Abstract
QnrB1 is a plasmid-encoded pentapeptide repeat protein (PRP) that confers a moderate degree of resistance to fluoroquinolones. Its gene was cloned into an expression vector with an N-terminal polyhistidine tag, and the protein was purified by nickel affinity chromatography. The structure of QnrB1 was determined by a combination of trypsinolysis, surface mutagenesis, and single anomalous dispersion phasing. QnrB1 folds as a right-handed quadrilateral β-helix with a highly asymmetric dimeric structure typical of PRP-topoisomerase poison resistance factors. The threading of pentapeptides into the β-helical fold is interrupted by two noncanonical PRP sequences that produce outward projecting loops that interrupt the regularity of the PRP surface. Deletion of the larger upper loop eliminated the protective effect of QnrB1 on DNA gyrase toward inhibition by quinolones, whereas deletion of the smaller lower loop drastically reduced the protective effect. These loops are conserved among all plasmid-based Qnr variants (QnrA, QnrC, QnrD, and QnrS) and some chromosomally encoded Qnr varieties. A mechanism in which PRP-topoisomerase poison resistance factors bind to and disrupt the quinolone-DNA-gyrase interaction is proposed.
Keywords: DNA Topoisomerase, Drug Resistance, Enzyme Mechanisms, Enzyme Structure, Protein-Protein Interactions, β-Helix, Fluoroquinolones, Repeat Proteins
Introduction
Qnr proteins are members of the pentapeptide repeat protein (PRP)2 family that bind to and protect DNA gyrase and topoisomerase IV from inhibition by quinolones, resulting in reduced susceptibility to this important class of antimicrobial agents (1–3). They have been found encoded by multiresistant plasmids in isolates of Enterobacteriaceae from around the world and may also be identifiable genes on the bacterial chromosome (4). Although quinolone resistance conferred by Qnr proteins is modest, their presence promotes selection of higher levels of resistance in vitro and in vivo (5–8). Five Qnr families (A, B, C, D, and S) are currently recognized (5, 9–12), with QnrB having the highest prevalence, the greatest number of alleles (more than 30), and the earliest documented discovery (13, 14). QnrB is also unique in being under control by the SOS system so that DNA damage produced by quinolones, such as ciprofloxacin, induces its expression by relief of binding to a LexA recognition site upstream from qnrB genes (12, 15).
Other PRPs protect against different topoisomerase poisons. For example, AlbG protects the sugarcane pathogen Xanthomonas albilineans against the albicidin family of antibiotics that it produces and that, like quinolones, are potent inhibitors of DNA gyrase supercoiling (16). McbG is a PRP made for self-protection by producers of microcin B17, a protein topoisomerase poison (17, 18). Finally, MfpA is a PRP encoded on the chromosome of Mycobacterium tuberculosis and other mycobacteria. Deletion of MfpA increases quinolone susceptibility, and augmenting its expression by cloning on a multicopy plasmid reduces susceptibility (19), although in a cell-free system, MfpA lacks quinolone protective activity and only inhibits DNA gyrase at concentrations between 1 and 5 μm (20, 21).
The crystal structure of MfpA suggested a model for its activity (20). MfpA is a dimer with each monomer almost entirely in the form of a right-handed β-helix stabilized by hydrogen bonding between backbone atoms of neighboring coils and with a negative electrostatic surface potential. It thus has features similar to DNA and therefore was proposed to dock against the highly cationic saddle region at the gyrase A2 dimer interface displacing DNA. MfpA and by inference other PRPs were therefore proposed to act by inhibiting the formation of the quinolone DNA-gyrase covalent complex, preventing DNA damage.
In contrast to MfpA, QnrB1 protects DNA gyrase from ciprofloxacin at concentrations as low as 5 pm and only inhibits DNA gyrase at high concentrations (>25 μm) (10). QnrB1 is a superior model system for study of PRP-topoisomerase-poison resistance factors (TPRFs) because its in vitro activities are consistent with the in vivo protective effects of PRP-TPRFs and are similar to the majority of PRP-TPRFs. We report here the structure of QnrB1 by x-ray crystallography and propose a model to explain its protective and inhibitory actions on topoisomerases.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
QnrB1 was PCR-amplified utilizing plasmid pMG298 (10) as a template, 5′-GGGAATTCCATATGACGCCATTACTGTATAAAAAAACAGGTA-3′ and 5′-CGCGGATCCCTAACCAATCACCGCGAT-3′ as primers and subsequently cloned into the NdeI, BamHI site of pET28a (Novagen). QnrB1 mutants were produced using QuikChange mutagenesis (Stratagene) and plasmid pET28a:QnrB1. For the M102R mutant, the primers were 5′-CGCGGCGCAAGCTTTAGGAATATGATCACCACG-3′ and 5′-CGTGGTGATCATATTCCTAAAGCTTGCGCCGCG-3′, and for R167E, the primers were 5′-CGACTTTCGAACTGGGAAGCAGCAGCAAACTTCAC-3′ and 5′-GTGAAGTTTGCTGCTTCCCAGTCGAAAGTCG-3′. QnrB1 was expressed by autoinduction using standard protocols (22, 23). Briefly, plasmid QnrB1 was transformed into BL21DE3 and plated on LB-Agar containing 100 μg ml−1 kanamycin. Four to six colonies were transferred to 75 ml of LB containing the same concentration of kanamycin and shaken (300 rpm) overnight at 37 °C. The overnight culture was used to inoculate ten 2-liter baffled flasks, each containing 400 ml of autoinduction medium (23). The flasks were shaken (300 rpm) for 6 h at 37 °C, then at 23 °C for an additional 24–36 h. The cells were harvested by centrifugation and stored at −80 °C. Cell paste was resuspended to three times the volume with buffer A (50 mm Tris, pH 8.0, 200 mm (NH4)2SO4, 10% glycerol, 20 mm imidazole) supplemented with 1 mg ml−1 lysozyme, 0.1 mg ml−1 DNase, and 0.5% Triton X-100. Following sonication and clarification by centrifugation (both at 4 °C), the supernatant was applied to a 2.5 × 15-cm Ni-NTA column (maintained at 20 °C) that had been equilibrated against buffer A. The column was washed extensively with buffer A, and proteins were eluted with buffer B (50 mm Tris, pH 8.0, 200 mm (NH4)2SO4, 10% glycerol, 300 mm imidazole). The collected elutant was immediately aliquoted, snap frozen in liquid N2, and stored at −80 °C.
