Abstract
The SPP1 siphophage uses its long non-contractile tail and tail tip to recognize and infect the Gram-positive bacterium Bacillus subtilis. The tail-end cap and its attached tip are the critical components for host recognition and opening of the tail tube for genome exit. In the present work, we determined the cryo-electron microscopic (cryo-EM) structure of a complex formed by the cap protein gp19.1 (Dit) and the N terminus of the downstream protein of gp19.1 in the SPP1 genome, gp211–552 (Tal). This complex assembles two back-to-back stacked gp19.1 ring hexamers, interacting loosely, and two gp211–552 trimers interacting with gp19.1 at both ends of the stack. Remarkably, one gp211–552 trimer displays a “closed” conformation, whereas the second is “open” delineating a central channel. The two conformational states dock nicely into the EM map of the SPP1 cap domain, respectively, before and after DNA release. Moreover, the open/closed conformations of gp19.1-gp211–552 are consistent with the structures of the corresponding proteins in the siphophage p2 baseplate, where the Tal protein (ORF16) attached to the ring of Dit (ORF15) was also found to adopt these two conformations. Therefore, the present contribution allowed us to revisit the SPP1 tail distal-end architectural organization. Considering the sequence conservation among Dit and the N-terminal region of Tal-like proteins in Gram-positive-infecting Siphoviridae, it also reveals the Tal opening mechanism as a hallmark of siphophages probably involved in the generation of the firing signal initiating the cascade of events that lead to phage DNA release in vivo.
Keywords: Bacteriophage, Electron Microscopy, Protein Structure, Viral Protein, Virus Structure, Image Processing, SPP1, Cryo-EM, Single Particle Reconstruction
Introduction
Bacteriophages represent the most abundant group of naturally occurring organisms in the biosphere (1). The vast majority of them, ∼96%, consists of an icosahedral capsid and a tail constituting the Caudovirales order (2). Their tail morphology defines three different families (3): Podoviridae, Myoviridae, and Siphoviridae. The tail is a multiprotein nanomachine responsible for specific host cell recognition and efficient phage DNA delivery. It is attached to the portal vertex of the phage icosahedral head, and its distal extremity carries the adsorption apparatus (4). Following host cell attachment, a signal is transmitted along the tail to trigger opening of the connector followed by phage DNA ejection from the capsid, through the tail tube, to reach the bacterial cytoplasm (5).
The combination of x-ray crystallography and EM has proven to be the method of choice to characterize locations and interactions of bacteriophage tail proteins, thus making it possible to determine the cascade of molecular events sustaining viral infection. In the case of Siphoviridae, structural insights into infection mechanism and virion assembly were reported for the coliphages T5 (6, 7) and lambda (8, 9), the Bacillus subtilis phage SPP1 (10), and the Lactococcus lactis phage p2 (11). Despite the diversity of infection mechanisms displayed by Siphoviridae, using surface proteins and/or peptidoglycan-associated sugars as receptors (12–14), the structure of their non-contractile tail is rather conserved. It is characterized by a long non-contractile tube, assembled by stacking several tens of homo-hexameric major tail protein (MTP)3 rings, and a central core formed by a few copies of the tape measure protein, which determines phage tail length. At the proximal tail end, the homo-hexameric terminator that stops tube elongation during assembly is found, whereas the distal tail end is characterized by the presence of the tail adsorption apparatus (10, 11, 15–17).
Apart from the recently described phage p2 and TP901-1 baseplate structures (11, 18) and the SPP1 tip (10), little structural information is available on the Gram-positive-infecting phage adsorption device. Although SPP1 and the lactococcal phages Tuc2009 and TP901-1 display different host-recognition devices with either an elongated tail tip or a bulky baseplate (10, 11, 17, 19, 20), they share sequence identity at the level of the Dit (distal tail) and the Tal (tail fiber) proteins. These two components, belonging to the adsorption apparatus, have been demonstrated to be essential both during tail morphogenesis and for the infection mechanism (17, 19, 21). The structures of Dit and Tal in phage p2 (ORF15 and ORF16, respectively) have been determined by x-ray crystallography (11). They form the inner part of the baseplate, and their structures reveal dramatic conformational changes probably involved in the infection mechanism (11).
