Abstract
Proteins interacting with membranes via a single hydrophobic segment can be classified as either monotopic or bitopic. Here, we probe the topology of a membrane-attached enzyme, the ϵ isoform of human diacylglycerol kinase (DGKϵ), when inserted into rough microsomes and compare it with the monotopic membrane protein mouse caveolin-1. In contrast to previous findings, the N-terminal hydrophobic stretch in DGKϵ attains a bitopic rather than a monotopic topology in our experimental system. In addition, we find that charged flanking residues as well as proline residues embedded in the hydrophobic segment are important determinants of monotopic versus bitopic topology.
Keywords: Caveolae, Membrane Bilayer, Membrane Enzymes, Membrane Proteins, Protein Structure
Introduction
A fundamental characteristic of all membrane proteins is their topology, i.e. the number of times they span the membrane and their overall orientation relative to the membrane. According to the terminology introduced by Blobel in 1980, proteins that integrate into membranes with a single “re-entrant” hydrophobic segment, entering and exiting on the same side of the membrane, are called monotopic; those that span the membrane once are termed bitopic; and polytopic proteins span the membrane multiple times (1). Unfortunately, current bioinformatics tools distinguish poorly between monotopic and bitopic proteins (2), and not many experimental studies address the sequence determinants for the two kinds of topologies.
One well studied example of a protein with a single hydrophobic segment is the ϵ isoform of diacylglycerol kinase (DGKϵ),3 an enzyme that catalyzes the phosphorylation of diacylglycerol to phosphatidic acid (3) and plays a role in modulating neuronal signaling linked to synaptic activity in mammals (4). The enzyme has one predicted N-terminal transmembrane (TM) segment and should therefore be bitopic, but experimental data suggest a monotopic topology: a peptide mimicking the hydrophobic segment of DGKϵ shows only a moderate degree of helicity when embedded in a phospholipid bilayer (5), and the intact enzyme seems only loosely attached to membranes because it can be extracted by a mild treatment with KCl (6, 7). Furthermore, a FLAG epitope fused to the N terminus of DGKϵ expressed in NIH-3T3 cells is only accessible from the inside of the cells upon permeabilization with detergent. Modeling of the conformational preference of the hydrophobic segment in a membrane-like environment suggests that the peptide can take up two different stable conformations, both a straight (corresponding to a TM) helix and a bent, monotopic form (7).
A well studied example of a monotopic protein is caveolin, known for its role in forming caveolae, invaginations in the plasma membrane involved in important cellular processes such as vesicular trafficking, cholesterol homeostasis, signal transduction, tumor suppression, and as buffers against acute mechanical stress (8, 9). Due to the prominent role of caveolin in forming caveolae, the nature of the membrane-integrated state is likely of great importance, and this is supported by the high degree of conservation in the primary sequence of hydrophobic segments from different caveolins (10). Several studies establish a monotopic topology for caveolins (11–14), in agreement with in silico analysis that suggests a single stable, U-shaped, conformation of the hydrophobic segment, entering and exiting on the same side of the membrane (14).
Although the available evidence points to a similar membrane-embedded structure for both the DGKϵ and caveolin hydrophobic segments, there is no obvious sequence similarity between the two (7), except that both sequences feature a central proline residue that is conserved within the two different protein families. Proline is well known for its helix-breaking propensity in soluble proteins, and although this has been shown to be less prominent in a membrane environment (15), two recent studies suggest a crucial role for prolines in defining the monotopic topology of DGKϵ and caveolin-1 (7, 14). In these studies, single mutations turning the membrane-embedded prolines into alanines converted both DGKϵ and caveolin-1 into bitopic proteins.
Here, using in vitro translation in the presence of dog pancreas rough microsomes (RMs), we explore the conditions that contribute to the stabilization of a monotopic membrane protein topology and address the relative impact on topology of prolines versus the residues that flank the hydrophobic segments in DGKϵ and caveolin. In contrast to previous results, our data strongly support a bitopic topology for wild-type DGKϵ, and we additionally show that flanking residues can have a large impact on whether the protein adopts a mono- or bitopic topology, as can the proline residue located near the middle of the hydrophobic segment.
EXPERIMENTAL PROCEDURES
DNA Constructs
pGEMu, pGEMuLep-H3, pGEMuLep-H2, and pGEMuLep-H1 plasmids were prepared as previously described (16). Human DGKϵ, DGKϵ (P33A), mouse caveolin-1, and P110A caveolin-1 DNA were amplified from the corresponding p3XFLAG constructs (7) with PfuX7 DNA polymerase as described previously (17). DNA encoding the DGKϵ wild-type and P33A hydrophobic segment was amplified with the oligonucleotides DGKϵLEPins5′ (5′-ACCAGGdUGGCCTGTTTGCGGAC-3′) and DGKϵLEPins3′ (ACCCGGdUCCCCGGCGCGACC) and cloned into pGEMuLep-H2 and pGEMuLep-H3 by uracil-excision (17, 18).