Structure-based Loop Deletion Mutants
Loop A deletion (ΔY46-Q51) mutant, loop B deletion (ΔM104-S113) mutant, and loop AB double deletion (ΔY46-Q51/ΔM104-S113) mutant were constructed using overlap extension PCR, cloned, expressed, and purified as described above. Desired deletions and the absence of any other mutations were confirmed by the DNA sequencing of the cloned constructs.
Trypsin Treatment
Protein (at 10–20 mg ml−1 in buffer B) was thawed from −80 °C storage and dialyzed overnight against storage buffer C (20 mm Tris, pH 8.0, 10 glycerol, 50 mm arginine) with the addition of trypsin (Sigma T-1005, 1:500 w/w ratio). Retention of an oligomeric state and protein mass was monitored by size exclusion chromatography on a 2.5 × 100-cm Sephadex column (Pharmacia) with samples compared against gel filtration standards. Trypsin-treated protein was immediately utilized for crystallization or snap frozen in liquid N2 and stored at −80 °C.
Crystallization
Wild type
Protein was dialyzed overnight against storage buffer C at a protein concentration <1 mg ml−1. Protein was concentrated by ultracentrifugation to 4 mg ml−1 and utilized immediately for crystallization. Crystallization was by vapor diffusion under oil utilizing 2 + 2 μl (reservoir + protein) drops under 150 μl of silicon oil (Fisher) in 96-well plates stored open to room humidity. Wild type QnrB1 crystallized as hexagonal plates in 100 mm sodium/potassium phosphate, pH 4.5, 2 m NaCl at 4 °C.
M102R
Trypsin-treated M102R (10 mg ml−1) crystallized as thick tetragonal rods in 100 mm citrate/phosphate, pH 4.5, 1 m (NH4)2SO4 at 20 °C.
R167E
Trypsin-treated R167E (10 mg ml−1) crystallized as irregular wedges in 100 mm BisTris/propane/citrate, pH 7.5, 15% PEG 3350 at 20 °C.
Structure Determination
M102R
Crystals of M102R were soaked in 100 mm sodium acetate, pH 4.5, 1 m (NH4)2SO4, 28% glycerol, 100 mm diethylenetiamine pentaacetic acid GdIII for 5 min prior to vitrification in liquid N2. The data were generated on a RU-200/R-axis IV++ (Rigaku) and processed using MOSFLM (24) and CCP4 (25). The data were nonisomorphous with native data, so phases were calculated using single anomalous dispersion. A single diethylenetiamine pentaacetic acid GdIII-binding site was found by PHENIX (26), and the resultant single anomalous dispersion solvent-flattened phased map was submitted to autobuilding by ARP/WARP (27). The significantly built model (>75%) was completed by iterative cycles of fitting in COOT (28) and refinement in PHENIX. A high resolution M102R data set was collected on Beamline X25 at National Synchrotron Light Source and processed with HKL3000. There is a dimer in the asymmetric unit with a solvent content of 65%.
E167R
Crystals of E167R were cryoprotected in 100 mm BisTris/propane/citrate, pH 7.5, 30% PEG 3350 and vitrified by liquid N2. The data were collected on Beamline X25 at National Synchrotron Light Source and processed with MOSFLM and CCP4. The structure was determined by molecular replacement within PHENIX utilizing a single M102R subunit as the search model. There is a dimer per asymmetric unit with a solvent content of 42%.
Wild Type
Crystals of QNRB1WT were transferred to 100 mm potassium/sodium phosphate, 2 m NaCl, 30% glycerol for 20 min and vitrified by liquid N2. The data were generated on a RU-200/R-axis IV++ and processed using MOSFLM and CCP4. The structure was determined by molecular replacement within PHENIX utilizing the M102R dimer as the search model. There is a tetramer per asymmetric unit with solvent content of 48%. The data collection and refinement statistics are listed in Table 1. Computation of buried surface area at the dimer interface was calculated in PISA (29). Structure figures were synthesized within PyMOL. Atomic coordinates and experimental structure factors for QnrB1WT, QnrB1M102R, and QnrB1R167E have been deposited in the Protein Data Bank (codes 2xtw, 2xtx, and 2xty).
TABLE 1.
Data collection and refinement statistics
Statistics in parenthesis are for the highest resolution bin. DTPA, diethylenetiamine pentaacetic acid GdIII.
| Data set | Native | M102RTrypsn | DTPA derivative (M102RTrypsin) | R167ETrypsin |
|---|---|---|---|---|
| Space group | P212121 | I422 | I422 | P41212 |
| Unit cell (Å, °) | a = 38.1, b = 119.532, c = 231.725 | a = 101.1, c = 275.6 | a = 101.5, c = 275.6 | a = 55.3, c = 282.5 |
| Resolution (Å) | 40-2.8 (2.8-2.95) | 40-2.2 (2.24-2.2) | 40-2.5 (2.64-2.5) | 40-1.8 (1.9-1.8) |
| Completeness (%) | 99.0 (98.2) | 99.3 (99.5) | 100.0 (100.0) | 94.5 (75.3) |
| Redundancy | 7.8 (7.4) | 7.8 (7.7) | 12.5 (11.7) | 7.0 (2.7) |
| I (mean (S.D.)) | 18.7 (6.5) | 27.0 (6.0) | 19.4 (4.2) | 19.3 (2.8) |
| Rsym | 0.077 (0.277) | 0.060 (0.366) | 0.084 (0.557) | 0.061 (0.355) |
| Wilson B factor (Å2) | 39.6 | 36.0 | 56.4 | 15.7 |
| Model and refinement data | ||||
| Resolution (Å) | 40-2.8 (2.9-2.8) | 40-2.2 (2.2-2.28) | 40-1.8 (1.8-1.86) | |
| Unique reflections | 26120 (2734) | 35308 (3097) | 39399 (2492) | |
| Rcryst (%) | 22.5 (33.9) | 19.3 (24.3) | 18.0 (23.8) | |
| Rfree (%, 5% of data) | 30.1 (46.6) | 22.2 (32.0) | 22.1 (30.0) | |
| Contents of model | ||||
| Residues (1–x) | A1–210, B1–212, C4–212, D3–210 | A1–102, A112–213, B1–107, B109–214 | A1–102, A111–213, B1–101, B112–214 | |
| Waters | - | 190 | 323 | |
| Other | - | 10 (SO4 × 2) | - | |
| Atoms total | 6610 | 3505 | 3542 | |
| Average B-factor (Å2) | ||||
| Protein/waters | 38.1 | 40.5/64.2 | 17.9/26.1 | |
| RMSD | ||||
| Bond lengths (Å)/angles (°) | 0.008/1.19 | 0.007/0.978 | 0.010/1.18 | |
| MolProbity statistics | ||||
| Ramachandran | ||||
| Favored/outliers (%) | 90.3/0.24 | 97.3/0.0 | 99.0/0.0 | |
| Rotamer outliers (%) | 4.7 | 0.87 | 1.5 | |
| Clash scorea | 24.1 (81st percentile) | 5.6 (98th percentile) | 45.6 (97th percentile) | |
| Overall scorea | 2.92 (71st percentile) | 1.43 (99th percentile) | 1.36 (97th percentile) | |
a The scores are ranked according to structures of similar resolution as formulated in MolProbity.