Based on the EM three-dimensional reconstruction of the SPP1 tail, gp19.1 (Dit) and gp21 (Tal) have been attributed to the region of the tail tip proximal to the tail cap. The cap is localized at the tail tube end opposite to the viral capsid (10). However, the recently reported crystal structure of gp19.1, and its docking into the EM maps, led to the reassignment of gp19.1 to the cap structure (22). This finding highlighted the need for a precise localization and study of the interactions between the SPP1 tail proteins composing the adsorption apparatus. Enhancing our knowledge of the protein mapping along the tail is essential to get a comprehensive view of this model phage and to decipher the mechanisms underlying host infection and virion assembly. In the current work, we have investigated the complex formed by SPP1 gp19.1 and gp211–552 proteins using light scattering and mass spectrometry (MS) and determined its structure by cryo-EM. This study led to reassignment of the SPP1 tail cap architectural organization. It also suggests a closed to open conformational switch in Tal proteins acting as a trigger, common to Gram-positive-infecting siphophages, to initiate the domino-like cascade signaling pathway, thus culminating in viral DNA ejection (10).
EXPERIMENTAL PROCEDURES
Protein Production
Cloning, expression, and purification procedures applied to gp19.1 were described elsewhere (22). The sequence coding for the first 552 residues of gp21, referred as gp211–552, was cloned into the GatewayTM expression vector pETG-20A according to standard GatewayTM protocols. The expression and four-step purification protocols applied to gp19.1 and gp211–552 were followed as described (22), except that gp211–552 overexpression was performed at 17 °C. Following the TEV protease digestion and the last Ni2+-affinity chromatography step, gp19.1 and gp211–552 were mixed in a 1:1 ratio, and the complex was purified by size-exclusion chromatography (SEC) on a preparative Superdex 200 HR 26/60 column equilibrated in 10 mm Hepes, pH 7.5, 150 mm NaCl. The presence of both gp19.1 and gp211–552 in the eluted peak was confirmed by SDS-PAGE.
Molar Mass and Hydrodynamic Radius Determination by SEC/MALS/RI/UV/QELS
SEC was carried out on an Alliance 2695 HPLC system (Waters) using a KW804 column (Shodex) equilibrated in 10 mm Hepes, pH 7.5, 150 mm NaCl, 0.02% NaN3 at 0.5 ml/min. Multiangle static light scattering (MALS), UV spectrophotometry, quasielastic light scattering (QELS), and refractometry (RI) measurements were achieved with a MiniDawn Treos (Wyatt Technology), a Photo Diode Array 2996 (Waters), a DynaPro (Wyatt Technology), and an Optilab rEX (Wyatt Technology), respectively. Masses and radii calculations were done with ASTRA V (Wyatt Technology) using a dn/dc value of 0.185 ml/g for all proteins.
Non-covalent MS Analysis and Negative-staining EM
See the supplemental materials for non-covalent MS analysis and negative-staining EM.
Cryo-EM, Image Processing, and Three-dimensional Reconstructions
Three microliters of gp19.1-gp211–552 at a final concentration of 0.07 mg/ml in 10 mm HEPES (pH 7.5), 150 mm NaCl, was applied to glow-discharged Quantifoil R 2/2 grids (Quantifoil Micro Tools GmbH, Jena, Germany), blotted for 1 s, and then flash frozen in liquid ethane. Cryo-EM was carried out on a JEOL 2200FS operating at 200 kV under low dose conditions (total dose of 20 electrons/Å2) in the zero-energy-loss mode with a slit width of 20 eV. Images were taken at a nominal magnification of 50,000 × corresponding to a calibrated magnification of 45,591 × with defocus ranging from 1.4 to 2.5 μm. All micrographs were recorded on Kodak SO-163 films (Agar Scientific, Stansted, UK) and digitized on a Nikon 9000ED Cool Scan scanner at a sampling rate of 2.19 Å/pix at the specimen level. Particles, including side and top views, were semi-automatically extracted using Boxer (23). The defocus and astigmatism values of each micrograph were determined using CTFFIND3 (24). IMAGIC-5 (25) was used to flip phases and for the subsequent image-processing steps. Side and top views were processed independently. Particles were binned to 4.38 Å/pixel, band-pass filtered between 300 and 15 Å, normalized, and centered by iteratively alignments against their rotationally averaged sum. Particle orientations were refined in Multi-Reference Alignment cycles using two-dimensional projections of the initial model obtained by EM-negative staining (see supplemental materials). Side views were aligned using 420 projections with the Euler angle γ ranging from 0° to 15° (20 projections) for each β angle ranging from 90° to 30° (3° step), and top views were aligned using 330 projections with the Euler angle γ ranging from 0° to 30° (15 projections) for each β angle range 0°–10° and 170°–170°–180° (1° step). Rectangular or circular masks were applied onto the sums of aligned images, and both views were used to compute the three-dimensional reconstruction using C3 symmetry, and the Euler angles were assigned by projection matching. Fourier shell correlation (26) was used to estimate the resolution of the final structure at 26 Å with a 0.5 correlation cut-off without masking.
