Abstract
Chronic infection of Helicobacter pylori in the stomach mucosa with translocation of the bacterial cytotoxin-associated gene A (CagA) effector protein via the cag-Type IV secretion system (TFSS) into host epithelial cells are major risk factors for gastritis, gastric ulcers, and cancer. The blood group antigen-binding adhesin BabA mediates the adherence of H. pylori to ABO/Lewis b (Leb) blood group antigens in the gastric pit region of the human stomach mucosa. Here, we show both in vitro and in vivo that BabA-mediated binding of H. pylori to Leb on the epithelial surface augments TFSS-dependent H. pylori pathogenicity by triggering the production of proinflammatory cytokines and precancer-related factors. We successfully generated Leb-positive cell lineages by transfecting Leb-negative cells with several glycosyltransferase genes. Using these established cell lines, we found increased mRNA levels of proinflammatory cytokines (CCL5 and IL-8) as well as precancer-related factors (CDX2 and MUC2) after the infection of Leb-positive cells with WT H. pylori but not with babA or TFSS deletion mutants. This increased mRNA expression was abrogated when Leb-negative cells were infected with WT H. pylori. Thus, H. pylori can exploit BabA-Leb binding to trigger TFSS-dependent host cell signaling to induce the transcription of genes that enhance inflammation, development of intestinal metaplasia, and associated precancerous transformations.
Keywords: Adhesion, Bacteria, Cancer Tumor Promoter, Carbohydrate, Carbohydrate-binding Protein, Glycoconjugate, Signal Transduction, BabA, Helicobacter pylori, Type IV Secretion System
Introduction
Helicobacter pylori is the main causative agent of gastric and duodenal ulcers, gastric adenocarcinoma, and MALT lymphoma (1). Long term colonization of the gastric epithelium is the main pathogenic feature of H. pylori, and bacterial adherence is thought to play an important role in not only triggering colonization but also regulating the functional interplay with the host epithelium. Although H. pylori-induced gastric inflammation is induced by antigen presentation that is mediated by intestinal Peyer's patches when H. pylori is captured by dendritic cells, H. pylori must colonize the gastric epithelium to induce gastric inflammation (2). Approximately 4% of the H. pylori genome encodes outer membrane proteins (OMPs),2 some of which are thought to function as adhesins (3). The fucosylated ABO blood group antigens and their related carbohydrate structures, such as sialyl-Lewis x/a antigens, are one of the major groups of functional receptors for H. pylori adhesins (4–7). Many studies have reported that one adhesin, the blood group antigen-binding adhesin (BabA), binds to the Lewis b (Leb) carbohydrate determinant, FucαGalβ(Fucα4)GlcNAc-R (8), of the synthetic carbohydrate or on gastric sections from humans or Leb-expressing mice (9, 10). However, the biological importance of the BabA-Leb interaction on the pathogenic features of H. pylori is poorly understood.
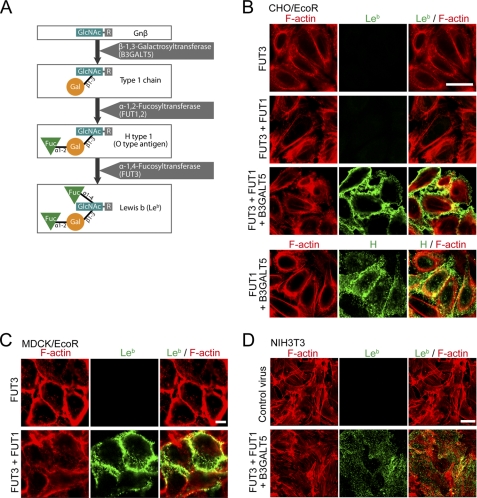
Leb is based on type 1 chains, which are synthesized by β-1,3-galactosyltransferases (11). It was reported that β-1,3-N-acetylglucosaminyltransferase 4, β-1,3-galactosyltransferase 5 (encoded by B3GALT5), β-1,2-fucosyltransferase I or II (encoded by FUT1 or FUT2), and β-1,3/4-fucosyltransferase III (encoded by FUT3) cDNAs are involved in naturally synthesizing Leb in mammalian cells (11) (Fig. 2A).
FIGURE 2.
Establishment of stable Leb-expressing cells. A, the Leb biosynthesis pathway in vivo. B–D, Leb-positive cell lines were established by infecting with a retrovirus encoding glycosyltransferases (FUT3, FUT1, and B3GALT5). CHO/EcoR (B), MDCK/EcoR (C), or NIH3T3 (D) cells that had been transduced with each glycosyltransferase-expressing retrovirus were fixed and stained with an anti-Leb antibody (green) or anti-H antibody (panels in B) and rhodamine-labeled phalloidin (F-actin; red). Scale bar, 20 μm.
The carcinogenic pathway of gastric cancer is mainly associated with persistent H. pylori colonization, followed by chronic inflammation, tissue damage, and regeneration. The attachment of H. pylori to the gastric mucosal surface results in a functional bacteria-host interaction that induces a marked inflammatory response with neutrophil infiltration, followed by the activation of T and B lymphocytes, plasma cells, and macrophages. Members of the chemokine supergene family, particularly the CXC chemokines, such as interleukin (IL)-8, and CC chemokines, such as regulated on activation normal T-cell-expressed and secreted (RANTES, CCL5), are thought to recruit these inflammatory cells into the gastric mucosa (12–14). During the regeneration process, cells deviate from the normal gastric differentiation pathway and change to an intestinal phenotype, which has been considered precancerous and is associated with the intestinal type of gastric cancer. Intestinal metaplasia is characterized by the transdifferentiation of gastric epithelial cells into an intestinal phenotype (15). The caudal type homeobox 2 (CDX2) transcription factor induces the early differentiation and maintenance of intestinal epithelial cells and is thought to be involved in inducing intestinal metaplasia of the stomach (16). CDX2 activates transcription of intestine-specific proteins, such as MUC2 (17).
H. pylori is a genetically diverse species, and various strains markedly differ in virulence. Strains from individuals with overt disease generally carry the cag pathogenicity island, which encodes a component of the Type IV secretion system (TFSS) and CagA (cytotoxin-associated gene A), a major virulence factor. CagA has versatile activities that highjack multiple host cell signaling pathways to stimulate epithelial cell proliferation, breach cell-cell junctions, and induce the inflammatory response (18). TFSS mediates the translocation of CagA, peptidoglycan, and possibly another unknown factor(s) into host cells, where they affect host cell signaling. These translocated factors affect the transcriptional activation of serum response element, serum response factor, nuclear factor-κB (NF-κB), AP-1, β-catenin, and nuclear factor of activated T cells (NFAT), which may result in chemokine production and lead to subsequent proinflammatory responses and a malignant pathology, including intestinal metaplasia (19–25).