Gyrase Assays
DNA supercoiling assays were performed using Escherichia coli gyrase assay kits (Inspiralis) according to the manufacturer's instructions. The reaction mixture containing 3 units (a unit is defined as the amount of gyrase required to convert 0.5 μg of relaxed pBR322 into completely supercoiled form at 37 °C in 30 min) of gyrase and 0.4 μg of relaxed DNA in a volume of 30 μl in gyrase assay buffer (35 mm Tris·HCl, pH 7.5, 24 mm KCl, 4 mm MgCl2, 2 mm DTT, 1.8 mm spermidine, 1 mm ATP, 6.5% glycerol, and 0.1 mg ml−1 BSA) was incubated at 37 °C for 30 min and QnrB1 (WT and deletion mutant forms); ciprofloxacin and novobiocin were included where appropriate. The reactions were terminated by the addition of 30 μl of chloroform/iso-amyl alcohol (24/1). The resulting topoisomers were separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light. For cleavage complex stabilization assays, 6 units of gyrase was used, and the assays were performed as described above. The reactions were terminated by the addition of 0.2% SDS, and the mixtures were further incubated with proteinase K (0.1 mg ml−1) for 30 min at 37 °C before chloroform/iso-amyl alcohol extraction and agarose gel electrophoresis. Inhibition and/or protection were calculated from band intensities using Syngene Bioimage analysis software.
RESULTS
QnrB1 was expressed with a thrombin-cleavable N-terminal His6 tag. Expression of soluble protein was very high with yields after Ni-NTA elution of ∼25 mg gm−1 of cell paste. QnrB1 required immediate dilution to <1 mg ml−1 in buffers containing 10% glycerol and 50 mm arginine (buffer C; see “Experimental Procedures”) to remain soluble. Alternatively, the Ni-NTA eluting fractions could be snap frozen and stored at −80 °C or diluted to 50% glycerol and stored at −20 °C. QnrB1 co-eluted with the 44-kDa gel filtration marker on size exclusion chromatography, consistent with QnrB1 being a molecular dimer (expected, 50 kDa).
QnrB1 with His6 tag, in buffer C, and briefly raised to 4 mg ml−1, formed crystals in a single condition at 4 °C (see “Experimental Procedures”). A data set was collected to 2.8 Å, but this crystal form was not consistently reproducible. Removal of the His6 tag produced thin needles in a number of conditions, but none produced useful diffraction.
Attempts were made to increase the solubility of QnrB1 by altering its surface characteristics and in parallel influence crystallization space. PRPs are excellent candidates for effective surface modification (31–33), because a priori to the structure we know the i−1, i+1, and i+2 residues are solvent-exposed (34). In preliminary experiments, hydrophobic residues were converted to arginines (M60R, L82R, M102R, C112R, V130R, L82R/M102R, and L82R/V130R) in an attempt to increase the surface charge. In another experiment, lysines and arginines were converted to glutamates (K52E, R87E, and R167E), because glutamates exhibit improved solvation properties (35). After overnight dialysis with 1 unit/mg thrombin in 20 mm Tris, pH 8.0, 200 mm ammonium sulfate, all of the hydrophobic-to-arginine mutants were less soluble than wild type. In contrast, all three lysine/arginine-to-glutamate mutants were at least as soluble as wild type, with the K52E and R167E stable for at least a week at 10–15 mg ml−1. Some of the mutants produced favorable improvement in the dimensions of the previously obtained needle-shaped crystal form, but they did not improve their diffraction characteristics.
Coincident with these experiments, it was observed that treatment of precipitated QnrB1 with trypsin resulted in its solubilization. SDS-polyacrylamide electrophoresis suggested that trypsin had removed the N-terminal tag and clipped the protein at least once, and possibly twice, within the main body of the protein. On size exclusion chromatography, however, the trypsin-treated protein remained a similarly sized dimer, suggesting that the clipping was at points that did not affect the cohesion of the protein structure. Crystal trials were reinitiated with trypsin-treated protein wherein trypsin was added to the various constructs in a 1 to 500 w/w ratio in overnight dialysis against buffer C. Two of the mutants, M102R and R167E, produced crystals that were suitable for structure determination. The R167E mutant diffracted to the highest resolution (1.8 Å) but was nonideal because of a long cell axis (282 Å); therefore, the M102R mutant (2.2 Å) was utilized, and its structure was determined by single anomalous dispersion from a gadolinium derivative. The high resolution structure of the R167E mutant and of uncleaved QnrB1 (2.8 Å) were determined by molecular replacement utilizing the M102R mutant as a search model.
