | 9.96E-03 | ADAMTS15, SEZ6 | CAGGTA |
| MAZ | 14 | 1.22E-02 | ZFX, DDAH2, SYNE1, DOCK4, ARRDC4, RUNX3, DUSP4, ADAMTS15, MDS1, HSPG2, JMJD2A, JPH4, DAB2IP, GSPT1 | GGGAGGRR |
| GABP | 2 | 1.32E-02 | GIT2, JPH4 | MGGAAGTG |
| LEF1 | 12 | 1.60E-02 | DOCK4, SEZ6, ANGPTL2, DUSP4, PPARGC1A, HOXB3, ODC1, KLF3, DAB2IP, NUP107, FARP1, ARHGEF4 | CTTTGT |
| NF1 | 2 | 1.81E-02 | DUSP4, HCN1 | TGCCAAR |
| SOX9 | 6 | 2.05E-02 | DOCK4, HSPG2, KLF3, MBP, PCDH12, ZNF502 | NNNNAACAATRGNN |
Array CGH Detects Copy Number Variations in MDS Leukocytes
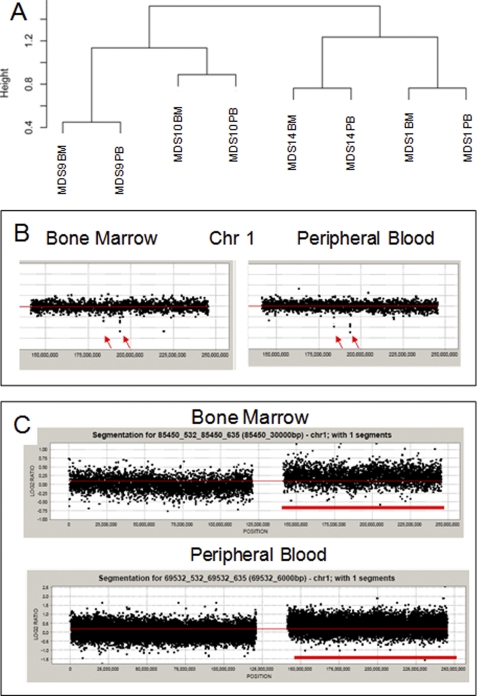
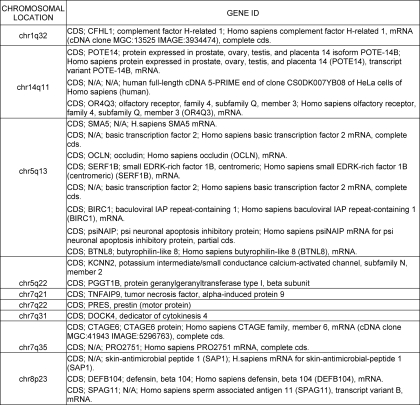
Because chromosomal deletions and amplifications have been seen in MDS bone marrow progenitors, we next wanted to determine whether these can also be seen in dysplastic leukocytes. We also wanted to test the potential of high resolution aCGH in detecting novel copy number variations in the peripheral blood. aCGH performed at a 6-kb resolution demonstrated that cytogenetic changes can be seen in peripheral blood leukocytes (Fig. 3 and Tables 4 and 5). The changes seen in peripheral blood are very similar to those seen in the bone marrow progenitors (Fig. 3A). Furthermore, both small and large chromosomal changes were successfully observed in peripheral leukocytes (Fig. 3, B and C). Next, we used aCGH data from 20 samples to uncover cryptic changes not seen by conventional karyotyping. We observed five common deletions and nine common chromosomal amplifications affecting 25% or more cases with our analysis (Tables 4 and 5). These included novel areas of deletion (1q32 and 14q11) and amplification (1q41–42, 15q11, 19q13, and 22q22) that were not seen by conventional karyotypic analysis. Interestingly, the 17q21-21 region found to be amplified in our analysis was also described as a novel MDS amplification in a recent report (18), thus confirming the applicability of our findings to other patient cohorts.