Studies have indicated that BabA-positive H. pylori is associated with severe gastric inflammation and an increased risk of peptic ulcer and gastric cancer in humans (26, 27). Some other reports have suggested that the babA status is closely related to the cagA status as well as the incidence and severity of gastric illness (28–31). A recent study has suggested that BabA contributes to severe mucosal injury in experimental infection of H. pylori with gerbils (32). On the other hand, patients infected with H. pylori strains poorly expressing BabA exhibited a higher level of mucosal injury than patients infected with BabA-expressing strains (33). Therefore, the precise role of BabA in the H. pylori functional interaction with gastric epithelium and the impact of BabA on H. pylori pathogenesis still remain partly unclear. Because no systematic study with Leb-positive and Leb-negative cell lines has been reported on the effect of BabA on H. pylori pathogenicity, in the present study, we have genetically modified Leb-negative cells into cell lines that stably express Leb. We used these cell lines to reproduce BabA binding to Leb in vitro and found that H. pylori can exploit the BabA-Leb interaction to increase TFSS-dependent host cell signaling and induce the transcription of genes that are involved in inflammation and intestinal metaplasia. We also demonstrate that BabA can strengthen TFSS-induced host responses in vivo in the Mongolian gerbil stomach.
EXPERIMENTAL PROCEDURES
Ethics Statement
This study was carried out in strict accordance with the University of Tokyo's regulations for Animal Care and Use protocol, which was approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo (approval number 20-62). The committee acknowledged and accepted both the legal and ethical responsibility for the animals, as specified in the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology, 2006 (Japan). All surgery was performed under carbon dioxide euthanasia, and all efforts were made to minimize suffering.
Bacterial Strains and Cell Cultures
Isogenic babA null mutants derived from ATCC 43504 and NCTC 11637 were constructed by insertional mutagenesis using aphA (which confers kanamycin resistance). To generate the GFP-expressing construct, GFP cDNA was amplified from pQBI63 (TaKaRa). The product was ligated between the cagA-upstream and -downstream sequences, which were PCR-amplified from the NCTC 11637 gene (20), in the pHel3 shuttle vector, resulting in pHel3-GFP. The H. pylori strains used in this study are listed in supplemental Table 1. All strains were cultured according to standard procedures (20). AGS cells were maintained in DMEM/F-12 (Ham's) (Invitrogen) containing 10% (v/v) fetal bovine serum (FBS), whereas Chinese hamster ovary (CHO), Madin-Darby canine kidney (MDCK), NIH3T3 (mouse embryonic fibroblast cell line), and mouse embryonic fibroblast (MEF) cells were maintained in DMEM containing 10% (v/v) FBS.
Reagents and Antibodies
The anti-Tyr(P)-CagA, anti-CagA, anti-UreA, and anti-FLAG rabbit antibodies were described previously (20). The anti-BabA, anti-VirB7, and anti-VirB9 rabbit antibodies were described previously (34, 35). The anti-H. pylori mouse antibody was from MONOSAN, the FITC-labeled anti-H. pylori antibody was from Abcam, and the anti-H antigen mouse IgG monoclonal antibody and anti-Leb mouse IgM clone T218 antibody were from SIGNET. The anti-human CD44H mouse monoclonal antibody and FITC-labeled anti-mouse IgM antibody were from R&D and Sigma-Aldrich, respectively. ISOGEN was from Wako. SYBR Premix Ex Taq was from TaKaRa, and reverse transcriptase ReverTra Ace was from TOYOBO.
Bacterial Infection
Cells were infected with H. pylori at the indicated multiplicity of infection (MOI). For Western blot analysis, equal amounts of protein from each sample were separated by SDS-PAGE and transferred to a nitrocellulose membrane. For immunohistochemical analysis, the cells were seeded onto coverslips, cultured until subconfluent, and then infected with H. pylori. The cells were fixed with 4% paraformaldehyde-PBS and subjected to immunohistochemical staining.
Real-time PCR
Total RNA was reverse transcribed into cDNA with random primers and then amplified and quantified using SYBR Green detection with a LightCycler DX400 (Roche Applied Science). Relative mRNA expression was calculated using canine GAPDH gene expression or Mongolian gerbil 18 S rRNA gene expression as an endogenous reference standard. Primer sequences were selected with Primer3 and BLAST. The primers used in this study are listed in supplemental Table 2.
Reporter Assays
AGS cells were seeded in 24-well plates and grown to ∼70% confluence. The cells were transiently transfected with 100 ng of reporter plasmids (pSRE-luc, pNFκB-luc, and pGL4.30[luc2P/NFAT-RE/Hygro] (which is referred to as pNFAT) from Promega and pAP1-luc from Agilent) and 4 ng of the Renilla luciferase control vector phRL-TK (Promega) using FuGENE6. After 16 h, the cells were infected with H. pylori at an MOI of ∼50 for 5 h and lysed, and then the Photinus and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay (Promega) with a luminometer. After normalizing to the Renilla luciferase levels, the Photinus luciferase activity was calculated and graphed as -fold induction compared with the control.
In Vitro Adherence Assay
96-well microtiter plates (Corning Costar) were coated with 50 ng/well of BSA-conjugated Leb (Dextra Laboratories Ltd.) or BSA diluted in 0.2 m carbonate buffer (pH 9.6). The plates were incubated for 16 h at 4 °C, and then the coating buffer was aspirated with a pipette, and 200 μl of blocking buffer (PBS containing 2% BSA) was added. After incubating for 1 h at room temperature, the plates were decanted without washing, 50 μl of a bacterial suspension in suspension buffer (PBS containing 0.2% BSA and 0.2% Tween 20) was added, and the plates were incubated for 1 h at room temperature. For the cellular adherence assay, the cells were seeded on 96-well plates and cultured to subconfluence. Then 50 μl of a bacterial suspension or purified proteins in Opti-MEM (Invitrogen) was added, and the cells were incubated for 1 h at 37 °C. Each well was washed three times with 100 μl of PBS. For the bacterial adherence assays, 4% paraformaldehyde in PBS was added to the plates, and after a 15-min incubation at room temperature, the plates were washed three times with PBS. Signals were detected using an anti-Helicobacter antibody (Abcam) for the bacterial adhesion assay or anti-FLAG antibody (Sigma-Aldrich) for the protein adhesion assay, followed by an HRP-anti-rabbit IgG antibody (Sigma-Aldrich) and 3,3',5,5'-tetramethylbenzidine (TMB) peroxidase solution (Bethyl) according to the manufacturer's instructions. Absorbance was quantified using a Varioskan Flash spectral scanning multimode reader (Thermo Scientific) at 450 nm and normalized to the controls. In each single experiment, all samples were tested in three coated wells along with three controls, and at least two independent assays were performed for each experiment.