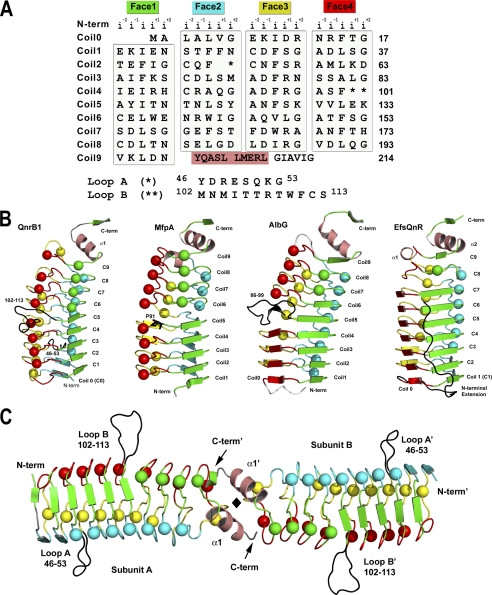
As suggested by its primary sequence (Fig. 1A), QnrB1 folds as a right-handed quadrilateral β-helix. Each of the repeats assumes one face of an approximately regular quadrilateral (coil), with each coil rising roughly 4.8 Å (Fig. 1B). There are a total of 10 coils, numbered 0 to 9, with full coils (four sides) for the body of the helix and partial coils at the termini (coils 0 and 9). The C terminus is capped by a dimerization module, whereas the N terminus is capped by nontypical PRP residues at the i−2 position (Glu-8 and Glu-18) with a salt bridge between Glu-8 and Arg-14 covering a portion of the bottom of β-helical coil 0. The dimerization module consists of a β/α/β structure common to all PRP-TPRFs (36). Dimerization is such that the β-helices are coaxial, producing a highly asymmetric cylindrical structure (15–30 Å diameter by 110-Å length) (Fig. 1C). The contact area is small and hydrophobic (∼ 730 Å2/subunit), allowing flexibility in the orientation of the two subunits. In contrast to the dimeric nature of the PRP-TPRFs, all PRP non-TPRFs determined to date are monomeric (37–39).
FIGURE 1.
Structure of QnrB1. A, structure-based PRP sequence diagram. The sequence of QnrB1 is segmented into four columns representing the four faces of the right-handed quadrilateral β-helix. The face name and color are represented at the top followed by the naming convention for the five residues of the pentapeptide repeats. Loops A and B are indicated by one and two asterisks, respectively, with their sequences indicated below. The N-terminal α-helix is blocked in a salmon color. B, monomer structures of QnrB1, MfpA, AlbG, and EfsQnr in a similar orientation and colored by face. C, dimeric structure of QnrB1. The A and B loops of QnrB1 are shown as a black trace. The molecular 2-fold is shown as a black diamond. Type II turn containing faces are shown as spheres, whereas type IV-containing faces are shown as strands.
The structure of any PRP β-helix can be roughly modeled by analysis of the primary sequence, because its general features are known. What cannot be determined a priori is the type of hydrogen bonding between coils (isolated β-bridges or full β-sheet) and the structures of excursions from the typical PRP sequence. These excursions often affect the β-helical axis and/or produce loops that alter the sequential stacking order of the pentapeptides (34, 37).
The structure of QnrB1 demonstrates that most of the coils are constructed from pentapeptides that take the type II turn conformation with an isolated β-bridge between coils (spheres in Fig. 1B). There are a total of seven pentapeptides that are in the opposing conformation with type IV turns and full hydrogen bonding between coils (strands in Fig. 1B). These are all located on faces 1 and 2 and are in the N-terminal portion of the β-helix. This theme is common among PRP-TPRFs in clustering of the type II-turn pentapeptides toward the C-terminal end. The type II-turn conformation condenses the pentapeptide by ∼0.5 Å, such that coils with a preponderance of type II turns have a smaller diameter (15 Å versus 30 Å).
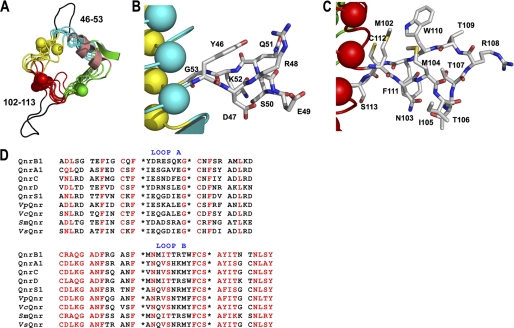
There are two loop excursions from the β-helix that protrude out into solvent: loop A, connecting face 2 to face 3 on coil 2 (residues 46–53), and loop B, which connects face 4 to face 1 of coils 4 and 5 (residues 102–113) (Figs. 1 and 2). The smaller A loop (8 versus 12 residues) is constructed with an isolated β-bridge (Asp-47 to Lys-52) and a 4-residue turn (Fig. 2, A and B), whereas the B loop is constructed of β-strands (Met-102–Asn-103 to Phe-111–Cys-112) with connecting residues labeled as a “bend” by the secondary structure detection program DSSP (40) (Fig. 2, A and C). The loops form highly extended structures with the A and B loop projecting ∼10 and 15 Å from the β-helix, respectively. In the QnrB1 wild type structure, there are four copies in the asymmetric unit, and in all four copies, the loops are in the same conformation, indicating that the observed conformation is encoded by the sequence and not an artifact of crystal packing. Loop A does not distort the interactions between coils 1, 2, and 3 on face 2 with only a 0.2–1.0 Å increase from the expected intercoil distances (∼4.8 Å). The conformation of loop A is partially supported by the packing of the side chain of Asp-47 against Phe-26, the i+1 residue of coil 1, and the packing of the side chain of Tyr-46 against Ser-72 and Met-73, the i+1 and i+2 residue of coil 3 (Fig. 2B). In contrast, loop B has an approximate 2-Å increase from expected between coils 4 and 5 on face 4 with the packing of Met-102 and Cys-112 against Val-130, the i−1 residue of coil 5, acting as a wedge. In addition, there is a hydrogen bond between the indole nitrogen of Trp-110 and the side chain of Glu-132, the i+1 residue of coil 5, that may anchor the loop conformation in relation to the β-helix. The wedging of the β-helix results in a change in the β-helical axis after the disruption and is reminiscent of a similar feature in the structures of MfpA (20) and AlbG (36). Examination of the trypsin treated M102R and the R167E structures suggest that the trypsinolysis occurred within loop B. There was continuous electron density in both structures for loop A, but missing electron density for residues in the B loop (M102R, A103–111, B108; R167E, A103–110, B103–110). The mutant structures also indicate that cleavage within the B loop did not affect the change in the β-helical axis between coils 4 and 5. Treatment of QnrB1 with trypsin resulted in a dramatic increase in protein solubility with trypsin-treated protein soluble to >100 mg ml−1. In addition, mutational deletion of either loop dramatically affected the ability of QnrB1 to rescue gyrase from fluoroquinolone inhibition (see below).
FIGURE 2.