DGKϵ DNA, including or excluding the 5′- and 3′-flanking residues, was amplified with the oligonucleotides DGKϵE2F (ACCAGGdUGAAGCGGAGAGGCGGC) and DGKϵD66R (ACCCGGdUCCGTCCGTGTCGCGCCACC); DGKϵG20F (ACCAGGdUGGGCACCTGATCTTGTGGAC) and DGKϵL42R (ACCCGGdUCCGAGGCTACACCAGAAGGTGATGAAC); DGKϵE2F and DGKϵL42R; or DGKϵG20F and DGKϵD66R. Similar, differently flanked versions of mouse caveolin-1 were made by combining the oligonucleotides mCavK5F (ACCAGGdUAAATACGTAGACTCCGAGGGACATC) and mCavE177R (ACCCGGdUCCCTCTTTCTGCGTGCTGATGCGGATG); mCavK96F (ACCAGGdUAAATATTGGTTTTACCGCTTGTTGTC) and mCavI139R (ACCCGGdUCCAATCAGGAAGCTCTTGATGCACG); mCavK5F and mCavI139R; or mCavK96F and mCavE177R. All of these PCR products were then cloned into pGEMuLep-H1 by uracil excision.
DNA of the different N-terminal versions of DGKϵ was constructed by PCR with the three oligonucleotides pGEMFLAGF (AGCCACCAdUGGACTACAAAGACCATGACGGTG), pGEMdgkF (AGCCACCAdUGGAAGCGGAGAGGCG), and pGEMglydgkF (AGCCACCAdUGGAAGCGAACTCCACAGAGAGGCGGCCG), in all combinations with the DGKϵD66R or DGKϵL42R oligonucleotides and cloned into pGemuLep-H1, or in combination with pGEMdgkR (AAGATGGCdUATTCAGTCGCCTTTATATCTTCTTGATC) and cloned into pGEMu. Full-length mouse caveolin-1 constructs were amplified with the oligonucleotides pGEMcavF (AGCCACCAdUGTCTGGGGGCAAATACGTAGAC) and pGEMglycavF (AGCCACCAXGTCTGGGAACTCCACAGGCAAATACGTAGACTCCGAG) in combination with pGEMcavR (AAGATGGCdUATATCTCTTTCTGCGTGCTGATG) and cloned into pGEMu.
Membrane Topology Assays
In vitro protein expression was performed using the TnT SP6 Quick Coupled SP6 Transcription/Translation System (Promega) in the presence or absence of RMs as described previously (19). Briefly, 9 μl of reticulocyte lysate was mixed with ∼200 ng of pGEM plasmid DNA, [35S]methionine, and 0.5 μl of RMs. The reaction was incubated for 30 min at 30 °C and then mixed with NuPage loading buffer (Invitrogen) and subjected to SDS-PAGE using NuPage 12% bis-tris gels (Invitrogen). Gels were dried and analyzed by exposure on PhosphorImager screens using standard techniques. All samples were run in triplicate, and relative amounts of singly and doubly glycosylated species were quantified using the ImageGauge Version 3.45 and the Qtiplot 0.9.3-rc2 softwares (20).
RESULTS
Insertion of the DGKϵ N-terminal Hydrophobic Segment into RMs
The membrane topology of human DGKϵ was previously assayed in cultured cells by determining the localization of a FLAG tag attached to the N terminus. This approach suggested a monotopic topology of the wild-type enzyme with both the N and C terminus in the cytosol (Ncyt-Ccyt), whereas a single proline-to-alanine (P33A) mutation in the predicted membrane-embedded segment converted the topology to bitopic with the N-terminal FLAG tag facing the outside of the cell (corresponding to the lumen in the endoplasmic reticulum; Nlum-Ccyt) (7).
Given the predicted bitopic topology of wild-type DGKϵ, we decided to study its membrane-insertion properties in more detail using in vitro translation in the presence of RMs (19). In a first set of experiments, the wild-type and P33A mutant N-terminal DGKϵ hydrophobic segments (including 5–7 hydrophilic flanking residues, Fig. 1A) were inserted into two different Escherichia coli leader peptidase (Lep) model membrane proteins (Fig. 1B). In these constructs, the DGKϵ hydrophobic segments (H-segments) are flanked by glycosylation sites that can be used as topological markers, because they can only be glycosylated by the oligosaccharyl transferase enzyme if translocated into the lumen of the RMs. The degree of membrane insertion of H-segments can therefore be quantified by comparing the ratios of singly and doubly glycosylated species after resolution of in vitro translated, 35S-labeled protein on SDS-polyacrylamide gels. The amount of unglycosylated species cannot be used as a quantitative measure of membrane integration properties because the efficiency of membrane targeting (i.e. the amount of unglycosylated versus glycosylated species) is different for different constructs and is somewhat sensitive to variations in the experimental conditions.