Detection of Tail Proteins in Infected Cells
B. subtilis YB888 was grown in LB medium at 37 °C to an optical density corresponding to 108 cfu/ml. The culture was supplemented with 10 mm CaCl2 and infected with SPP1 phages to an input multiplicity of five phages per bacterium. At 5- and 20-min post infection, a 2-ml culture aliquot was transferred to a tube and infected cells were collected by centrifugation (5 min, 4 °C, and 10,000 × g). The supernatant was carefully removed, the empty tube was re-centrifuged for 1 min, and the rest of the supernatant was discarded to remove input phages that would give background signal in Western blots of structural proteins. The infected cells pellet was stored at −20 °C (short term storage) or at −80 °C (long term storage). The pellet was resuspended in 1/20 of the initial culture volume in resuspension buffer (50 mm glucose, 1 mm EDTA, 50 mm Tris (pH 8.0), 1 mg/ml lysozyme, and protease inhibitors), incubated for 5 min at room temperature, and mixed with an equal volume of ice-cold lysis buffer (500 mm NaCl, 1% Nonidet P-40, 50 mm Tris, pH 8.0, 5 mm MgCl2, 1 μg/ml DNase, 100 μg/ml RNase). After incubation on ice for 30 min cell debris were removed by centrifugation (30 min, 4 °C, and 14,000 × g) followed by SDS-PAGE and Western blot analyses.
DNA Ejection in Vitro
A 10-μl reaction mixture containing SPP1 wild type (∼5 × 109 pfu/10 μl = 0.83 nm) in ejection buffer (300 mm NaCl, 100 mm Tris-HCl, pH 7.5, 10 mm MgCl2) and YueB780 (0.15 μg/10 μl = 91 nm YueB780 dimers in 300 mm NaCl, 50 mm HEPES, pH 7.0) was incubated on ice for 30 min and then transferred to a water bath at 37 °C for 1 h. Ejected DNA was eliminated during the procedure by the presence of 25 IU of Benzonase (Merck). Samples were kept at 4 °C until their loading into 10% SDS-PAGE gels followed by Western blot analyses.
Imaging of SPP1 Tail Cap after DNA Ejection
SPP1 phages incubated with purified YueB780 receptors were prepared as follow: 18 μl of phages (5 × 1010 pfu/ml) was mixed with 1 μl of DNase (final concentration of 1 μg/ml) for 15 min at 37 °C. Then, 1 μl of YueB780 from a stock at 100 μg/ml was added, and the mixture was incubated for 1 h at 37 °C to ensure full ejection. Frozen-hydrated specimens were prepared as described above with the difference that the samples were applied to Lacey carbon grids (Ted Pella Inc.). Images were recorded with an UltraScan4000 charge-coupled device camera (Gatan Inc.) under low dose conditions at a nominal magnification of 50,000×.
RESULTS
Light Scattering Characterization and Mass Measurements of the gp19.1-gp21 Complex
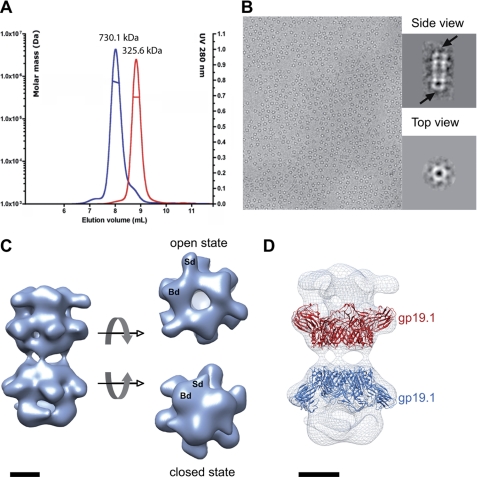
SPP1 gp19.1 and gp211–552 were overproduced and purified to homogeneity. We used SEC/MALS/RI/UV measurements to characterize the oligomeric state of the gp19.1-gp211–552 complex. Our previous data established that gp19.1 forms a dimer of hexamers in solution. Mixing gp19.1 and gp211–552 shifted the SEC elution peak (Fig. 1A), indicating that complex formation occurred. We measured a mass of 730.1 ± 3.7 kDa for the gp19.1-gp211–552 complex, which is consistent with the association of one dimer of Dit hexamers (22) with six gp211–552 subunits.