Plasmids and Recombinant Proteins
To generate glutathione S-transferase (GST)-FLAG, DNA fragments encoding GST were PCR-amplified using GST-F and GST-R primers (listed in supplemental Table 2) and cloned into the pET-21a(+) plasmid (Novagen) with a PreScission recognition site sequence at the C terminus of GST. The DNA fragments encoding amino acids 21–570 of BabA (jhp0833) from the H. pylori J99 strain (nucleotides 61–1710) were PCR-amplified using BabA-F and BabA-R primers (listed in supplemental Table 2) and then cloned into the plasmid above with a FLAG tag sequence at the C terminus of BabA to yield the pET-21a(+)-gst-babA-FLAG plasmid. The GST fusion proteins were purified using protocols supplied by the manufacturer. Briefly, the GST-FLAG protein was eluted from the glutathione-Sepharose resin (GE Healthcare) using a glutathione elution buffer. To prepare the BabA-FLAG protein, GST-BabA-FLAG protein bound to glutathione-Sepharose was digested with PreScission protease (GE Healthcare). Human FUT3, FUT1, or B3GALT5 cDNAs were amplified by RT-PCR from total AGS cellular RNA using the primers listed in supplemental Table 2 and cloned into the retroviral vector pMX-puro (36). To construct FUT3 and FUT1 cDNA-harboring pIRES (Clontech), human FUT3 and FUT1 cDNAs were amplified by RT-PCR from the total AGS cellular RNA and cloned into pBluescript II-SK(+). After digestion, each cDNA fragment was cloned into the pIRES plasmid.
Establishment of Stably Expressing Cell Lines
MDCK cells or CHO cells were transfected with pM5neo-murine ecotropic receptor plasmids (37), and stable transformants expressing the murine ecotropic receptor (MDCK/EcoR and CHO/EcoR, respectively) were isolated by selecting with G418. The pMX-puro or pMX-puro-FUT3, -FUT1, or -B3GALT5 plasmids were transfected into PLAT-E cells using Lipofectamine 2000 (Invitrogen), and the supernatants were harvested 48 h after transfection. CHO/EcoR, MDCK/EcoR, or NIH3T3 cells were infected with each combination of the supernatants in the presence of DOTAP (Roche Applied Science). At 24 h postinfection, the cells were cultured with DMEM containing 10% FBS and 2 μg/ml puromycin for 2 days, and the resulting cells were infected with H. pylori. To establish MEF cells expressing Leb, MEF cells were transfected with pIRES-FUT1-FUT3 using Lipofectamine 2000, and stable transformants (MEF/pIRES-FUT1-FUT3) were isolated by selecting with G418. Then the cells were transfected with the supernatant from PLAT-E cells that had been transfected with pMX-puro-B3GALT5 mentioned above. To generate MDCK cells stably expressing Leb, MDCK cells were transfected with pIRES-FUT1-FUT3 or the control pIRES, and stable transformants (MDCK/Leb and MDCK/pIRES, respectively) were isolated by selecting with G418.
Enzyme-linked Immunosorbent Assay (ELISA)
An ELISA system from R&D was used to analyze the canine IL-8 secretion of H. pylori-infected MDCK/Leb or MDCK/pIRES cells. Cells were infected with H. pylori at an MOI of ∼50 for 24 h. Following infection, the culture medium was centrifuged (5 min, 15,000 × g, 4 °C), and the IL-8 protein content was determined using the ELISA according to the manufacturer's instructions.
Immunoprecipitation Assay
MDCK/Leb or MDCK/pIRES cells cultured to about 70% confluence were infected with H. pylori for 24 h at an MOI of ∼50. The cells were washed twice with PBS and then lysed in 1 ml of ice-cold radioimmune precipitation assay buffer (25 mm Tris-HCl, 150 mm NaCl, 0.1% (v/v) Nonidet P-40, 3 mm Na3VO4, 100 μm Genistein, and protease inhibitors (Complete protease inhibitor mixture, Roche Applied Science), pH 7.5). The lysates were centrifuged at 10,000 × g for 10 min at 4 °C, and the supernatants were subjected to immunoprecipitation with anti-Tyr(P)-CagA antibody. Immunoprecipitates and whole-cell lysates were subjected to a Western blot analysis using anti-Tyr(P)-CagA and anti-actin, respectively.
H. pylori Gastric Infection of Mongolian Gerbils
Six-week-old male MGS/Sea Mongolian gerbils (Meriones unguiculatus; CLEA Japan Inc.) were intragastrically inoculated with a bacterial culture containing 109 cfu of H. pylori in Brucella broth containing 5% (v/v) FCS. Control animals were orally administered Brucella broth containing 5% (v/v) FCS. After 8 weeks, the stomach of each infected Mongolian gerbil was opened along the greater curvature. To isolate RNA, the tissue was immediately treated with RNAlater (Ambion). Sections for histological analysis were fixed in 4% paraformaldehyde in PBS. The gastric tissue embedded in tissue-freezing medium (Jung Tissue Freezing Medium, Leica Microsystems) was frozen in liquid nitrogen and sectioned with a Leica cryostat (model CM1900).
Immunofluorescence Microscopy
Anguilla anguilla agglutinin (Sigma-Aldrich), which was used to visualize pit cells, was labeled with Cy3 using the Cy3 monofunctional reactive dye (Amersham Biosciences). Immunofluorescence staining was performed as described previously (23). FITC- or TRITC-conjugated secondary antibodies were used to visualize the rabbit or mouse antibodies, DAPI (Sigma-Aldrich) was used to visualize DNA, and rhodamine-labeled phalloidin (Invitrogen) was used to visualize F-actin. The stained specimens were examined with a confocal laser-scanning microscope (LSM510, Carl Zeiss).
Statistical Analysis
Differences between the groups in the in vitro experiments were assessed using the paired, two-tailed Student's t test. The in vivo data were statistically analyzed using the Mann-Whitney U test for unpaired groups.
RESULTS
Establishment of Stable Leb-expressing Cell Lines
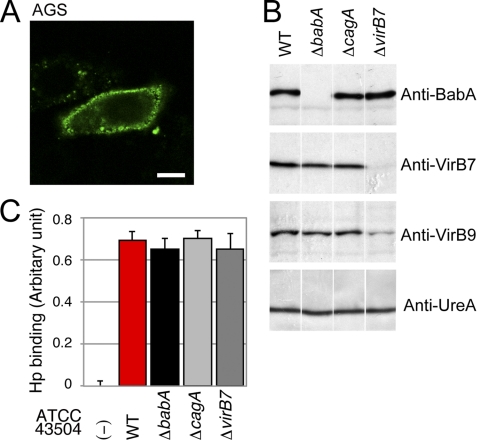
As reported previously (8), WT but not ΔbabA (BabA-deficient) H. pylori binds to Leb in a BabA-dependent manner (supplemental Fig. S1). To investigate the cellular response induced by a BabA-dependent binding of H. pylori to cells, we first examined Leb expression on the surface of the gastric epithelial cell line AGS, which is commonly used to study H. pylori-induced signaling pathways. Immunohistochemical staining showed that AGS cells express Leb epitopes on their surface (Fig. 1A). To determine whether H. pylori binds to AGS cells in a BabA-dependent manner, cells were infected with WT, ΔbabA, ΔcagA, or ΔvirB7 (TFSS-deficient) H. pylori derived from ATCC 43504 strain (Fig. 1B) and then examined for the amount of attached bacteria. As shown in Fig. 1C, H. pylori adhered to AGS cells in a BabA-independent manner, indicating that the binding between H. pylori and AGS is mediated by adhesin(s) other than BabA and that AGS cells are not suitable to study BabA-dependent adhesion.