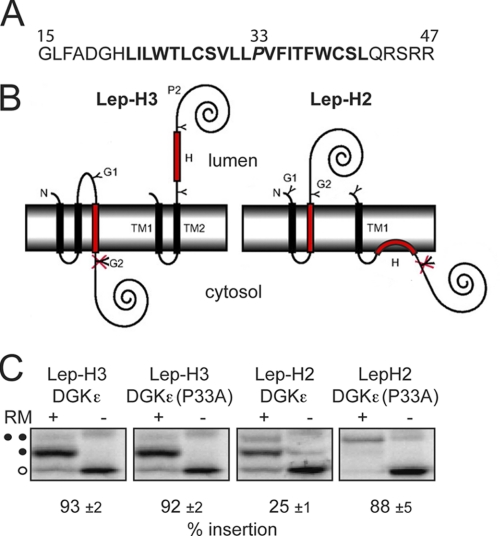
FIGURE 1.
Evaluation of the membrane integration properties of the hydrophobic segment of DGKϵ in the Lep-H3 and Lep-H2 constructs. A, residues 15–47 of the DGKϵ primary sequence and the P33A (residue italicized) DGKϵ mutant, termed the H-segment, were selected for analysis based on results from the ΔG predictor of transmembrane segments (27) (prediction highlighted in bold). B, H-segments were introduced into the Lep-H3 and Lep-H2 model proteins. In Lep-H3, two N-terminal TM regions (TM1, TM2) and a C-terminal soluble domain (P2) originate from E. coli leader peptidase (Lep). Membrane integration of the H-segment leads to a singly glycosylated species (a glycosylated site, G1 or G2, is indicated with a Y, an unglycosylated site is indicated with a crossed-over Y). Lep-H2 has only one Lep-derived transmembrane segment (TM1), and membrane-integrated H-segments lead to doubly glycosylated species. C, DNA constructs were transcribed in vitro and translated in the presence of RMs. Control reactions were performed in the absence of RMs. Unglycosylated proteins are indicated with an open circle. In Lep-H3, membrane insertion efficiency was quantified as the ratio gs/(gs+gd) where gs is the amount of singly glyvcosylated (one closed circle) and gd the amount of doubly glycosylated (two closed circles), [35S]Met-labeled proteins on SDS-polyacrylamide gels. In Lep-H2, insertion was quantified as the ratio gd/(gs+gd). Representative gels are shown. The percent insertion values are averages of three independent experiments with S.D.
In the Lep-H3 constructs (Fig. 1B, left), singly glycosylated proteins have their H-segment inserted in the membrane in an Nlum-Ccyt orientation, whereas doubly glycosylated proteins have a noninserted H-segment. Both the wild-type and P33A DGKϵ segments were >90% singly glycosylated (Fig. 1C), demonstrating that DGKϵ inserts as a TM helix in the Lep-H3 model system. In the Lep-H2 system (21) (Fig. 1B, right), membrane insertion of the H-segment leads to translocation of the C terminus and double glycosylation. In this orientation, the P33A mutation was necessary for efficient insertion as it increased the relative amount of doubly glycosylated protein from 25% to 88% (Fig. 1C). Possibly, the DGKϵ wild-type segment inserts as a re-entrant-loop structure (Ncyt-Ccyt) in the Lep-H2 context (Fig. 1B, right).
Flanking Residues Can Influence the Topology of the DGKϵ and Caveolin Membrane-interacting Segments
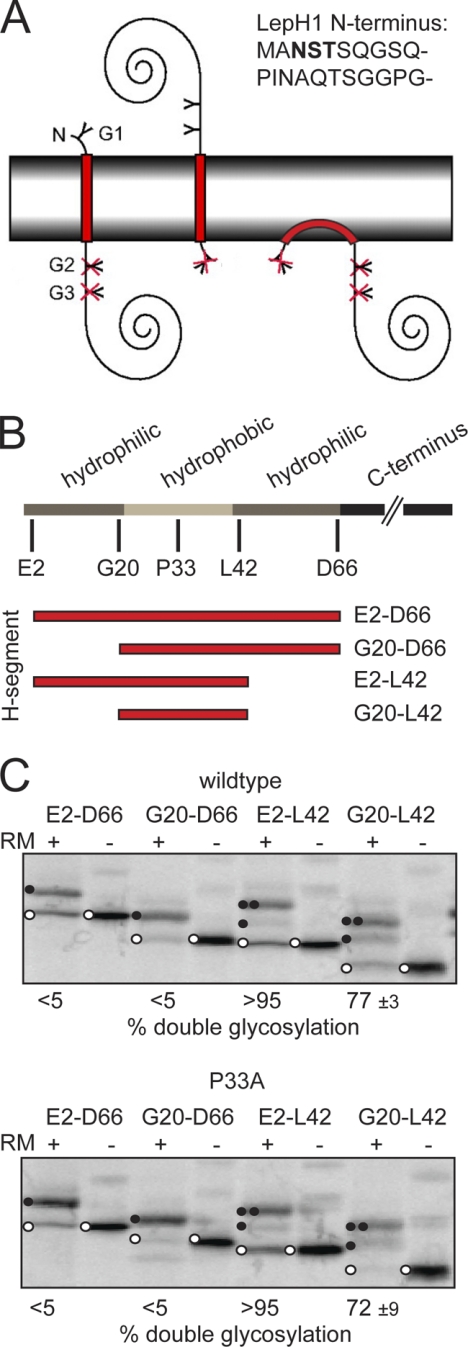
The efficient transmembrane insertion of the DGKϵ H-segment in the Nlum-Ccyt orientation encouraged us to assay its topology in the absence of Lep TM segments (Lep-H1) (21) (Fig. 2A), i.e. in a context similar to the DGKϵ wild-type protein. In Lep-H1, most of the Lep-derived N-terminal domain (including both transmembrane segments) has been omitted, and glycosylation sites have been engineered so that single-spanning proteins with an Nlum-Ccyt topology will be singly glycosylated whereas the opposite topology (Ncyt-Clum) will give a doubly glycosylated protein (Fig. 2A). Proteins inserting with an Ncyt-Ccyt re-entrant loop structure will not be glycosylated (Fig. 2A, right).