FIGURE 3.
Array CGH can detect copy number variations in MDS leukocytes. Unsupervised clustering of copy number analysis reveals similarity between matched marrow (BM) and peripheral blood (PB) samples from the same patient (A). Array CGH plots of chromosome 1 reveal small deletions seen in both bone marrow and peripheral blood samples from one patient (B). In another patient with amplification of the short arm of chromosome 1 in bone marrow cells, the amplification is also seen in peripheral blood (C).
TABLE 4.
Chromosomal regions commonly deleted in MDS
TABLE 5.
Chromosomal regions commonly amplified in MDS
Integrative Analysis Can Reveal Novel Pathogenic Genes
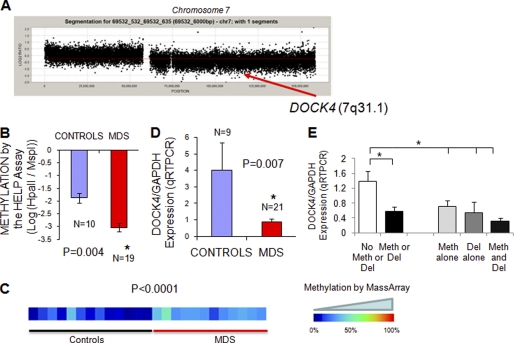
We hypothesized that genes silenced by both deletion and methylation are likely to be involved in disease pathogenesis as they are being silenced by distinct mechanisms in separate cases. Therefore, an integrative analysis of epigenetic and genetic lesions could prioritize candidate lesions for functional validation. Using this strategy, we selected five genes (DOCK4, PRES, KCNN2, PGGT1B, and TNFAIP9) that were targeted by both genetic deletion and epigenetic silencing in our dataset. These genes were selected on the basis of being deleted in at least 25% of cases and differentially methylated in the others. One of these genes, DOCK4 (dedicator of cytokinesis-4) has been postulated as a tumor suppressor (41) and is located on chromosome 7q31, a frequently deleted segment in MDS (Fig. 4A) (42). DOCK4 was found to be hypermethylated by the HELP assay (Fig. 4B), and the methylation was validated quantitatively by MassArray EpiTYPERTM analysis, demonstrating significantly increased methylation in MDS samples when compared with controls (Fig. 4C). To determine the effect of DOCK4 methylation on transcription, we measured its expression in these samples by quantitative RT-PCR and found it to be significantly reduced in the MDS leukocyte samples (Fig. 4D). Furthermore, DOCK4 expression was significantly down-regulated by both promoter methylation and 7q deletion in MDS samples illustrating that it is affected by both genetic and epigenetic alterations (Fig. 4E).
FIGURE 4.
Integrative analysis reveals DOCK4 to be silenced by both deletion and hypermethylation in MDS. The aCGH plot of chromosome 7 from an MDS patient with 7q deletion shows the location of the DOCK4 gene (A). Mean methylation from the HELP assay (depicted by log2(HpaII/MspI)) is significantly higher in MDS samples as evident from a more negative value (two-tailed t test) (B). Methylation analysis of the DOCK4 promoter by MassArray analysis reveals greater methylation in MDS samples as depicted in the heat map (C). quantitative RT-PCR was performed on RNA from MDS leukocytes and control samples and showed a significantly reduced expression in MDS (D). Means ± S.E. with p value were calculated by two-tailed t test. Expression of DOCK4 was evaluated in MDS samples with or without deletions (Del) and promoter methylation (Meth) and shows significant reduction with either genetic or epigenetic silencing. Means ± S.E. with p value were calculated by two-tailed t test (E).