FIGURE 1.
H. pylori binds to AGS cells in a BabA-independent manner. A, AGS cells express Leb. AGS cells were stained with an anti-Leb antibody (green). Scale bar, 20 μm. B, Western blot analysis of WT H. pylori ATCC 43504 and its mutants. Whole-cell lysates of H. pylori ATCC 43504 WT (WT), ΔbabA, ΔcagA, or ΔvirB7 were separated by SDS-PAGE and subjected to Western blot analysis using the noted antibodies, including an antibody recognizing VirB9, which is another TFSS component. UreA served as a loading control. C, H. pylori (Hp) binds to AGS cells in a BabA-independent manner. AGS cells were seeded in 96-well plates and incubated with the indicated H. pylori strain at an MOI of 50 for 4 h. After fixation, adherent H. pylori was detected using an anti-H. pylori antibody, HRP-labeled anti-rabbit IgG, and TMB substrate. The results represent the average of three separate experiments (each n = 3). Data are presented as the means ± S.D. (error bars).
Next, we examined other cell lines, such as those that have low levels of H. pylori adherence and lack expression of the Leb epitopes, in order to generate stable Leb-expressing cells by transfecting in various glycosyltransferase cDNAs. We tested several cell lines for H. pylori binding using an adherence assay and found that MDCK, CHO, NIH3T3, and MEF cells had a reduced bacterial binding efficiency compared with AGS cells and undetectable Leb expression by immunohistochemical staining (data not shown). For transfection, we established MDCK and CHO cells that stably express the ecotropic viral receptor (MDCK/EcoR and CHO/EcoR, respectively) for retroviral transduction (supplemental Fig. S2). Next, we transduced the cells with retroviruses encoding a combination of B3GALT5, FUT1, and FUT3, and analyzed Leb expression by immunostaining with an anti-Leb antibody. As shown in Fig. 2, B–D, and supplemental Fig. S3, transducing MDCK/EcoR cells with both FUT3 and FUT1 or transducing CHO/EcoR, NIH3T3, or MEF cells with FUT3, FUT1, and B3GALT5 resulted in Leb expression on the surface of the cell. Transducing CHO/EcoR cells with FUT1 and B3GALT5 resulted in H antigen expression (Fig. 2B) but not Leb expression (data not shown), further confirming the glycosylation pathway depicted in Fig. 2A.
H. pylori BabA Binds to Leb-expressing Cells
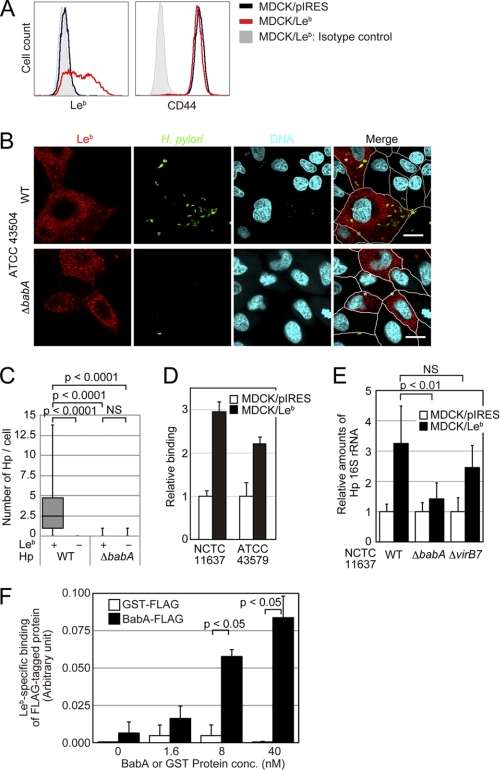
Because MDCK cells form a confluent, polarized monolayer that can be used in functional H. pylori adherence assays, we used MDCK cells stably expressing Leb (MDCK/Leb) and control Leb-negative cells (MDCK/pIRES) for further analyses. Leb expression was confirmed by flow cytometry using an anti-Leb antibody (Fig. 3A). To evaluate the Leb-dependent binding ability of H. pylori on these cells, WT or ΔbabA derived from the ATCC 43504 strain were cocultured with MDCK/Leb or MDCK/pIRES cells. As shown in Fig. 3, B and C, WT but not ΔbabA H. pylori bound more efficiently to Leb-expressing MDCK cells than Leb-negative cells. The same results were obtained with GFP-expressing ATCC 43579 H. pylori cocultured with Leb-expressing MDCK, NIH3T3, and CHO cells or the respective control cells (supplemental Figs. S4–S6). The adhesion assay showed that MDCK/Leb cells had 2.1–2.9-fold higher numbers of bound WT H. pylori of NCTC 11637 or ATCC 43579 compared with MDCK/pIRES cells (Fig. 3D). To confirm that the binding of H. pylori to MDCK/Leb cells was BabA-dependent, the cells were infected with WT, ΔbabA, or ΔvirB7 H. pylori derived from NCTC 11637 strain, and the amount of bacteria attached to the cells was measured by real-time PCR. The RT-PCR analysis of H. pylori 16 S rRNA levels showed that WT and ΔvirB7 attached at ∼2.2- and 1.9-fold higher levels than ΔbabA (Fig. 3E). To further demonstrate that the BabA protein directly binds to Leb on cultured cells, we purified a recombinant FLAG-tagged BabA protein and the control FLAG-tagged GST protein. These recombinant proteins were subjected to binding assays, and the recombinant BabA protein bound to MDCK/Leb cells but not to MDCK/pIRES cells in a dose-dependent manner (Fig. 3F). Collectively, these findings demonstrate that H. pylori binds to Leb-expressing cells in a BabA- and Leb-dependent manner.
FIGURE 3.