The extruding loops of QnrB1. A, looking down the β-helix from the C terminus to the N terminus showing the position and the extended conformation the A and B loops take from the β-helix. B, stick representation of loop A. C, stick representation of loop B. The residues are colored by atom type. D, sequence alignment of loop A (top panel) and loop B (bottom panel) sequences of plasmid encoded (QnrA-S) and chromosomally encoded Qnr proteins. The sequences are highlighted based on a much larger sequence alignment created with the sequence conservation program CONSURF (30, 55). Sequence positions that scored from 7 to 9 (of a possible 10) and that were labeled as conserved are highlighted in red. Outside of the loop structures (bounded by the asterisks), the sequences are sectioned as pentapeptides.
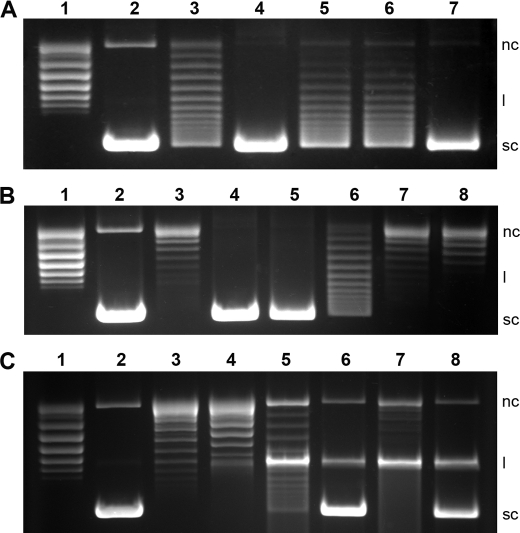
QnrB1 purified for this study did not inhibit the DNA supercoiling activity of E. coli gyrase up to 5 μm concentrations, whereas it protected gyrase against ciprofloxacin similar to the results previously obtained for the in vitro activity of QnrB1 (10). 20 nm QnrB1 completely protected gyrase against 5 μm ciprofloxacin, and partial protection was observed at 10 μm ciprofloxacin (Fig. 3A). However, increased concentrations of QnrB1 (up to 200 nm) failed to provide complete protection to gyrase against ciprofloxacin concentrations above 5 μm (data not shown). The ability of the structure-based loop A deletion mutant (ΔY46-Q51) to rescue gyrase from ciprofloxacin decreased drastically. It partially rescued fluoroquinolone inhibition of gyrase (∼50% protection compared with wild type) at a concentration of 2 μm. Structure-based loop B deletion mutant (ΔM104-S113) and double deletion mutant (ΔY46-Q51/ΔM104-S113) QnrB1 completely lost the ability to protect gyrase against the inhibitory effects of ciprofloxacin (Fig. 3B). None of the mutants inhibited the gyrase at concentrations as high as 5 μm (data not shown). QnrB1 failed to rescue novobiocin-mediated ATPase activity inhibition of GyrB (Fig. 3C). Up to 1 μm QnrB1 could not provide any detectable protection against 2 μm novobiocin (data not shown). In a cleavage complex stabilization assay, 4 μm ciprofloxacin blocked more than 90% of gyrase activity, whereas 16 μm completely blocked the gyrase activity with an increase in cleaved forms of DNA (Fig. 3C). The presence of QnrB1 (200 nm), as determined by the formation of supercoiled DNA, reversed the stabilization by ∼80 and 30% against 4 and 16 μm ciprofloxacin, respectively (Fig. 3C).
FIGURE 3.
Inhibition of DNA gyrase by ciprofloxacin/novobiocin and protection by QnrB1. A, QnrB1-mediated protection of gyrase (supercoiling activity) against ciprofloxacin. Lane 1, relaxed plasmid pBR322 alone; lane 2, relaxed pBR322 plus gyrase; lanes 3–7, relaxed pBR322, gyrase, and 2 μm ciprofloxacin in the presence of 20 nm QnrB1WT (lane 4), 2 μm loop-AB double deletion (lane 5), loop B deletion (lane 6), and loop A deletion (lane 7) mutants. B, concentration-dependent inactivation of gyrase (supercoiling activity) by ciprofloxacin in the presence of QnrB1. Lane 1, relaxed plasmid pBR322; lane 2, relaxed pBR322 plus gyrase; lane 3, relaxed pBR322, gyrase, and 10 μm ciprofloxacin in the absence of QnrB1WT; lanes 4–8, 2 (lane 4), 5 (lane 5), 10 (lane 6), 25 (lane 7), and 50 μm (lane 8) ciprofloxacin in the presence of 40 nm QnrB1WT. C, effect of QnrB1 on ATPase inhibition and cleavage complex stabilization. Lane 1, relaxed plasmid pBR322; lane 2, relaxed pBR322 plus gyrase; lanes 3 and 4, relaxed pBR322, gyrase and 2 μm novobiocin in the absence of QnrB1WT (lane 3) and in the presence of 200 nm QnrB1WT (lane 4); lanes 5–8, cleavage complex stabilization assays using 4 μm (lanes 5 and 6) and 16 μm ciprofloxacin (lanes 7 and 8) in the absence of QnrB1 (lanes 5 and 7), and in the presence of 200 nm QnrB1 (lanes 6 and 8), respectively. nc, l, and sc indicate the nicked circular, linear, and supercoiled forms of pBR322, respectively.