DOCK4 Is Hypermethylated and Reduced in Expression in MDS Bone Marrows in Independent Datasets
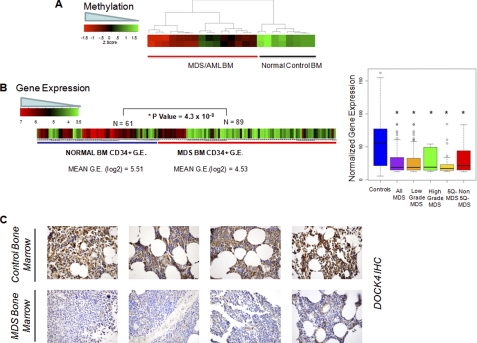
To validate these findings in bone marrow samples, we examined DOCK4 methylation in an independent cohort of 15 MDS and secondary AML patients enrolled in a clinical trial (38). Analysis of these HELP DNA methylation profiles revealed striking hypermethylation of the DOCK4 promoter in MDS/AML patients when compared with normal bone marrow controls (t test, p value <0.01) (Fig. 5A). To further test DOCK4 expression in a larger set of MDS-derived bone marrow CD34+ cells, we utilized a recently constructed meta-analytical data base of MDS bone marrow gene expression profiles (32, 33). DOCK4 was significantly underexpressed in 89 MDS CD34+ cell samples when compared with 61 normal bone marrow CD34+ cells (p value = 4.3 × 10−8, t test) Significantly reduced levels of DOCK4 were seen in all subtypes of MDS examined (Fig. 5B, right panel, box plots), thus demonstrating a potential important role in the pathobiology of this disease. Finally, we also determined DOCK4 protein expression in bone marrow biopsies by immunohistochemistry in 9 cases of MDS and compared these with 19 cases of age-matched controls with anemia due to various other etiologies (chronic disease, nutrient deficiency, and HIV). Only a minority of MDS samples (2/9, 22%) showed strong expression of DOCK4 in the bone marrow progenitors as compared with most of the controls (19/22, 86% with strong staining, p value = 0.001, Fisher's exact test) as is shown in representative cases in Fig. 5C. These data obtained from different laboratories support a potential role of DOCK-4 in MDS pathogenesis.
FIGURE 5.
Validation in independent cohorts demonstrate reduction in DOCK4 in marrow samples from MDS/AML. Methylation values obtained from the HELP assay performed on marrow (BM) samples in an independent cohort of patients (38) show hypermethylation of the promoter in MDS/AML samples (A). Gene expression values from various studies on MDS and normal bone marrow-derived CD34+ cells were obtained and normalized. Mean expression of DOCK4 was significantly reduced in 89 MDS cases when compared with 61 controls (two-tailed t test) (B, left panel); box plots of MDS subtypes show significantly reduced levels of DOCK4 in all subtypes of MDS (B, right panel). Bone marrow biopsy samples were stained with DOCK4 antibody and show decreased expression in four representative cases of MDS when compared with controls (C).
DOCK4 Knockdown Leads to Ineffective Hematopoiesis
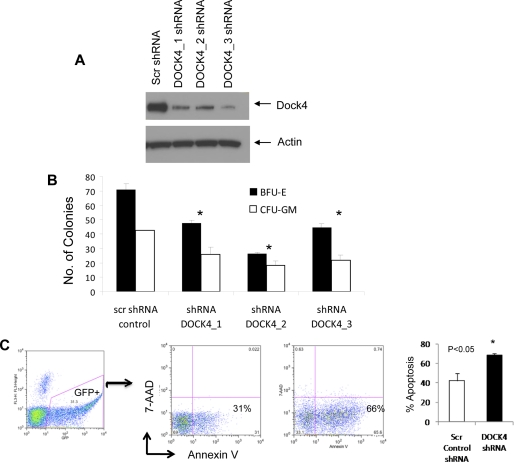
MDS is characterized by ineffective hematopoiesis. Increased progenitor and stem cell apoptosis coupled with dysplastic maturation of blood cells is seen in MDS. To determine the functional role of DOCK4 in hematopoiesis, we tested three different shRNAs against DOCK4 and demonstrated specific knockdown of the gene with all three constructs (Fig. 6A). These were then used to knock down DOCK4 in primary bone marrow-derived CD34+ stem cells that were subsequently used for hematopoietic colony assays. DOCK4 shRNA led to significantly decreased erythroid and myeloid colony formation demonstrating an important role in hematopoiesis. Furthermore, DOCK4 knockdown led to significant increase in apoptosis of CD34+ cells, demonstrating similarity with phenotypic changes seen in MDS bone marrows and validating the potential of our integrative platform in gene discovery in this disease.