FIGURE 2.
Membrane integration properties of Lep-H1-DGKϵ constructs with different flanking residues. A, schematic outline of the Lep-H1-based membrane insertion assay. Lep-H1 constructs with H-segments inserting with the N terminus in the lumen become singly glycosylated, due to the NST site residing in the Lep-H1 N terminus (sequence shown in upper right corner), whereas the opposite topology leads to double glycosylation of two NST sites residing in the Lep P2 soluble domain (a glycosylation site, G1, G2, or G3, is illustrated with a Y in the panel). Monotopically inserted H-segments will not be glycosylated (crossed-over Y). B, schematic representation of the DGKϵ primary sequence highlights the hydrophobic segment and flanking sequences and the different positions fused to Lep-H1. The predicted hydrophobic segment is in light gray, hydrophilic flanking regions in dark gray, and the different H-segments in red. C, glycosylation assay of DGKϵ (upper panel) and the P33A mutant (lower panel) with different flanking residues fused to the Lep-H1 model protein is shown. Controls were performed in the absence of RMs. Unglycosylated proteins are marked with an open circle, singly glycosylated proteins with one closed circle, and doubly glycosylated proteins with two closed circles. Representative gels are shown. The percent double glycosylation values are averages of three independent experiments with S.D.
We noted that the predicted N-terminal TM segment in DGKϵ is flanked by a number of charged residues and speculated that these could be important determinants of the membrane topology as previously shown for multispanning membrane proteins (22). Therefore, several versions of the DGKϵ N-terminal segment, including or excluding flanking charged residues (Fig. 2B; cf. Fig. 6), were engineered into the Lep-H1 construct. As noted above for the Lep-H2 and Lep-H3 constructs, the amount of unglycosylated protein cannot be used as a quantitative measure of membrane integration, and the Lep-H1 derived constructs were mainly used for a qualitative assessment of topology.
FIGURE 6.
Primary amino acid sequences of DGKϵ and caveolin-1. Membrane-integrated hydrophobic regions, predicted with the ΔG predictor software (27), are highlighted in light gray. Positively and negatively charged residues within a distance of 50 residues flanking the hydrophobic segments are highlighted in light blue and red, respectively. The flanking regions included in the Lep-based model constructs are highlighted in dark gray. A cryptic glycosylation site in DGKϵ, predicted with the NetNGlyc software (24), is highlighted in blue.
When including the C-terminal hydrophilic residues (constructs E2-D66 and G20-D66, Fig. 2B), both the wild-type and the P33A mutant were nearly 100% singly glycosylated (versus doubly glycosylated) and therefore inserted almost exclusively with Nlum-Ccyt topologies (Fig. 2C). In contrast, including the N terminus, but excluding the flanking C-terminal residues (E2-L42) led mainly to double glycosylation and thus an Ncyt-Clum topology for both wild-type and P33A. Finally, the absence of both flanking sequences led to mixed topologies for both wild-type and P33A, though with a clear preference (77% for wild-type and 72% for P33A) for the Ncyt-Clum orientation. These results show that the intact DGKϵ N-terminal segment E2-D66 is inserted with an Nlum-Ccyt topology. In addition, flanking residues on both sides of the hydrophobic segment are essential topology determinants: when the C-terminal flanking residues 43–66 are removed, the protein inverts to the Ncyt-Clum topology, and this is also the case when both flanking regions (residues 2–19 and 43–66) are removed. The Nlum-Ccyt topology is preserved upon removal of residues 2–19 alone. Finally, the results obtained with the P33A mutant versions are indistinguishable from the wild-type constructs in all cases.