H. pylori and recombinant BabA protein bind to Leb-expressing cells. A, characterization of stable Leb-expressing cells. MDCK cells stably expressing Leb (MDCK/Leb) or control cells (MDCK/pIRES) were established by introducing plasmids encoding FUT1 and FUT3 (pIRES-FUT1-FUT3) or a control plasmid (pIRES). The Leb expression levels on the cell surface were analyzed by flow cytometry. An anti-CD44 antibody was used as a positive control for a protein that is expressed on the cell surface. Black lines, MDCK/pIRES; red lines, MDCK/Leb; shaded areas, isotype control. The figure is representative of three separate experiments. B and C, H. pylori (Hp) binds to MDCK/Leb but not to MDCK/pIRES cells in a BabA-dependent manner. MDCK/Leb and MDCK/pIRES cells were mixed and infected with ATCC 43504 H. pylori WT or ΔbabA for 24 h at an MOI of 100. After fixation, the cells were stained with an anti-Leb antibody (red), an anti-H. pylori antibody (green), and DAPI (DNA; blue). B, scale bar, 20 μm. C, the numbers of bacteria bound to the cells were counted. In each case, at least 180 cells were measured. All plots show the 25th to 75th percentiles (boxes) and the 5th to 95th percentiles (whiskers). The line in each box represents the median. D, H. pylori binds to cells in a Leb-dependent manner. MDCK/Leb or MDCK/pIRES cells seeded in 96-well plates were incubated with the indicated H. pylori strain at an MOI of 200 for 24 h. After fixation, adherent H. pylori was detected with an anti-H. pylori antibody, HRP-labeled anti-rabbit IgG-HRP, and TMB substrate. The results represent the average of three separate experiments (each n = 3). Data are presented as the means ± S.D. E, H. pylori binds to MDCK/Leb cells in a BabA-dependent manner. MDCK/Leb or MDCK/pIRES cells were infected with the indicated H. pylori for 4 h at an MOI of 200. The cells were washed, and the bacteria levels were quantitated by real-time PCR. The results represent the average of three separate experiments (each n = 3). Data are presented as the means ± S.D. F, the BabA protein binds to MDCK/Leb cells in a dose-dependent manner. MDCK/Leb or MDCK/pIRES cells seeded in 96-well plates were incubated with the FLAG-tagged J99-derived BabA recombinant protein (BabA-FLAG) or FLAG-tagged GST protein (GST-FLAG) for 1 h. After washing, the adherent proteins were detected with an anti-FLAG antibody, HRP-labeled anti-rabbit IgG, and TMB substrate. The results represent the average of three separate experiments (each n = 3). Data are presented as the means ± S.D. (error bars). NS, not significant.
The BabA-Leb Interaction Enhances the TFSS-dependent Transcriptional Activity in H. pylori-infected Cells
Several reports have indicated that most of the canonical host transcriptional responses to H. pylori, including the expression of innate immune and signal transduction genes, require the TFSS effector CagA, peptidoglycan, and the TFSS structural component (18, 21, 25, 38, 39). To examine whether BabA can stimulate the host transcriptional response, we performed reporter assays for NF-κB, NFAT, AP-1, and serum response element, the major transcriptional factors that are induced by H. pylori in infected AGS cells, which bind to H. pylori in a BabA-independent manner (Fig. 1C). H. pylori ΔbabA but not the ΔvirB7 mutant transcriptionally activated AGS cells and was similar to WT H. pylori (supplemental Fig. S7), indicating that BabA does not directly induce transcriptional activity in these cells, as previously reported (34).
To further analyze whether BabA contributes to the intracellular CagA-induced cell scattering/hummingbird phenotype, which is the most prominent feature of infected AGS cells, the cells were infected with H. pylori, and morphological changes were examined by confocal microscopy. As shown in supplemental Fig. S8, H. pylori ΔbabA but not the ΔvirB7 mutant induced the scattering phenotype of AGS cells and was similar to WT H. pylori, showing that BabA does not directly induce cell scattering phenotype in these cells.
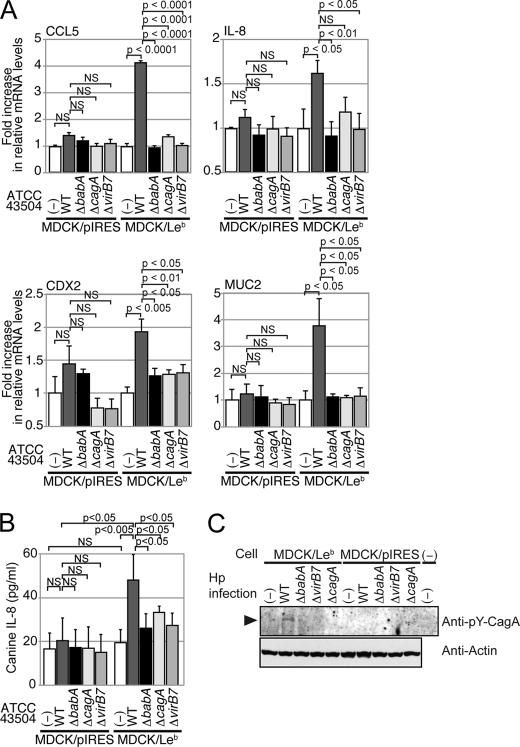
To investigate whether BabA has an essential role in promoting the TFSS-dependent activation of transcriptional factors to induce proinflammatory cytokines, MDCK/Leb or MDCK/pIRES cells were infected with the WT strain ATCC 43504 or its ΔbabA, ΔcagA, or ΔvirB7 mutants, and the CCL5 and IL-8 mRNA levels were measured. An RT-PCR analysis showed that infecting MDCK/pIRES cells with WT or ΔbabA induced an ∼1.4- or 1.2-fold increase in CCL5 mRNA levels compared with the uninfected control or ΔvirB7-infected cells (Fig. 4A). On the other hand, the CCL5 mRNA levels in MDCK/Leb cells that were infected with WT, but not ΔbabA, were ∼4.0-fold higher than the levels in uninfected cells (Fig. 4A). The IL-8 mRNA as well as protein levels in MDCK/Leb cells were also increased upon WT but not ΔbabA, ΔcagA, or ΔvirB7 infection compared with uninfected cells or infected MDCK/pIRES cells (Fig. 4, A and B).
FIGURE 4.
The BabA-Leb interaction enhances the TFSS-dependent activity. A, MDCK/Leb or MDCK/pIRES cells were either left uninfected (−) or infected with the noted H. pylori strain for 8 h. The expression levels of CCL5, IL-8, CDX2, and MUC2 were determined by real-time PCR. The results represent the average of six separate experiments (each n = 3). Data are presented as the means ± S.D. (error bars). B, MDCK/Leb or MDCK/pIRES cells were either left uninfected (−) or infected or infected with the noted H. pylori strain for 24 h. The supernatants were subjected to ELISA for the detection of IL-8 (n = 4). Data are presented as the means ± S.D. C, MDCK/Leb or MDCK/pIRES cells were either left uninfected (−) or infected with the noted H. pylori (Hp) strain for 24 h. Cell lysates were subjected to immunoprecipitation with anti-Tyr(P)-CagA (anti-pY-CagA) antibody. Immunoprecipitates and whole-cell lysates were subjected to Western blot analysis using anti-Tyr(P)-CagA or anti-actin, respectively. NS, not significant.