FIGURE 6.
DOCK4 knockdown leads to ineffective hematopoiesis in vitro. DOCK4 protein expression was reduced by three lentiviral mediated shRNA constructs (A). Primary bone marrow CD34+ stem cells with DOCK4 shRNAs produced fewer erythroid (erythroid burst-forming units (BFU-E)) and myeloid (CFU-GM) colonies (means ± S.E.; t test, p value< 0.05) (B). DOCK4 shRNA was able to increase apoptosis significantly in GFP+-sorted CD34+ cells (t test, p value< 0.05). Three independent experiments shown as means ± S.E. (C).
DISCUSSION
MDS is a stem cell disorder that responds to treatment with cytosine analogues, azacytidine and decitabine, agents that deplete DNA methyltransferases, suggesting a role of aberrant methylation in the pathobiology of this disease. Even though most studies have looked at the methylation status of selected genes in MDS, recent studies have started exploring epigenetic aberrations in an unbiased manner across the genome (9). Most of these studies have focused on marrow samples that are hard to obtain and frequently limited by poor quality and quantity of derived nucleic acid. We used an unbiased global assay to look for epigenomic disturbances in peripheral blood cells in MDS. Our aim was to evaluate whether high resolution assays would be able to reveal epigenetic and genetic disturbances in these cells and thus could be used for future gene discovery and biomarker studies. Our studies revealed aberrations in DNA methylation that clearly distinguished MDS from normal controls, even when total leukocyte populations were used for analysis. These results also suggest that aberrant methylation marks are stable and can be seen at the level of differentiated and heterogeneous cell populations, when examined by high resolution assays. Furthermore, by combining epigenomic assays with genetic assays, we could find novel genes that may play roles in the pathogenesis of this disease.
Our epigenetic studies were based on the HELP assay that examines cytosine methylation at CCGG (HpaII) sites, some of which lie outside of CpG islands (13). In fact, we found that the majority of common differentially hypermethylated cytosines in MDS samples are not located in CpG islands. Recent work has also shown that non-CpG island cytosine methylation can be important in controlling gene transcription and can be involved in normal development and carcinogenesis (20, 43). These findings will be important for future predictive biomarker studies in MDS and underscore the importance of using unbiased high resolution assays not restricted to CpG islands for these studies.
A problem with genomic assays is the large number of candidate genes that are discovered during analysis. It is difficult to rank these targets by their functional importance, and it is thus challenging to conclude which of these are the actual drivers of disease pathophysiology. We tried to use our integrative platform to address this issue by hypothesizing that genuinely important pathogenic genes may be disrupted by different mechanisms in different patient samples. Using this approach, we found five genes that were targeted by hypermethylation and deletions in different MDS samples. One of these genes, DOCK4, happens to reside within the chromosome 7q31 region that has been found to be a common region deleted in poor prognosis MDS (42). DOCK4 is a member of family of guanine exchange factors that can activate GTPases Rap and Rac (34). DOCK4 is a multidomain protein and is a part of the DOCK superfamily of 11 unconventional guanine exchange factors, characterized by the presence of DHR1 and DHR2 (DOCK homology regions 1 and 2) domains. DOCK4 deletions have been seen in murine tumor models, and missense mutations have been described in prostate and ovarian cancer cell lines (41). DOCK4 is required for Rap GTPAse activation that controls formation and maintenance of adherens junctions. Loss of DOCK4 function leads to loss of cell adherence and can support tumorigenicity, implicating it as a tumor suppressor gene. GTPases such as Rac and Rap also play important roles in cytokine signaling during hematopoiesis (6, 44, 45), and so modulation of their activation can impact this process. Additionally, DOCK4 has also been shown to interact molecularly with the β-catenin pathway, specifically with GSK-3, pathways that play important roles in regulating stem cell function in hematopoiesis (34). Chromosome 7 is frequently deleted in MDS and leads to a worse prognosis in this disease. Studies have shown that 7q31 may be the commonly deleted segment in this disease (36). Our identification of DOCK4 in an unbiased manner using our integrative platform shows the potential of combining different genomic assays to prioritize identification of important genes in this heterogeneous disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants RO1HL082846, RO1AG02913801, and CA009173. This work was also supported by American Cancer Society, Partnership for Cures grant and the Gabrielle Angel Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1 and 2.