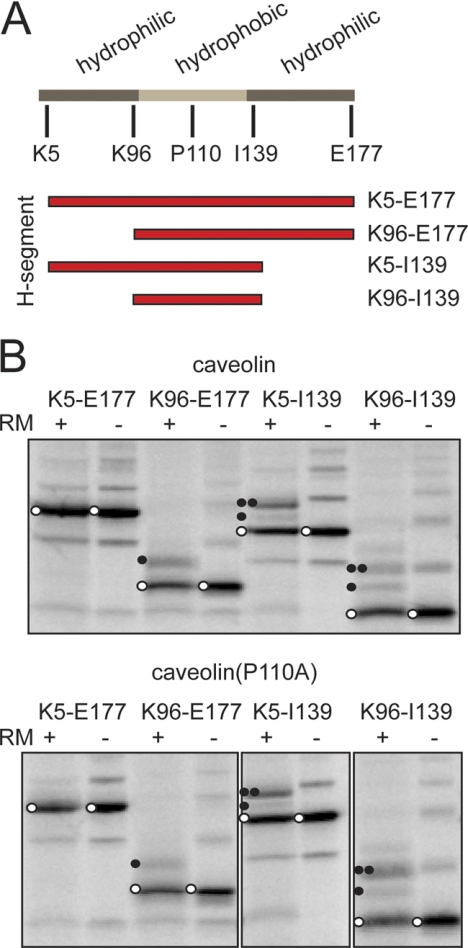
For comparison, similar experiments were performed with the monotopic membrane protein caveolin. The membrane-interacting segment of caveolin-1 is located near the middle of the protein. Similar to DGKϵ, it is flanked by hydrophilic regions (Fig. 3A; cf. Fig. 6). We tested differently flanked caveolin segments in Lep-H1 essentially as outlined for DGKϵ. In contrast to DGKϵ, all caveolin-derived constructs were poorly glycosylated (compare glycosylated with unglycosylated proteins in Fig. 2C versus Fig. 3B), possibly due to a predominant monotopic Ncyt-Ccyt topology in all cases, although inefficient membrane targeting cannot be ruled out. Caveolin is known to form multimers (11), and this, combined with the hydrophobic and hairpin-like nature of the protein, may cause the ladder-like migration behavior of the in vitro synthesized products observed in Fig. 3B, as previously described for similar protein segments (23). Due to the low degree of glycosylation and the complex patterned gels, the assay can in this case be used only as a qualitative indicator of topology. However, in clear contrast to DGKϵ, glycosylation was completely absent when expressing the near-full-length wild-type or P110A caveolin-1 constructs (K5-E177) that include the flanking sequences on both sides of the hydrophobic segment (Fig. 3B). Upon removal of N-terminal hydrophilic residues (construct K96-E177), a part of the synthesized protein became singly glycosylated and therefore has an Nlum-Ccyt topology, whereas removal of the C-terminal hydrophilic residues (construct K5-I139) led primarily to a doubly glycosylated protein with an Ncyt-Clum topology. When only the hydrophobic segment was included (construct K96-I139), both singly and doubly glycosylated proteins were produced, indicative of a mixed topology. As observed with DGKϵ, there were no significant differences between the wild-type and the P110A mutant constructs. These experiments show that when residues that flank a monotopic hydrophobic segment are removed, the segment can be converted into a bitopic TM segment.
FIGURE 3.
Membrane integration properties of Lep-H1-caveolin-1 constructs with different flanking residues. A, schematic representation of the DGKϵ primary sequence, highlighting the hydrophobic segment and flanking sequences and the different positions fused to Lep-H1. The predicted hydrophobic segment is in light gray and the hydrophilic flanking regions in dark gray. B, glycosylation assay of caveolin (upper panel) and the P110A mutant (lower panel) with different flanking residues fused to the Lep-H1 model protein. Controls were performed in the absence of RMs. Unglycosylated proteins are marked with open circles, singly glycosylated with one closed circle, and doubly glycosylated with two closed circles. All experiments were done in triplicate, and representative gels are shown.
A Peptide Tag Can Change the Topology of a Membrane Protein
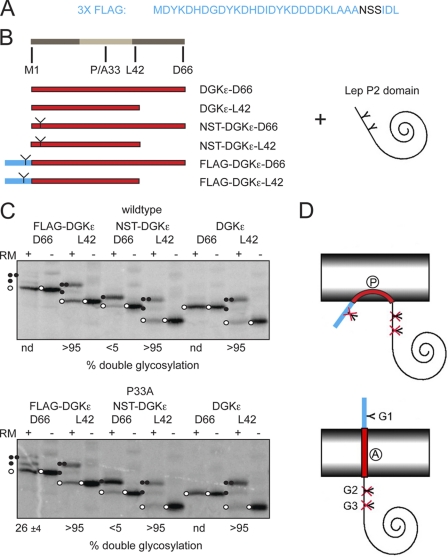
The results obtained with the Lep-H1 system on the topology of the N-terminal DGKϵ membrane-interacting segment are in contrast to the previously reported monotopic membrane topology (7). We noted that in the previous study a highly charged FLAG tag was fused to the N terminus of DGKϵ (Fig. 4A) to assess the membrane topology, and we speculated that this might have caused the conflicting results. We therefore interchanged the Lep-H1-derived N terminus of the constructs described above with either the native DGKϵ N terminus, a native DGKϵ N terminus with an engineered NST consensus N-glycosylation site, or with the FLAG tag used previously (7) (Fig. 4B). Importantly, the FLAG tag contains a consensus glycosylation site (NSS) (Fig. 4A). These constructs again allowed us to assess the membrane topology by comparing singly and doubly glycosylated species. The FLAG-tagged, wild-type DGKϵ Lep-H1 construct with C-terminal flanking residues (FLAG-DGKϵ-D66) was only very inefficiently glycosylated, indicating either absent membrane insertion or a monotopic topology (Fig. 4C, upper panel). In contrast, when introducing the single P33A mutation, both the singly and doubly glycosylated species were produced (Fig. 4C, lower panel), although the latter to a lesser degree (26%), indicating a preference for the Nlum-Ccyt orientation. Again, deleting the C-terminal flanking residues (all L42 constructs) imparted an Ncyt-Clum orientation, both for the wild-type and P33A mutants and independent of the nature of the N terminus, as indicated by the dominating doubly glycosylated species. The two constructs (with and without the P33A mutation) with a wild-type-like N terminus harboring an engineered glycosylation site and, including the C-terminal flanking residues (NST-DGKϵ-D66), showed a clear preference for the singly glycosylated Nlum-Ccyt orientation, which was confirmed by the lack of glycosylation of the corresponding controls with no N-terminal glycosylation site (DGKϵ-D66).