DISCUSSION
PRPs can currently be classified into three groups. The largest number of PRPs originates from cyanobacterial genomes, and several of their structures have been determined (37–39). These proteins conform fairly strictly to the PRP consensus sequence, are highly symmetrical being almost entirely composed of pentapeptides in the isolated β-bridge/type II-turn conformation, and have so far been found to be monomeric in structure. Versions with C-terminal α-helical capping (Rfr32 (37)) and N-terminal α-helical capping (RfR23 (38), NP275/276 (41), and HetL (42)) have been observed, and several of these proteins contain loop excursions (HetL and Rfr23), although none drastically change the pitch of the β-helix. Their function has yet to be determined, although Rfr23 and HglK have been found to be involved in manganese uptake and heterocyst formation, respectively (42, 43). The remaining PRPs fall into two groups, both of which have a topoisomerase poison resistance factor activity. In the first group are a number of chromosomally encoded enzymes that either have a known poison for which the PRP acts as a resistance factor or an unknown function with a side activity of reducing fluoroquinolone susceptibility. For example, the proteins McbG and AlbG engender a self-resistance to the topoisomerase II poisons microcin B17 and albicidin (16, 17), respectively, whereas the PRP proteins MfpA and Qnr from Enterococcus faecalis (EfsQnr) confer fluoroquinolone resistance but have no known cellular function (19, 44). The structure of MfpA from M. tuberculosis, AlbG from X. alibilines, and EfsQnr have been determined (Fig. 1B) (20, 36, 45). Their unique features over the cyanobacterial PRPs include divergence from the consensus sequence, inclusion of more β-strand interactions between coils (type IV turns), alterations in the pitch of the β-helical axis, and inclusion of a dimerization module at the C terminus. Despite their similarity in structure and the underlying constraints of the PRP consensus sequence, they exhibit low sequence identity (<25%). In the second group are a number of plasmid-based PRP-TPRFs that have collectively been called Qnr proteins. These proteins include QnrA, QnrB, QnrC, QnrD, and QnrS, with sequence identities of >30% between them and >70% for allelic variants (4, 13). Qnr proteins were originally recognized on resistance plasmids (5). Some plasmid-based PRP-TPRFs appear to have originated from the chromosomal pool of PRP-TPRFs. For example, the chromosomally encoded protein Qnr from Shewanella algae (SaQnr) has >98% amino acid identity with QnrA1 (46, 47). The structure of QnrB1 determined here is the first from the plasmid-based PRP-TPRFs and highlights their similarity to the chromosomally encoded group. Like MfpA, AlbG, and EfsQNR, QnrB1 is a highly asymmetric dimer of similar dimensions, with a mix of type II and type IV turn pentapeptides. Like MfpA and AlbG, there is a helical kink between coils 4 and 5 on face 4, and in the case of AlbG, there is a similarly located loop excursion.
Conservation of Loops A and B
Prior to the determination of the structure of QnrB1, it had been noted that a characteristic feature of Qnr proteins is that they are formed by two domains of pentapeptide repeats separated by a single amino acid, glycine, followed by a cysteine (1). Additional analysis of the predicted pentapeptide preceding the glycine reveals that there are nontypical PRP residues at the internal positions (i−2 and i). For example, in QnrB1, the i−2 residue is an arginine (Arg-48), and the i residue is a serine (Ser-50). A similar analysis of later repeats notes similar discontinuities for the i−2 and i residues of repeats approximately midway through the sequence, for example, the predicted i−2 residue Met-104 and i residue Thr-106 of QnrB1. However, this particular discontinuity is easily overlooked because there is not a disruption in the frame of 5-residue elements between clearly defined pentapeptides. The structure of QnrB1 can now explain these discontinuities, with the former resulting in an 8-residue loop (Loop A) and the later a 12-residue loop (Loop B). Examination of all Qnr variants, including those chromosomally encoded PRPs with >35% similarity with QnrB1, demonstrates the preservation of the A and B loops. However, only the B loop has any significant sequence conservation within the loop for QnrA, B, C, D, and S with a consensus sequence of FXNX(I/V)(S/T)XXX(W/F/Y)FCX(A/V)(Y/F/H)(I/L/M), with the B-loop underlined and the i residue of the adjoining pentapeptides in bold (Fig. 2D). There are residues that are interestingly positioned in the loop, suggesting a conserved structural conformation and or possible contact surface with topoisomerases. The side chains of Asn-103 and Thr-106 are hydrogen-bonded going into the bend and appear to hold Ile-105 out into solvent. A similarly positioned hydrophobic residue is held outwards in the loop structure of AlbG (Val-94, Protein Data Base code 2xt2). The run of residues from Phe-111 to Ile-116 is highly conserved, but it is difficult to determine whether these are genuine activity-based conservations or are structurally conserved to maintain proper re-entry into the β-helix. The higher conservation within the B loop is mirrored by the larger effect on deletion of this loop, with the B-loop deletion abrogating any protection of gyrase from fluoroquinolones, whereas the A-loop deletion has a lesser impact.
A Rescue Model for PRP-TPRFs
Previously an inhibition model was proposed for the manner in which the chromosomally encoded pentapeptide repeat protein MfpA was able to engender a fluoroquinolone resistance phenotype (20). This model was based on biophysical data in which MfpA inhibited E. coli gyrase supercoiling with an IC50 of 1.2 μm, an inability of MfpA to rescue gyrase from fluoroquinolone inhibition, and the structure of MfpA, which had charge and shape characteristics reminiscent of B-form DNA. A convincing model of MfpA bound to the G-segment DNA-binding saddle of gyrase was generated, suggesting that MfpA was outcompeting the generation of the initial gyrase-DNA complex. The binding of gyrase by MfpA would inhibit formation of the gyrase-covalent DNA-fluoroquinolone complex and thereby block the bacteriostatic (stalled ribosomes) and bactericidal (double strand breaks) effects of fluoroquinolones. The question can be asked, however, how the cell can function with an inhibited gyrase, and in such an inhibition model, how are PRPs regulated such that they are generated only when needed? Further problems with this model arose with the determination of the structures of several topoisomerase II enzymes with DNA bound to the DNA gate. In these structures, the DNA segment takes a highly bent conformation, with a central region located deep within the saddle to react with the active site tyrosines exiting at a 150° angle along the topoisomerase II tower domains (48–52). It is doubtful that MfpA could be bent in a similar fashion as to make similar contacts with the DNA saddle.
The accumulation of data for a diverse set of PRP-TPRFs suggests that the assay data for MfpA may be atypical of the family. QnrA1 at 320 nm was found to reverse 50% of gyrase inhibition by 1.5 μm ciprofloxacin, with no inhibition of gyrase at the highest QnrA1 concentrations tested (2.01 μm) (1–3). QnrB4 increased the IC50 of ciprofloxacin 5-fold at 0.5 μm and had protective effects as low as 100 nm (21). QnrB4 did not inhibit E. coli DNA gyrase supercoiling unless concentrations were at least as high as 30 μm. Similarly, QnrB1 at 0.5 nm was found to reverse 50% of ciprofloxacin (6 μm) inhibition of supercoiling with some protection even at 5 pm. Some inhibition of supercoiling was observed at the highest concentration tested (25 μm) (10). The chromosomally encoded PRP-TPRF, EfsQnr was found to protect gyrase against ciprofloxacin inhibition partially, and partial protection was observed even at the lowest tested concentration of 20 nm (44, 45). The determined IC50 value for gyrase inhibition by ciprofloxacin was 0.25 μm, whereas in the presence of 0.2 μm EfsQnr, the IC50 increased to 1.4 μm. Purified EfsQnr inhibited the ATP-dependent DNA supercoiling activity of E. coli gyrase with a calculated IC50 value of 1.2 μm. Finally, purified AlbG (0.65 μm) protected gyrase from the effects of albicidin in supercoiling assays, increasing the IC50 for albicidin by 2–4-fold, with little to no effect on the sensitivity to ciprofloxacin (16). AlbG partially inhibited the supercoiling activity of DNA gyrase in the absence of albicidin with an IC50 of 6 μm.