We further analyzed the mRNA levels of intestinal metastasis-related factors, CDX2 and MUC2. The levels of CDX2 and MUC2 upon WT infection, but not ΔbabA, ΔcagA, or ΔvirB7 infection, were increased in MDCK/Leb cells compared with MDCK/pIRES cells (Fig. 4A). To further analyze whether BabA contributes to the delivery of CagA effector protein into the infected cells, MDCK/Leb or MDCK/pIRES cells were infected with the WT strain ATCC 43504 or its ΔbabA, ΔcagA, or ΔvirB7 mutants. When CagA is directly injected from the bacteria into the cell via TFSS, it undergoes tyrosine phosphorylation in the host cells by Src/Abl kinases. As shown in Fig. 4C, a higher level of tyrosine-phosphorylated CagA was detected when MDCK/Leb cells but not MDCK/pIRES cells were cocultured with H. pylori WT. These results indicated that the BabA-Leb interaction has a functional role in enhancing the production of inflammatory cytokines and intestinal metaplasia-related factors upon TFSS-mediated H. pylori infection.
The BabA-Leb Interaction Contributes to Proinflammatory Cytokine Production in Vivo
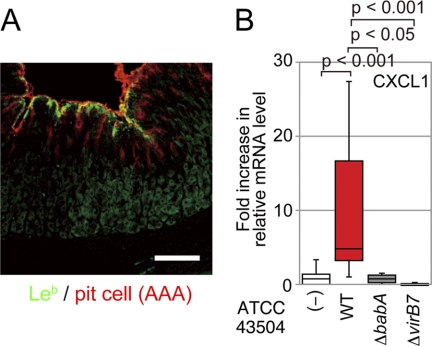
The Mongolian gerbil is widely used as an experimental model for H. pylori infection of the human gastric stomach. Inoculated H. pylori colonizes the stomach of Mongolian gerbils, which eventually develop severe gastritis, ulceration, and adenocarcinoma, simulating human gastric diseases (40–42). Immunohistochemical analysis with an anti-Leb antibody revealed that Leb is expressed in the gastric pit of Mongolian gerbils (Fig. 5A), as was previously reported (43). To investigate whether the BabA-Leb interaction strengthens gastric responses during TFSS-mediated H. pylori infection in vivo, Mongolian gerbils were inoculated with WT, ΔbabA, or ΔvirB7, and the mRNA levels of CXCL1/KC, the functional IL-8 homologue in rodents, in the stomach were measured by RT-PCR after 8 weeks. CXCL1 mRNA was increased more than 10-fold in WT-infected animals compared with the uninfected controls and ΔbabA- or ΔvirB7-infected animals (Fig. 5B). These results further support the notion that BabA potentiates TFSS-dependent proinflammatory cytokine production, which may lead to malignant gastric diseases.
FIGURE 5.
H. pylori induces CXCL1 expression in the gastric region of H. pylori-infected Mongolian gerbils. A, Leb is expressed in the gastric pit of Mongolian gerbils. Gastric sections from animals were immunostained with an anti-Leb antibody (green) and Cy3-conjugated A. anguilla agglutinin (red). Scale bar, 100 μm. B, H. pylori up-regulated CXCL1 mRNA levels in the stomach. Mongolian gerbils were inoculated with H. pylori for 8 weeks. CXCL1 gene expression in the stomach was determined by isolating total stomach RNA and then quantifying CXCL1 mRNA by real-time PCR (n = 9). All plots show the 25th to 75th percentiles (boxes) and 5th to 95th percentiles (whiskers). The line in each box represents the median. Error bars, S.D.
DISCUSSION
Virulence factors must be efficiently secreted from the secretion machinery in order for pathogenic bacteria to establish a successful infection. To our knowledge, this is the first study that formally demonstrates the importance of the BabA adhesin-host Leb interaction in potentiating TFSS-dependent host cell responses in vitro and in vivo.
Approximately 20% of H. pylori in the stomach adheres to the surface of mucus epithelial cells (44), where H. pylori sustains persistent infection. A whole genome analysis revealed that H. pylori has over 30 genes that encode OMPs, and several of these have been classified as adhesins or related proteins, strongly suggesting that the bacteria uses multiple and redundant or variable modes to attach to cell surfaces and to adapt to inflamed environments. To date, the most well studied H. pylori adhesins are BabA, which binds to Leb, and SabA, which binds to sialyl-LeX (5). Several other adhesins have also been identified, including HopZ, HopH (OipA), and AlpA/AlpB, although their receptors are still unknown (45–47). In addition to OMPs, recent papers reported that components of the TFSS, CagL and CagY, have the potential to bind to β1-integrins, which are mammalian adhesion receptors, to mediate cell-cell or cell-extracellular matrix interactions (48, 49). However, in natural, uninjured epithelial tissue, the epithelium has apical-basolateral polarity, and β1-integrin expression is restricted to the basolateral surface of the intact gastric epithelium. Thus, β1-integrin is separated from the H. pylori-accessible apical surface through tight junctions between adjacent cells (50). We previously reported that CagA delivered by the H. pylori TFSS into the gastric epithelium suppressed apoptosis by up-regulating the expression of the antiapoptotic protein MCL1, which greatly contributed to bacterial colonization of the gut epithelium by inhibiting gastric epithelium self-renewal (23). CagA is the only known effector protein that is translocated directly from H. pylori via the TFSS into the epithelial cell cytoplasm, where CagA interacts with various host signaling proteins to elicit three distinct phenotypes. (i) The cytoskeleton is reorganized and cells are elongated, resulting in a scattering cell phenotype in nonpolarized epithelial cells. (ii) CagA associates with tight junction proteins in polarized epithelial cells, which disrupts tight junctions and disintegrates cell polarity. (iii) Several transcription factors that control cell proliferation, inflammation, and survival in vitro and in vivo are activated (18). Therefore, it is possible that at the very early phase of infection, H. pylori binds intimately to the apical epithelial surface via BabA, whose ligand Leb is abundantly expressed on gastric epithelial cells, as shown in Fig. 5A, and delivers CagA into the attached cells. Then CagA induces cellular transcription to produce several proinflammatory cytokines (IL-8 and CCL5), antiapoptotic proteins (MCL1) (23), and precancerous stage-related factors (CDX2 and MUC2). Although we examined the induction of two intestine-specific genes, CDX2 and MUC2, in H. pylori-infected ectopic kidney cell lines (MDCK; Fig. 4A), our data demonstrated that H. pylori is able to induce the expression of these genes in a BabA-Leb-dependent manner. At the same time, CagA-mediated disruption of tight junctions may allow H. pylori to better access normally buried integrins and thereby allow the TFSS to interact with integrins. Although the data presented here do not preclude the possibility that TFSS-delivered factors other than CagA might also contribute to the TFSS-dependent transcriptional activity, we provide clear evidence that the adherence of H. pylori to gastric epithelial cells via BabA and Leb is important to efficiently initiate TFSS-dependent pathogenesis.