FIGURE 4.
Effect of adding a FLAG tag on the topology of DGKϵ-Lep-P2 constructs. A, sequence of the 3XFLAG tag. The NSS glycan acceptor site is highlighted. B, FLAG tag shown in A (highlighted in blue) fused to the DGKϵ N-terminal segment, with and without C-terminal flanking residues (highlighted in dark gray at the right side of the light gray hydrophobic segment) and to the Lep P2 soluble domain. The FLAG-tagged constructs were compared with constructs with a native-like DGKϵ N terminus, with and without an engineered NST glycosylation site in position 3 (illustrated with a Y). C, glycosylation assay of DGKϵ and the P33A mutant with the different N termini and different C-terminal flanking residues fused to the C-terminal P2 part of the Lep-H1 model protein. In the assay, noninserted and monotopic proteins are unglycosylated, Nlum-Ccyt orientated proteins are singly glycosylated (except for the native controls without an N-terminal glycosylation site), and Ncyt-Clum orientated proteins are doubly glycosylated. Controls were performed in the absence of RMs. Unglycosylated proteins are marked with an open circle, singly glycosylated proteins with one closed circle, and doubly glycosylated proteins with two closed circles. Representative gels are shown. The percent double glycosylation values are averages of three independent experiments with S.D. D, schematic interpretation of the effect of the FLAG tag (highlighted in blue) and the P33A mutation. Upper, FLAG tag preventing translocation of the N terminus leading to an unglycosylated protein. Lower, P33A mutation allowing the FLAG tag to be translocated. The protein becomes singly glycosylated.
In summary, and in agreement with the previous cell culture experiments (7), the results indicate that the FLAG-tagged, DGKϵ N-terminal segment adopts a monotopic topology but that a fraction of the molecules span the membrane with an Nlum-Ccyt orientation in the P33A mutant version (Fig. 4D). In contrast, DGKϵ with a native-like N terminus appears to attain a bitopic Nlum-Ccyt topology. The experiment also demonstrates, in agreement with previous observations (7, 14, 19), that proline residues buried in hydrophobic segments can significantly affect membrane topology.
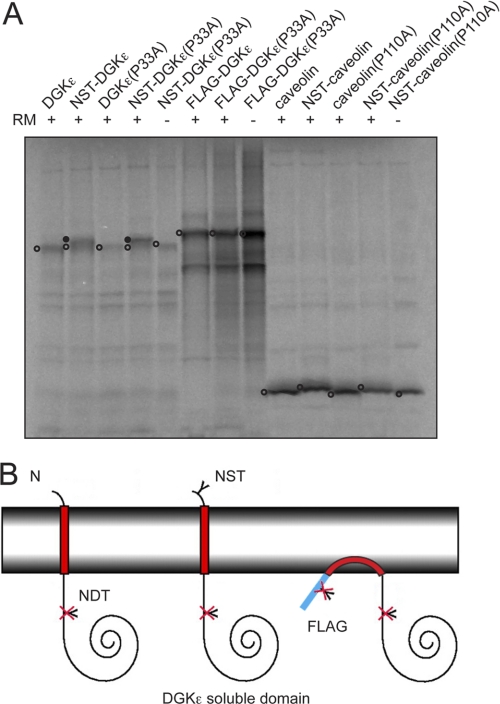
Probing the Topology of Full-length DGKϵ and Caveolin
Finally, we studied the native, full-length DGKϵ sequence with the three different N-terminal modifications described above. Expressing both the wild-type and the P33A mutant version with an N-terminal, engineered glycosylation site resulted primarily in a species shifted in mobility on the gel, corresponding to a glycosylated protein, although an unglycosylated species was also visible (Fig. 5A, constructs NST-DGKϵ and NST-DGKϵ(P33A)). Translation in the absence of RMs and of the glycosylation-site-less constructs served as controls and showed no mobility shift. If DGKϵ were to adopt an Ncyt-Clum topology, the potential NDT glycosylation site downstream of the hydrophobic segment (Fig. 5A; cf. Fig. 6) is predicted to be glycosylated (24), but this was not observed (Fig. 5A constructs DGKϵ and DGKϵ(P33A)). In contrast to the native-like versions of DGKϵ, the corresponding FLAG-tagged constructs showed no differences in mobility on the gel. These results again suggest that wild-type DGKϵ attains a bitopic Nlum-Ccyt topology but that translocation of the N terminus of the hydrophobic segment is inhibited by the addition of the highly charged FLAG tag (Fig. 5B). We also noted that the native-like DGKϵ constructs expressed poorly in our in vitro system compared with, e.g. the FLAG-tagged proteins and caveolin (Fig. 5A), which we have also observed in mammalian cell culture before. We speculate that this phenomenon may be caused by the very high GC content in the 5′-region of the DGKϵ nucleotide sequence (data not shown).