Taken together, these results suggest that the main protective effect, which manifests in the phenotypic resistance to gyrase poisons, arises from the interaction of PRP-TPRFs at submicromolar concentrations with the topoisomerase-poison-cleavage complex. In the new model, the PRP-TPRFs act by binding to and destabilizing the topoisomerase-poison-cleavage complex, causing release of the poison and allowing religation and release of DNA. In this model, religation of the DNA would drive the release of the PRP-TPRF and regeneration of a catalytically active form of the topoisomerase. Inhibition of gyrase at higher concentrations of PRP-TPRFs would be the byproduct of a weaker binding to the apo or DNA-bound topoisomerase. Type II toposiomerases contain two major catalytic components; energy transduction resulting from the ATP hydrolysis and DNA strand cleavage/religation that are carried out by different subunits. Topoisomerase inhibitors primarily inhibit either ATP hydrolysis or strand cleavage/religation. Fluoroquinolones primarily inhibit the strand cleavage/religation with a secondary ATPase inhibition, whereas novobiocin and other aminocoumarins inhibit the ATPase activity. The experimental data obtained here for QnrB1-mediated rescue of gyrase activity further support the above model. The inability of QnrB1 to rescue the gyrase from novobiocin inhibition rules out the possibility of QnrB1 protecting the gyrase from ATPase inhibition by quinolones, although the reversal of quinolone-mediated cleavage complex stabilization by QnrB1 suggests that the gyrase protection activity of QnrB1 emanates from the destabilization of quinolone-induced stabilization of the cleavage complex.
In general, topoisomerase II-DNA complexes remain predominantly in the closed, uncleaved state, only progressing to the cleaved state upon interaction with a T-segment DNA (53). As such, the conformation of the topoisomerase that the PRP-TPRFs observe would not accumulate to levels sufficient for PRP-TPRF binding if PRP-TPRF levels are low. When topoisomerase II enzymes interact with topoisomerase poisons, the DNA-cleavage state is stabilized and accumulates to high levels such that low constitutively expressed PRP-TPRFs can act. We envision that the PRP-TPRFs are tuned to the specific conformation driven by the topoisomerase-poison pair, such that there is both species specificity and poison specificity. PRP-TPRFs are ideal for evolutionary selection against protein-DNA interfaces, because they have an extended modular structure, with few if any restrictions on the surface residues (i−1, i+1, and i+2). AlbG and EcMcbG are the only chromosomally encoded PRP-TPRFs for which the corresponding topoisomerase poisons (albicidin and microcin B17, respectively) are known. In these cases, the genes for the PRP-TPRFs are part of the chromosomal operon for the construction of the poison itself and act as self-immunity factors. In other examples of chromosomally encoded PRP-TPRFs that appear not to be encoded in an operon, we envision that they are constitutively expressed and act against cellular metabolites that cross-react to stabilize the topoisomerase-DNA cleavage complex. In this model, the plasmid-based fluoroquinolone resistance PRPs were selected from the large pool of chromosomally based PRPs based on the ability to destabilize a similar topo-DNA-poison conformation. The structural determination of PRP-TPRFs does not yet highlight the exact method by which PRP-TPRFs would destabilize the topoisomerase-DNA-poison cleavage complex, but their structures do suggest they interact with topoisomerase in a similar fashion.
Upon final preparation of this manuscript the structure of a chromosomally encoded Qnr, Qnr from Aeromonas hydrophila (AhQnr), was published (54). Like QnrB1, two similarly positioned surface loops project from the surface of the AhQnr β-helix. QnrB1 and AhQnr exhibit 41% sequence identity, with 63 and 42% sequence identity in loop A and loop B, respectively. Similarly, deletion of AhQnr-loop A resulted in a diminution of its protective effect against fluoroquinolone inhibition of DNA gyrase supercoiling activity, whereas deletion of AhQnr-loop B or both loops completely eliminated the protective effect. The authors produced a molecular model of AhQnr bound to the central DNA-binding cleft of gyrA and propose that loop A interacts with the gyrA “tower” and loop B with the gyrB TOPRIM domains. The AhQnr:gyrase molecular model is conceptually analogous to the molecular model produced for the binding of MfpA to the gyrA59 fragment (20). Although these molecular models are visually enticing, we now believe they are inconsistent with the in vitro data, which indicates that PRP-TPRFs are not inhibitory to DNA gyrase supercoiling at physiologically relevant PRP concentrations. For example, neither AhQnr nor the AhQnr loop deletion mutants showed any detectable inhibition of supercoiling activity at AhQnr concentrations of up to 40 μm. We therefore are proposing an alternative model in which PRP-TPRFs act by preferentially binding to and destabilize the topoisomerase-DNA-poison cleavage complex. Details of this interaction await further biophysical protein-protein mapping and/or a topoisomerase-PRP-TPRF complex structure.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI033696 (to J. S. B.) and R01AI057576 (to D. C. H. and G. A. J.).