FIGURE 1.
Characterization of gp19.1-gp211–552 complexes. A, SEC elution profile of gp19.1 (Dit) (red curve) and gp19.1 in the presence of gp211–552 (blue curve). The displacement of the elution volume indicates that gp211–552 interacts with gp19.1. The computed masses are indicated above each peak. B, image of frozen-hydrated gp19.1-gp211–552 complexes revealing two preferential orientations in amorphous ice. Insets display class averages corresponding to side and top views, in which arrows show structural differences observed at the extremities of gp19.1-gp211–552 particles. C, EM reconstruction of frozen-hydrated gp19.1-gp211–552 complexes at 26-Å resolution at 0.5 FSC (Fourier Shell Correlation) cut-off. Two different conformations were observed at the complex extremities, corresponding to open and closed states. The structure extremities consist of a trimer of bilobed densities named Broad (Bd) and Sharpened (Sd) densities, respectively. D, fit of the gp19.1 atomic structure in a cryo-EM map. Two hexamers of gp19.1 have been fitted independently in the cryo-EM reconstruction. Scale bars, 5 nm.
We used non-native mass spectra to reinforce the validity of the stoichiometries determined by light scattering. The spectrum obtained for gp19.1 indicated the presence of a dodecamer with a mass of 342,121 Da. Unfortunately, the gp19.1-gp211–552 complex did not produce spectrum for a reliable estimation of its mass. However, when using a smaller gp21 fragment (1–404) known to host residues involved in gp19.1 interplay, the gp19.1-gp211–404 complex spectrum yielded a mass of 618,937 Da, which agrees with the theoretical mass of 618,796 Da (supplemental Fig. S1) for a stoichiometry of one Dit dodecamer and six gp211–404. Consequently, according to the 2-fold symmetry of dodecameric Dit, results showed that three gp21subunits are likely associated with one Dit hexamer.
Structure of the gp19.1-gp211–552 Complex
The gp19.1-gp211–552 complex was then imaged by EM either after negative staining (supplemental Fig. S2) or from frozen-hydrated specimens (Fig. 1B). Images revealed the presence of homogeneous particles with two predominant views: a side view showing elongated ladder-like particles and a top view displaying round star-like particles. The ratio between side and top particle views depended on specimen preparation conditions. Consequently, with respect to the difference in particle size, top and side views were extracted from images and processed separately as described under “Experimental Procedures.”
The three-dimensional reconstruction of gp19.1-gp211–552 after negative staining showed two back-to-back rings of hexameric Dit surrounded by six extra-densities (supplemental Fig. S2). This starting model was used to process images of the frozen-hydrated gp19.1-gp211–552 complex. Hence, 10,796 side views were extracted from 100 digitized micrographs and combined to 11,002 top views. We failed to compute a reliable model consistent with our data with the angular reconstitution method and the enforced D3 symmetry that was previously used for processing of images of gp19.1 negatively stained particles (22). The successful strategy has consisted of assigning Euler angles with the projection matching method using restrained projection angles for the side and top views (see “Experimental Procedures”) and applying a C3 symmetry. The reconstruction of C3 symmetry after one Multi-Reference Alignment cycle revealed one-half well resolved volume while the other half volume was blurred and not improved after iterative cycles of images alignment and three-dimensional reconstruction. Interestingly, after a second Multi-Reference Alignment cycle based on an artificial model assembled with two well-resolved parts, both halves of the particles were resolved, and the three-dimensional reconstruction resulted in a volume that clearly diverged from the reference model. After five iterative cycles of images alignment and three-dimensional reconstructions, image processing provided a stable reconstruction of gp19.1-gp211–552 at 26-Å resolution as presented in Fig. 1C. The final three-dimensional EM map reveals a particle of ∼22 nm long and 14 nm wide. It is composed of two non-identical back-to-back units that interact loosely together. Each unit is composed of a hexameric part with six domains pointing outward and topped by a trimer of bilobed densities. The main differences between the two units result from the conformation of the trimer. Indeed, one trimer is open and delineates a 2 nm central channel compatible with DNA transit, whereas the opposite trimer is closed and compact. These two conformations will be referred from hereafter as the open and closed states, respectively.