- MDS
- myelodysplastic syndrome
- CGH
- comparative genomic hybridization
- aCGH
- array CGH
- GSEA
- gene set enrichment analysis
- ES
- enrichment score
- AML
- acute myeloid leukemia.
REFERENCES
- 1. Pellagatti A., Jädersten M., Forsblom A. M., Cattan H., Christensson B., Emanuelsson E. K., Merup M., Nilsson L., Samuelsson J., Sander B., Wainscoat J. S., Boultwood J., Hellström-Lindberg E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11406–11411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pellagatti A., Cazzola M., Giagounidis A. A., Malcovati L., Porta M. G., Killick S., Campbell L. J., Wang L., Langford C. F., Fidler C., Oscier D., Aul C., Wainscoat J. S., Boultwood J. (2006) Blood 108, 337–345 [DOI] [PubMed] [Google Scholar]
- 3. Jones P. A., Baylin S. B. (2002) Nat. Rev. Genet. 3, 415–428 [DOI] [PubMed] [Google Scholar]
- 4. Christiansen D. H., Andersen M. K., Pedersen-Bjergaard J. (2003) Leukemia 17, 1813–1819 [DOI] [PubMed] [Google Scholar]
- 5. Aggerholm A., Holm M. S., Guldberg P., Olesen L. H., Hokland P. (2006) Eur. J. Haematol. 76, 23–32 [DOI] [PubMed] [Google Scholar]
- 6. Cancelas J. A., Lee A. W., Prabhakar R., Stringer K. F., Zheng Y., Williams D. A. (2005) Nat. Med. 11, 886–891 [DOI] [PubMed] [Google Scholar]
- 7. Issa J. P., Garcia-Manero G., Giles F. J., Mannari R., Thomas D., Faderl S., Bayar E., Lyons J., Rosenfeld C. S., Cortes J., Kantarjian H. M. (2004) Blood 103, 1635–1640 [DOI] [PubMed] [Google Scholar]
- 8. Kantarjian H., Oki Y., Garcia-Manero G., Huang X., O'Brien S., Cortes J., Faderl S., Bueso-Ramos C., Ravandi F., Estrov Z., Ferrajoli A., Wierda W., Shan J., Davis J., Giles F., Saba H. I., Issa J. P. (2007) Blood 109, 52–57 [DOI] [PubMed] [Google Scholar]
- 9. Jiang Y., Dunbar A., Gondek L. P., Mohan S., Rataul M., O'Keefe C., Sekeres M., Saunthararajah Y., Maciejewski J. P. (2009) Blood 113, 1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esteller M. (2003) Adv. Exp. Med. Biol. 532, 39–49 [DOI] [PubMed] [Google Scholar]
- 11. Schumacher A., Kapranov P., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Petronis A. (2006) Nucleic Acids Res. 34, 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weber M., Davies J. J., Wittig D., Oakeley E. J., Haase M., Lam W. L., Schübeler D. (2005) Nat. Genet. 37, 853–862 [DOI] [PubMed] [Google Scholar]
- 13. Figueroa M. E., Lugthart S., Li Y., Erpelinck-Verschueren C., Deng X., Christos P. J., Schifano E., Booth J., van Putten W., Skrabanek L., Campagne F., Mazumdar M., Greally J. M., Valk P. J., Löwenberg B., Delwel R., Melnick A. (2010) Cancer Cell 17, 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khulan B., Thompson R. F., Ye K., Fazzari M. J., Suzuki M., Stasiek E., Figueroa M. E., Glass J. L., Chen Q., Montagna C., Hatchwell E., Selzer R. R., Richmond T. A., Green R. D., Melnick A., Greally J. M. (2006) Genome Res. 16, 