The atomic coordinates and structure factors (code 2xtw, 2xtx, and 2xty) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PRP
- pentapeptide repeat protein
- PRP-TPRF
- pentapeptide repeat protein topoisomerase-poison resistance factors
- EfsQnr
- E. faecalis Qnr
- AhQnr
- A. hydrophila Qnr
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Tran J. H., Jacoby G. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5638–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tran J. H., Jacoby G. A., Hooper D. C. (2005) Antimicrob. Agents Chemother. 49, 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tran J. H., Jacoby G. A., Hooper D. C. (2005) Antimicrob. Agents Chemother. 49, 3050–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. (2009) Clin. Microbiol. Rev. 22, 664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martínez-Martínez L., Pascual A., Jacoby G. A. (1998) Lancet 351, 797–799 [DOI] [PubMed] [Google Scholar]
- 6. Rodríguez-Martínez J. M., Velasco C., García I., Cano M. E., Martínez-Martínez L., Pascual A. (2007) Antimicrob. Agents Chemother. 51, 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robicsek A., Sahm D. F., Strahilevitz J., Jacoby G. A., Hooper D. C. (2005) Antimicrob. Agents Chemother. 49, 3001–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H. B., Park C. H., Kim C. J., Kim E. C., Jacoby G. A., Hooper D. C. (2009) Antimicrob. Agents Chemother. 53, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hata M., Suzuki M., Matsumoto M., Takahashi M., Sato K., Ibe S., Sakae K. (2005) Antimicrob. Agents Chemother. 49, 801–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacoby G. A., Walsh K. E., Mills D. M., Walker V. J., Oh H., Robicsek A., Hooper D. C. (2006) Antimicrob. Agents Chemother. 50, 1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavaco L. M., Hasman H., Xia S., Aarestrup F. M. (2009) Antimicrob. Agents Chemother. 53, 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang M., Jacoby G. A., Mills D. M., Hooper D. C. (2009) Antimicrob. Agents Chemother. 53, 821–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jacoby G., Cattoir V., Hooper D., Martínez-Martínez L., Nordmann P., Pascual A., Poirel L., Wang M. (2008) Antimicrob. Agents Chemother. 52, 2297–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jacoby G. A., Gacharna N., Black T. A., Miller G. H., Hooper D. C. (2009) Antimicrob. Agents Chemother. 53, 1665–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Da Re S., Garnier F., Guérin E., Campoy S., Denis F., Ploy M. C. (2009) EMBO Rep. 10, 929–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hashimi S. M., Wall M. K., Smith A. B., Maxwell A., Birch R. G. (2007) Antimicrob. Agents Chemother. 51, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garrido M. C., Herrero M., Kolter R., Moreno F. (1988) EMBO J. 7, 1853–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heddle J. G., Blance S. J., Zamble D. B., Hollfelder F., Miller D. A., Wentzell L. M., Walsh C. T., Maxwell A. (2001) J. Mol. Biol. 307, 1223–1234 [DOI] [PubMed] [Google Scholar]
- 19. Montero C., Mateu G., Rodriguez R., Takiff H. (2001) Antimicrob. Agents Chemother. 45, 3387–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hegde S. S., Vetting M. W., Roderick S. L., Mitchenall L. A., Maxwell A., Takiff H. E., Blanchard J. S. (2005) Science 308, 1480–1483 [DOI] [PubMed] [Google Scholar]
- 21. Mérens A., Matrat S., Aubry A., Lascols C., Jarlier V., Soussy C. J., Cavallo J. D., Cambau E. (2009) J. Bacteriol. 191, 1587–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Studier F. W. (2005) Protein Expression Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 23. Fox B. G., Blommel P. G. (2009) Curr. Protoc. Protein Sci., Chapter 5, Unit 5.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project, no. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25. Evans P. (2006) Acta Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) J. Synchrotron Rad. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 27. Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. Sect. D 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. Sect. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 30. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
- 31. Derewenda Z. S. (2004) Structure 12, 529–535 [DOI] [PubMed] [Google Scholar]
- 32. Derewenda Z. S. (2004) Methods 34, 354–363 [DOI] [PubMed] [Google Scholar]
- 33. Longenecker K. L., Garrard S. M., Sheffield P. J., Derewenda Z. S. (2001) Acta Crystallogr. Sect. D 57, 679–688 [DOI] [PubMed] [Google Scholar]
- 34. Vetting M. W., Hegde S. S., Fajardo J. E., Fiser A., Roderick S. L., Takiff H. E., Blanchard J. S. (2006) Biochemistry 45, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trevino S. R., Scholtz J. M., Pace C. N. (2007) J. Mol. Biol. 366, 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vetting M. W., Hegde S. S., Zhang Y., Blanchard J. S. (2011) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buchko G. W., Ni S., Robinson H., Welsh E. A., Pakrasi H. B., Kennedy M. A. (2006) Protein Sci. 15, 2579–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchko G. W., Robinson H., Pakrasi H. B., Kennedy M. A. (2008) J. Struct. Biol. 162, 184–192 [DOI] [PubMed] [Google Scholar]
- 39. Ni S., Sheldrick G. M., Benning M. M., Kennedy M. A. (2009) J. Struct. Biol. 165, 47–52 [DOI] [PubMed] [Google Scholar]
- 40. Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 41. Vetting M. W., Hegde S. S., Hazleton K. Z., Blanchard J. S. (2007) Protein Sci. 16, 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chandler L. E., Bartsevich V. V., Pakrasi H. B. (2003) Biochemistry 42, 5508–5514 [DOI] [PubMed] [Google Scholar]
- 43. Black K., Buikema W. J., Haselkorn R. (1995) J. Bacteriol. 177, 6440–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arsène S., Leclercq R. (2007) Antimicrob. Agents Chemother. 51, 3254–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hegde S. S., Vetting M. W., Mitchenall L. A., Maxwell A., Blanchard J. S. (2011) Antimicrob. Agents Chemother. 55, 110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Poirel L., Liard A., Rodriguez-Martinez J. M., Nordmann P. (2005) J. Antimicrob. Chemother. 56, 1118–1121 [DOI] [PubMed] [Google Scholar]
- 47. Poirel L., Rodriguez-Martinez J. M., Mammeri H., Liard A., Nordmann P. (2005) Antimicrob. Agents Chemother. 49, 3523–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laponogov I., Pan X. S., Veselkov D. A., McAuley K. E., Fisher L. M., Sanderson M. R. (2010) PLoS One 5, e11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laponogov I., Sohi M. K., Veselkov D. A., Pan X. S., Sawhney R., Thompson A. W., McAuley K. E., Fisher L. M., Sanderson M. R. (2009) Nat. Struct. Mol. Biol. 16, 667–669 [DOI] [PubMed] [Google Scholar]
- 50. Schmidt B. H., Burgin A. B., Deweese J. E., Osheroff N., Berger J. M. (2010) Nature 465, 641–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dong K. C., Berger J. M. (2007) Nature 450, 1201–1205 [DOI] [PubMed] [Google Scholar]
- 52. Wohlkonig A., Chan P. F., Fosberry A. P., Homes P., Huang J., Kranz M., Leydon V. R., Miles T. J., Pearson N. D., Perera R. L., Shillings A. J., Gwynn M. N., Bax B. D. (2010) Nat. Struct. Mol. Biol. 17, 1152–1153 [DOI] [PubMed] [Google Scholar]
- 53. Gubaev A., Hilbert M., Klostermeier D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiong X., Bromley E. H., Oelschlaeger P., Woolfson D. N., Spencer J. (2011) Nucleic Acids Res. 39, 3917–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]