Many studies have suggested that there is a relationship between babA-positive H. pylori and clinical outcomes, whereas some papers have reported that there is no correlation between babA2 and intense cellular mucosal inflammation (33). Some studies have reported that the cagA status is related to the Leb-binding activity or the babA gene (8). BabA expression is thought to be highly variable due to several regulatory mechanisms, including transcription, translation, and phase variation (51). Moreover, aside from BabA, H. pylori can utilize many adherence mechanisms, as shown by its ability to bind to AGS cells (Fig. 1C). Therefore, it is likely that the cooperation between BabA and TFSS, as shown here, is functionally coordinated during persistent infection of H. pylori.
We have demonstrated that transducing Leb-negative cells with two (FUT3 and FUT1) or three (B3GALT5, FUT3, and FUT1) relevant glycosyltransferase genes results in Leb expression (Fig. 2 and supplemental Fig. S3). Löfling et al. (52) established Leb-expressing CHO-K1 cells by co-transfecting expression vectors encoding B3Gn-T5, B3GALT5, FUT2, and FUT3. Although we have not yet examined each endogenous glycosyltransferase in native cells, our results suggest that only the FUT1 and FUT3 genes are required for MDCK cells, whereas three genes, including FUT1, FUT3, and BGALT5, are requisite for CHO, NIH3T3, and MEF cells to express Leb on the cell surface. As we demonstrated, these cells are a good model to analyze host cell responses to Leb-mediated bacterial pathogenesis in vitro and will be useful to establish an easy screening system to study the pathogenesis of H. pylori clinical isolates as well as identify drugs relevant to bacterial adhesion.
In summary, we have obtained the first evidence that the BabA-Leb interaction is important not only for H. pylori to adhere to the stomach surface but also to anchor the bacterial secretion system to the host cell surface so that bacterial factors can be effectively injected into the host cell cytosol. Using genetically modified cell lines expressing Leb, isogenic H. pylori mutants defective in babA and virB7, which is a component of the TFSS, and an animal model, we have clearly shown that the binding between BabA on H. pylori and Leb on the host cell surface plays an important role in potentiating TFSS-mediated secretion, resulting in inflammation and intestinal metaplasia, the prominent features of the cag TFSS.
Supplementary Material
Acknowledgments
We thank Yoshiko Kawahara M.S. and Dr. Piao Zhenzi for technical help, Dr. Anna Shevtsova for technical help and discussions, and Dr. Toshio Kitamura for providing pMX-puro vector and PLAT-E cells. We also grateful to Dr. Douglas E. Berg for valuable discussions and all members of the Sasakawa laboratory for helpful discussions and technical advice.
This work was supported by Grant-in-aid for Scientific Research (B) 23390102 (to H. M.), (S) 20229006 (to C. S.), Grant-in-aid for Challenging Exploratory Research 23659220 (to H. M.), Grant-in-aid for Scientific Research on Priority Areas 18073003 (to C. S.), Grant-in-aid for Young Scientists (B) 23790472 (to H. A.), and the Contract Research Fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases (to C. S.), from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. This work was also supported in part by grants from the Naito Foundation (to H. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S8.
- OMP
- outer membrane protein
- TFSS
- Type IV secretion system
- Leb
- Lewis b
- NF-κB
- nuclear factor-κB
- NFAT
- nuclear factor of activated T cells
- MOI
- multiplicity of infection
- MDCK
- Madin-Darby canine kidney
- MEF
- mouse embryonic fibroblast
- TRITC
- tetramethylrhodamine isothiocyanate
- TMB
- 3,3',5,5'-tetramethylbenzidine.
REFERENCES
- 1. Blaser M. J., Atherton J. C. (2004) J. Clin. Invest. 113, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagai S., Mimuro H., Yamada T., Baba Y., Moro K., Nochi T., Kiyono H., Suzuki T., Sasakawa C., Koyasu S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alm R. A., Bina J., Andrews B. M., Doig P., Hancock R. E., Trust T. J. (2000) Infect. Immun. 68, 4155–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borén T., Falk P., Roth K. A., Larson G., Normark S. (1993) Science 262, 1892–1895 [DOI] [PubMed] [Google Scholar]
- 5. Mahdavi J., Sondén B., Hurtig M., Olfat F. O., Forsberg L., Roche N., Angstrom J., Larsson T., Teneberg S., Karlsson K. A., Altraja S., Wadström T., Kersulyte D., Berg D. E., Dubois A., Petersson C., Magnusson K. E., Norberg T., Lindh F., Lundskog B. B., Arnqvist A., Hammarström L., Borén T. (2002) Science 297, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montecucco C., Rappuoli R. (2001) Nat. Rev. Mol. Cell Biol. 2, 457–466 [DOI] [PubMed] [Google Scholar]
- 7. Peek R. M., Jr., Blaser M. J. (2002) Nat. Rev. Cancer 2, 28–37 [DOI] [PubMed] [Google Scholar]
- 8. Ilver D., Arnqvist A., Ogren J., Frick I. M., Kersulyte D., Incecik E. T., Berg D. E., Covacci A., Engstrand L., Borén T. (1998) Science 279, 373–377 [DOI] [PubMed] [Google Scholar]
- 9. Falk P. G., Bry L., Holgersson J., Gordon J. I. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guruge J. L., Falk P. G., Lorenz R. G., Dans M., Wirth H. P., Blaser M. J., Berg D. E., Gordon J. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3925–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holgersson J., Löfling J. (2006) Glycobiology 16, 584–593 [DOI] [PubMed] [Google Scholar]
- 12. Crabtree J. E., Peichl P., Wyatt J. I., Stachl U., Lindley I. J. (1993) Scand. J. Immunol. 37, 65–70 [DOI] [PubMed] [Google Scholar]
- 13. Kikuchi T., Kato K., Ohara S., Sekine H., Arikawa T., Suzuki T., Noguchi K., Saito M., Saito Y., Nagura H., Toyota T., Shimosegawa T. (2000) J. Pathol. 192, 243–250 [DOI] [PubMed] [Google Scholar]