FIGURE 5.
Probing the topology of full-length DGKϵ and caveolin-1 with engineered glycosylation sites. A, glycosylation assay of DGKϵ and caveolin. Native and proline-to-alanine mutants of DGKϵ and caveolin were tested with and without an added NST glycosylation site in position 3, and DGKϵ was tested with and without an N-terminal 3XFLAG tag. In the assay, noninserted and monotopic proteins are unglycosylated, and Nlum-Ccyt orientated proteins are singly glycosylated. Controls were performed in the absence of RMs. Unglycosylated proteins are marked with an open circle and singly glycosylated proteins with a closed circle. The gel is representative of three independent experiments. B, graphic representation of DGKϵ full-length sequences with different N termini illustrating the topologies observed in A. The native-like N terminus with an engineered NST glycosylation site becomes glycosylated, suggesting that the wild-type protein attains an Nlum-Ccyt TM topology. In contrast, in the native protein context, the FLAG tag prevents membrane translocation even in the presence of the P33A mutation.
Similar to DGKϵ, the caveolin-1 membrane topology was recently probed with a FLAG-tag accessibility assay (14), showing that the hairpin loop structure could be turned into a TM segment by mutation of a single proline residue in the hydrophobic segment. We decided to test whether we obtained similar results in our experimental system. Thus, essentially as described above for DGKϵ, native and P110A caveolin were expressed in the presence or absence of engineered N-terminal glycosylation sites (Fig. 5A). In contrast to results obtained with DGKϵ, none of the different caveolin constructs showed any sign of glycosylation, and thus, in our experimental system, the P110A mutation does not result in a detectable level of protein with a bitopic topology.
DISCUSSION
In some cases, the topology of a membrane protein can depend on a delicate balance between multiple topological determinants in the sequence and can be significantly affected by a single mutation, even for multispanning membrane proteins like the small multidrug transport protein EmrE (25). Thus, it is not surprising that a highly charged epitope like the FLAG tag can have a strong impact on the topology of a protein, as demonstrated here.
Our results show that the N-terminal hydrophobic segment in wild-type DGKϵ inserts with an Nlum-Ccyt bitopic topology in RMs and that the P33A mutation in the hydrophobic segment only marginally affects the insertion and topology in the context of the native protein. This is in contrast to previous results obtained with N-terminally FLAG-tagged DGKϵ constructs expressed in mammalian cell lines (7). However, when probing the topology of a similar FLAG-tagged construct in our in vitro system we see a monotopic topology for FLAG-DGKϵ. Therefore, even though we cannot completely rule out that wild-type DGKϵ attains different topologies when expressed in vitro and in vivo, DGKϵ is most likely a bitopic protein. A monotopic topology was also suggested in a study of a DGKϵ-derived hydrophobic peptide flanked by several lysine residues (5), but it is likely that the added charged residues affected the observed topology in a similar way as the FLAG tag.
The contrasting results between the topology of the caveolin P110A mutant expressed in cell culture and in vitro is more puzzling. In cell culture the mutant version is bitopic, whereas a monotopic topology is seen in our in vitro system. One possible explanation is that the topology of P110A caveolin is indeed different in different expression systems. Another possible explanation is more technical: in the glycosylation assay, the unglycosylated, singly and doubly glycosylated products serve as internal controls for each other, whereas the epitope-tagging strategy compares the signal from the antigenic tag before and after permeabilization with detergent, i.e. under different experimental conditions.
In the experiments described here, wild-type DGKϵ and caveolin have different topologies in the membrane. What could be the reasons for this? The most obvious differences between the two proteins are (i) that the length of the N-terminal tail upstream of the hydrophobic region is much shorter and contains fewer charged residues in DGKϵ, and (ii) that the hydrophobic region is longer in caveolin (Fig. 6). Presumably, the short N-terminal tail in DGKϵ translocates more easily across the membrane than the long, highly charged N- and C-terminal tails in caveolin. Moreover, the hydrophobic segment in caveolin is about 39 residues long compared with only about 21 residues in DGKϵ. The caveolin segment is sufficiently long to form a tight helical hairpin in the membrane (26), leading to a monotopic topology, whereas the DGKϵ segment is only long enough to span the membrane once, favoring a bitopic topology.