According to the stoichiometry estimation, i.e. one gp19.1 dodecamer with six gp211–552 subunits, and the gp19.1 dodecamer shape (22), we concluded that each unit is composed of an hexameric ring of gp19.1 and a trimer of gp211–552. This was confirmed by the unambiguous fit of the gp19.1 hexamer crystallographic structure into the cryo-EM map (Fig. 1D). It is important to note that the gp19.1 dodecamer could not be straightforwardly positioned into the EM map despite obvious structural features. Instead, each hexamer of gp19.1 had to be positioned independently to obtain the result show in Fig. 1D. It implies that the interface between the two gp19.1 hexamers has been modified when compared with the compact dodecameric gp19.1 conformation observed both in an EM-negative stain and in crystals (22). The weak interaction between gp19.1 hexamers results in bent gp19.1-gp211–552 complexes displayed in cryo-EM images. This flexibility impacted on the resolution of the final cryo-EM reconstruction. At 26-Å resolution, both conformational states readily superimposed onto each other with a correlation coefficient of 0.76. The two opposite gp19.1 hexamers are quasi-identical indicating that the main differences occur within gp211–552-associated densities. In conclusion, the cryo-EM gp19.1-gp211–552 map exhibits two different conformations mainly associated with structural reorganization of gp211–552 densities that form a central channel in the open state and a dome-like structure in the closed state (Fig. 1C). Two domains can be distinguished within each gp211–552 density, a broad domed domain (Bd) that interacts with gp19.1 and a sharpened domain (Sd) pointing outwards from the C3 axis. These densities appear to be more elongated and more curved in the open state than in the compact closed state. The two gp211–552 conformational states were distinguishable in the class averages of aligned images (Fig. 1B) strengthening the reliability of our EM reconstruction.
Identification of SPP1 Tail Cap Proteins
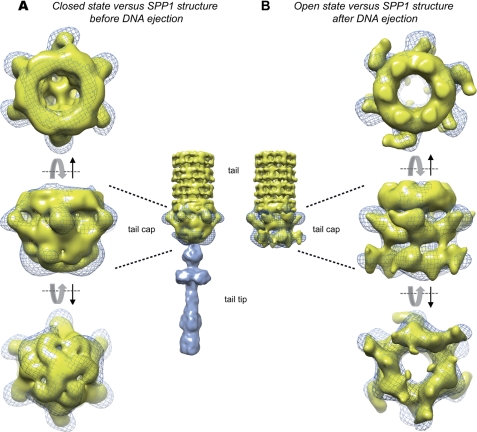
Reconstructions of the SPP1 tail, tail cap, and tail tip were previously reported before and after DNA ejection (10). The subsequent determination of the gp19.1 crystallographic structure assigned it to the cap, but an unambiguous positioning in this region of the tail was not possible (22). To establish the cap organization, we compared the open/closed states of the gp19.1-gp211–552 cryo-EM map with SPP1 tail maps (Fig. 2). The only way to fit the gp19.1-gp211–552 complex was to place gp19.1 inside the last ring of densities previously assigned to the MTP (Fig. 3). At this position, the closed and open maps of gp19.1-gp211–552 match perfectly to the in virus tail cap densities, respectively, before and after DNA ejection. In addition, in both states, gp19.1 fits perfectly well into the upper part of the cap domain with its six protruding densities clearly corresponding to the gp19.1 C-terminal domain pointing toward the cap region. The tail cap before DNA release is a dome-like structure with a trimeric organization resembling that of a triskelion motif. The cryo-EM map of the gp19.1-gp211–552 closed state superimposes well to the tail cap before DNA ejection. The Bd and Sd encompass the rounded structure and one end of the triskelion, respectively (Fig. 2A). In the gp19.1-gp211–552 open state, some structural features are also straightforwardly recognizable (Fig. 2B). The direction, shape, and curvature of the gp211–552 densities closely outline the lower part of the tail cap after DNA ejection. In conclusion, our data unambiguously demonstrate that the so-called SPP1 tail cap is composed of gp19.1 and of the N-terminal half of gp21, whose C terminus must extend to the tail tip region.