1046–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson R. F., Reimers M., Khulan B., Gissot M., Richmond T. A., Chen Q., Zheng X., Kim K., Greally J. M. (2008) Bioinformatics 24, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gondek L. P., Tiu R., O'Keefe C. L., Sekeres M. A., Theil K. S., Maciejewski J. P. (2008) Blood 111, 1534–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohamedali A., Gäken J., Twine N. A., Ingram W., Westwood N., Lea N. C., Hayden J., Donaldson N., Aul C., Gattermann N., Giagounidis A., Germing U., List A. F., Mufti G. J. (2007) Blood 110, 3365–3373 [DOI] [PubMed] [Google Scholar]
- 18. Starczynowski D. T., Vercauteren S., Telenius A., Sung S., Tohyama K., Brooks-Wilson A., Spinelli J. J., Eaves C. J., Eaves A. C., Horsman D. E., Lam W. L., Karsan A. (2008) Blood 112, 3412–3424 [DOI] [PubMed] [Google Scholar]
- 19. Figueroa M. E., Reimers M., Thompson R. F., Ye K., Li Y., Selzer R. R., Fridriksson J., Paietta E., Wiernik P., Green R. D., Greally J. M., Melnick A. (2008) PLoS ONE 3, e1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Figueroa M. E., Wouters B. J., Skrabanek L., Glass J., Li Y., Erpelinck-Verschueren C. A., Langerak A. W., Löwenberg B., Fazzari M., Greally J. M., Valk P. J., Melnick A., Delwel R. (2009) Blood 113, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deleted in proof.
- 22. Olshen A. B., Venkatraman E. S., Lucito R., Wigler M. (2004) Biostatistics 5, 557–572 [DOI] [PubMed] [Google Scholar]
- 23. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgar R., Domrachev M., Lash A. E. (2002) Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Breit S., Nees M., Schaefer U., Pfoersich M., Hagemeier C., Muckenthaler M., Kulozik A. E. (2004) Br. J. Haematol. 126, 231–243 [DOI] [PubMed] [Google Scholar]
- 28. Eckfeldt C. E., Mendenhall E. M., Flynn C. M., Wang T. F., Pickart M. A., Grindle S. M., Ekker S. C., Verfaillie C. M. (2005) PLoS Biol. 3, e254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oswald J., Steudel C., Salchert K., Joergensen B., Thiede C., Ehninger G., Werner C., Bornhäuser M. (2006) Stem Cells 24, 494–500 [DOI] [PubMed] [Google Scholar]
- 30. Sternberg A., Killick S., Littlewood T., Hatton C., Peniket A., Seidl T., Soneji S., Leach J., Bowen D., Chapman C., Standen G., Massey E., Robinson L., Vadher B., Kaczmarski R., Janmohammed R., Clipsham K., Carr A., Vyas P. (2005) Blood 106, 2982–2991 [DOI] [PubMed] [Google Scholar]
- 31. Li Y., Sassano A., Majchrzak B., Deb D. K., Levy D. E., Gaestel M., Nebreda A. R., Fish E. N., Platanias L. C. (2004) J. Biol. Chem. 279, 970–979 [DOI] [PubMed] [Google Scholar]