In summary, for the two proteins we have studied there appears to be a fine balance between the stability of the monotopic and bitopic topologies so that relatively small changes, such as addition/removal of charges in the flanking regions or replacement of Pro for Ala in FLAG-DGKϵ, can favor one topology over the other.
This work was supported by Human Frontiers Science Program Grant RGP0039/2007, European Research Council Grant ERC-2008-AdG 232648, grants from the Swedish Foundation for Strategic Research and the Swedish Research Council, the Swedish Cancer Foundation (to G. v. H.), and Natural Sciences and Engineering Research Council of Canada Grant 9848 (to R. M. E.).
- DGKϵ
- ϵ isoform of diacylglycerol kinase
- bis-tris
- bis(2-hydroxyethyl)aminotris(hydroxymethyl)methane
- cyt
- cytosolic
- Lep
- leader peptidase
- lum
- luminal
- RM
- dog pancreas rough microsome
- TM
- transmembrane.
REFERENCES
- 1. Blobel G. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernsel A., Von Heijne G. (2005) Protein Sci. 14, 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Topham M. K., Epand R. M. (2009) Biochim. Biophys. Acta 1790, 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodriguez de Turco E. B., Tang W., Topham M. K., Sakane F., Marcheselli V. L., Chen C., Taketomi A., Prescott S. M., Bazan N. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4740–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glukhov E., Shulga Y. V., Epand R. F., Dicu A. O., Topham M. K., Deber C. M., Epand R. M. (2007) Biochim. Biophys. Acta 1768, 2549–2558 [DOI] [PubMed] [Google Scholar]
- 6. Dicu A. O., Topham M. K., Ottaway L., Epand R. M. (2007) Biochemistry 46, 6109–6117 [DOI] [PubMed] [Google Scholar]
- 7. Decaffmeyer M., Shulga Y. V., Dicu A. O., Thomas A., Truant R., Topham M. K., Brasseur R., Epand R. M. (2008) J. Mol. Biol. 383, 797–809 [DOI] [PubMed] [Google Scholar]
- 8. Williams T. M., Lisanti M. P. (2004) Genome Biol. 5, 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinha B., Köster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R. V., Butler-Browne G., Vedie B., Johannes L., Morone N., Parton R. G., Raposo G., Sens P., Lamaze C., Nassoy P. (2011) Cell 144, 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Epand R. M., Sayer B. G., Epand R. F. (2005) J. Mol. Biol. 345, 339–350 [DOI] [PubMed] [Google Scholar]
- 11. Sargiacomo M., Scherer P. E., Tang Z., Kübler E., Song K. S., Sanders M. C., Lisanti M. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okamoto T., Schlegel A., Scherer P. E., Lisanti M. P. (1998) J. Biol. Chem. 273, 5419–5422 [DOI] [PubMed] [Google Scholar]
- 13. Schlegel A., Schwab R. B., Scherer P. E., Lisanti M. P. (1999) J. Biol. Chem. 274, 22660–22667 [DOI] [PubMed] [Google Scholar]
- 14. Aoki S., Thomas A., Decaffmeyer M., Brasseur R., Epand R. M. (2010) J. Biol. Chem. 285, 33371–33380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S. C., Goto N. K., Williams K. A., Deber C. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nørholm M. H., Cunningham F., Deber C. M., von Heijne G. (2011) J. Mol. Biol. 407, 171–179 [DOI] [PubMed] [Google Scholar]
- 17. Nørholm M. H. (2010) BMC Biotechnol. 10, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A. (2006) Nucleic Acids Res. 34, e122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. (2005) Nature 433, 377–381 [DOI] [PubMed] [Google Scholar]
- 20. Hedin L. E., Ojemalm K., Bernsel A., Hennerdal A., Illergård K., Enquist K., Kauko A., Cristobal S., von Heijne G., Lerch-Bader M., Nilsson I., Elofsson A. (2010) J. Mol. Biol. 396, 221–229 [DOI] [PubMed] [Google Scholar]
- 21. Lundin C., Kim H., Nilsson I., White S. H., von Heijne G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15702–15707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lerch-Bader M., Lundin C., Kim H., Nilsson I., von Heijne G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4127–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. (2004) Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 25. Seppälä S., Slusky J. S., Lloris-Garcerá P., Rapp M., von Heijne G. (2010) Science 328, 1698–1700 [DOI] [PubMed] [Google Scholar]
- 26. Monné M., Nilsson I., Elofsson A., von Heijne G. (1999) J. Mol. Biol. 293, 807–814 [DOI] [PubMed] [Google Scholar]
- 27. Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. (2007) Nature 450, 1026–1030 [DOI] [PubMed] [Google Scholar]