- 14. Shimoyama T., Everett S. M., Dixon M. F., Axon A. T., Crabtree J. E. (1998) J. Clin. Pathol. 51, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuasa Y. (2003) Nat. Rev. Cancer 3, 592–600 [DOI] [PubMed] [Google Scholar]
- 16. Bai Y. Q., Yamamoto H., Akiyama Y., Tanaka H., Takizawa T., Koike M., Kenji Yagi O., Saitoh K., Takeshita K., Iwai T., Yuasa Y. (2002) Cancer Lett. 176, 47–55 [DOI] [PubMed] [Google Scholar]
- 17. Yamamoto H., Bai Y. Q., Yuasa Y. (2003) Biochem. Biophys. Res. Commun. 300, 813–818 [DOI] [PubMed] [Google Scholar]
- 18. Mimuro H., Berg D. E., Sasakawa C. (2008) BioEssays 30, 515–520 [DOI] [PubMed] [Google Scholar]
- 19. Isomoto H., Mizuta Y., Miyazaki M., Takeshima F., Omagari K., Murase K., Nishiyama T., Inoue K., Murata I., Kohno S. (2000) Am. J. Gastroenterol. 95, 2768–2776 [DOI] [PubMed] [Google Scholar]
- 20. Mimuro H., Suzuki T., Tanaka J., Asahi M., Haas R., Sasakawa C. (2002) Mol. Cell. 10, 745–755 [DOI] [PubMed] [Google Scholar]
- 21. Viala J., Chaput C., Boneca I. G., Cardona A., Girardin S. E., Moran A. P., Athman R., Mémet S., Huerre M. R., Coyle A. J., DiStefano P. S., Sansonetti P. J., Labigne A., Bertin J., Philpott D. J., Ferrero R. L. (2004) Nat. Immunol. 5, 1166–1174 [DOI] [PubMed] [Google Scholar]
- 22. Murata-Kamiya N., Kurashima Y., Teishikata Y., Yamahashi Y., Saito Y., Higashi H., Aburatani H., Akiyama T., Peek R. M., Jr., Azuma T., Hatakeyama M. (2007) Oncogene 26, 4617–4626 [DOI] [PubMed] [Google Scholar]
- 23. Mimuro H., Suzuki T., Nagai S., Rieder G., Suzuki M., Nagai T., Fujita Y., Nagamatsu K., Ishijima N., Koyasu S., Haas R., Sasakawa C. (2007) Cell Host Microbe 2, 250–263 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki M., Mimuro H., Kiga K., Fukumatsu M., Ishijima N., Morikawa H., Nagai S., Koyasu S., Gilman R. H., Kersulyte D., Berg D. E., Sasakawa C. (2009) Cell Host Microbe 5, 23–34 [DOI] [PubMed] [Google Scholar]
- 25. Backert S., Naumann M. (2010) Trends Microbiol. 18, 479–486 [DOI] [PubMed] [Google Scholar]
- 26. Yamaoka Y., Kikuchi S., el-Zimaity H. M., Gutierrez O., Osato M. S., Graham D. Y. (2002) Gastroenterology 123, 414–424 [DOI] [PubMed] [Google Scholar]
- 27. Yamaoka Y., Ojo O., Fujimoto S., Odenbreit S., Haas R., Gutierrez O., El-Zimaity H. M., Reddy R., Arnqvist A., Graham D. Y. (2006) Gut 55, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerhard M., Lehn N., Neumayer N., Borén T., Rad R., Schepp W., Miehlke S., Classen M., Prinz C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pride D. T., Meinersmann R. J., Blaser M. J. (2001) Infect. Immun. 69, 1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rad R., Gerhard M., Lang R., Schöniger M., Rösch T., Schepp W., Becker I., Wagner H., Prinz C. (2002) J. Immunol. 168, 3033–3041 [DOI] [PubMed] [Google Scholar]
- 31. Kodama K., Sumii K., Kawano M., Kido T., Nojima K., Sumii M., Haruma K., Yoshihara M., Chayama K. (2002) Helicobacter 7, 9–13 [DOI] [PubMed] [Google Scholar]
- 32. Ohno T., Vallström A., Rugge M., Ota H., Graham D. Y., Arnqvist A., Yamaoka Y. (2011) J. Infect. Dis. 203, 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujimoto S., Olaniyi Ojo O., Arnqvist A., Wu J. Y., Odenbreit S., Haas R., Graham D. Y., Yamaoka Y. (2007) Clin. Gastroenterol. Hepatol. 5, 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Odenbreit S., Kavermann H., Püls J., Haas R. (2002) Int. J. Med. Microbiol. 292, 257–266 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka J., Suzuki T., Mimuro H., Sasakawa C. (2003) Cell Microbiol. 5, 395–404 [DOI] [PubMed] [Google Scholar]
- 36. Morita S., Kojima T., Kitamura T. (2000) Gene. Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 37. Baker B. W., Boettiger D., Spooncer E., Norton J. D. (1992) Nucleic Acids Res. 20, 5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guillemin K., Salama N. R., Tompkins L. S., Falkow S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15136–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snider J. L., Allison C., Bellaire B. H., Ferrero R. L., Cardelli J. A. (2008) J. Biol. Chem. 283, 13952–13963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirayama F., Takagi S., Kusuhara H., Iwao E., Yokoyama Y., Ikeda Y. (1996) J. Gastroenterol. 31, 755–757 [DOI] [PubMed] [Google Scholar]
- 41. Watanabe T., Tada M., Nagai H., Sasaki S., Nakao M. (1998) Gastroenterology 115, 642–648 [DOI] [PubMed] [Google Scholar]
- 42. Sugiyama A., Maruta F., Ikeno T., Ishida K., Kawasaki S., Katsuyama T., Shimizu N., Tatematsu M. (1998) Cancer Res. 58, 2067–2069 [PubMed] [Google Scholar]
- 43. Osawa H., Sugano K., Iwamori M., Kawakami M., Tada M., Nakao M. (2001) Dig. Dis. Sci. 46, 69–74 [DOI] [PubMed] [Google Scholar]
- 44. Hessey S. J., Spencer J., Wyatt J. I., Sobala G., Rathbone B. J., Axon A. T., Dixon M. F. (1990) Gut 31, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peck B., Ortkamp M., Diehl K. D., Hundt E., Knapp B. (1999) Nucleic Acids Res. 27, 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dossumbekova A., Prinz C., Mages J., Lang R., Kusters J. G., Van Vliet A. H., Reindl W., Backert S., Saur D., Schmid R. M., Rad R. (2006) J. Infect. Dis. 194, 1346–1355 [DOI] [PubMed] [Google Scholar]
- 47. de Jonge R., Durrani Z., Rijpkema S. G., Kuipers E. J., van Vliet A. H., Kusters J. G. (2004) J. Med. Microbiol. 53, 375–379 [DOI] [PubMed] [Google Scholar]
- 48. Kwok T., Zabler D., Urman S., Rohde M., Hartig R., Wessler S., Misselwitz R., Berger J., Sewald N., König W., Backert S. (2007) Nature 449, 862–866 [DOI] [PubMed] [Google Scholar]
- 49. Jiménez-Soto L. F., Kutter S., Sewald X., Ertl C., Weiss E., Kapp U., Rohde M., Pirch T., Jung K., Retta S. F., Terradot L., Fischer W., Haas R. (2009) PLoS. Pathog. 5, e1000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 51. Yamaoka Y. (2008) World J. Gastroenterol. 14, 4265–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Löfling J., Diswall M., Eriksson S., Borén T., Breimer M. E., Holgersson J. (2008) Glycobiology 18, 494–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.