- 32. Sohal D., Yeatts A., Ye K., Pellagatti A., Zhou L., Pahanish P., Mo Y., Bhagat T., Mariadason J., Boultwood J., Melnick A., Greally J., Verma A. (2008) PLoS ONE 3, e2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou L., Nguyen A. N., Sohal D., Ying Ma J., Pahanish P., Gundabolu K., Hayman J., Chubak A., Mo Y., Bhagat T. D., Das B., Kapoun A. M., Navas T. A., Parmar S., Kambhampati S., Pellagatti A., Braunchweig I., Zhang Y., Wickrema A., Medicherla S., Boultwood J., Platanias L. C., Higgins L. S., List A. F., Bitzer M., Verma A. (2008) Blood 112, 3434–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Upadhyay G., Goessling W., North T. E., Xavier R., Zon L. I., Yajnik V. (2008) Oncogene 27, 5845–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Figueroa M. E., Abdel-Wahab O., Lu C., Ward P. S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H. F., Tallman M. S., Sun Z., Wolniak K., Peeters J. K., Liu W., Choe S. E., Fantin V. R., Paietta E., Löwenberg B., Licht J. D., Godley L. A., Delwel R., Valk P. J., Thompson C. B., Levine R. L., Melnick A. (2010) Cancer Cell 18, 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greenberg P., Cox C., LeBeau M. M., Fenaux P., Morel P., Sanz G., Sanz M., Vallespi T., Hamblin T., Oscier D., Ohyashiki K., Toyama K., Aul C., Mufti G., Bennett J. (1997) Blood 89, 2079–2088 [PubMed] [Google Scholar]
- 37. Alvarez H., Opalinska J., Zhou L., Sohal D., Fazzari M., Yu Y., Montagna C., Montgomery E., Canto M., Dunbar K., Wang J., Roa J., Mo Y., Bhagat T., Ramesh K., Cannizzaro L., Mollenhauer J., Thompson R., Suzuki M., Meltzer S., Melnick A., Greally J. M., Maitra A., Verma A. (2011) PLoS Genet. 7, e1001356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Figueroa M. E., Skrabanek L., Li Y., Jiemjit A., Fandy T. E., Paietta E., Fernandez H., Tallman M. S., Greally J. M., Carraway H., Licht J. D., Gore S. D., Melnick A. (2009) Blood 114, 3448–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soderholm J., Kobayashi H., Mathieu C., Rowley J. D., Nucifora G. (1997) Leukemia 11, 352–358 [DOI] [PubMed] [Google Scholar]
- 40. Eaton S. A., Funnell A. P., Sue N., Nicholas H., Pearson R. C., Crossley M. (2008) J. Biol. Chem. 283, 26937–26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yajnik V., Paulding C., Sordella R., McClatchey A. I., Saito M., Wahrer D. C., Reynolds P., Bell D. W., Lake R., van den Heuvel S., Settleman J., Haber D. A. (2003) Cell 112, 673–684 [DOI] [PubMed] [Google Scholar]
- 42. Liang H., Castro P. D., Ma J., Nagarajan L. (2005) Cancer Genet. Cytogenet. 162, 151–159 [DOI] [PubMed] [Google Scholar]
- 43. Irizarry R. A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., Ji H., Potash J. B., Sabunciyan S., Feinberg A. P. (2009) Nat. Genet. 41, 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Verma A., Deb D. K., Sassano A., Kambhampati S., Wickrema A., Uddin S., Mohindru M., Van Besien K., Platanias L. C. (2002) J. Immunol. 168, 5984–5988 [DOI] [PubMed] [Google Scholar]
- 45. Verma A., Deb D. K., Sassano A., Uddin S., Varga J., Wickrema A., Platanias L. C. (2002) J. Biol. Chem. 277, 7726–7735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.