FIGURE 2.
Comparison of cryo-EM structures of gp19.1-gp211–552 in its closed and open conformation with SPP1 tail maps before and after DNA ejection. SPP1 tail and tail cap maps are in yellow and the tail tip map is in blue. The cryo-EM structures of gp19.1-gp211–552 in its open and closed states are in blue mesh. A, closed state versus phage tail structure before DNA ejection. Gp19.1 fits well in the upper part of tail cap, which contacts the tail tube, whereas gp211–552 fits well in the lower part of the tail cap. B, open state versus tail phage structure after DNA ejection. The opening of the tail cap is equivalent to that observed in the open conformation map of gp19.1-gp211–552 complex. Common characteristic features with the open conformation map of gp19.1-gp2l1–552 complex can be identified. Hence, the six protruding domains of gp19.1 can be observed in the upper part of the tail cap map, whereas the lower part presents domains that superimpose well with Bd and Sd domains of the open conformation map of the gp19.1-gp211–552 complex.
FIGURE 3.
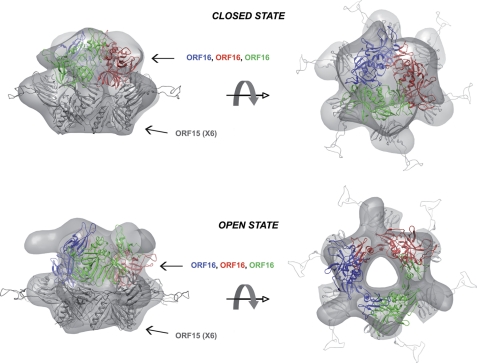
Docking of bacteriophage p2 ORF16-ORF15 open and closed structures in the gp19.1-gp211–552 maps. The ORF15-ORF16 complex with ORF16 in its closed conformation has been fitted into a cryo-EM map of gp19.1-gp211–552 complex in its closed state. In the same way, the ORF15-ORF16 complex with ORF16 in its open conformation has been fitted into the cryo-EM map of gp19.1-gp211–552 complex in its open state. For a better visualization, hexameric ORF15 has been shaded gray, whereas the trimer of ORF16 has been colored in blue, red, and green.
Gp21, a Key Protein Involved in DNA Channel Opening
PSI-BLAST searches on the N-terminal 400-amino acid sequences of Tal revealed that it is well conserved across Siphoviridae (>28% sequence identity) and that it is classified in the protein Pfam06605 family. To date, the only structural representative of this conserved family is the prophage tail protein gp18 from Listeria monocytogenes Egd-E (PDB: 3GS9). Nevertheless, a Dali search for similar structures using gp18 as query returned several hits with significant Z-scores, including the 43-kDa tail protein from the prophage MUSO2 (PDB: 3CDD, Z-score: 16.6), the baseplate's protein gp27 from the coliphage T4 (27) (PDB: 1K28, Z-score: 14.9), and ORF16 from the lactococcal phage p2 (11) (PDB: 2WZP, Z-score:10.5). These proteins are mainly composed of β-strands and superimpose quite well their four domains onto each other (root mean square deviation of 3.7 Å on the Cα backbone on average), with the exception of the “activated” conformation of ORF16 in which the third domain points outward from the compact core (11). Based on these findings and on the structural similarity between SPP1 gp19.1 and p2 ORF15, we then tried to fit either the closed or open conformations of p2 ORF15-ORF16 complexes in our cryo-EM map (Fig. 3). The fit was done manually without modifying any part of ORF15-ORF16 complexes and refined only as a single rigid body using Chimera (28). Clearly, ORF15-ORF16 in its closed conformation fits quite well with our map of the closed state. ORF15 readily fits into the EM density of the closed state map except for its extension involved in griping the receptor-binding proteins of phage p2 (ORF18) that are missing in gp19.1 and thus not found in the SPP1 cap density, whereas ORF16 only needs a slight rotation to be perfectly accommodated. The ORF15-ORF16 complex in its open conformation fits equally well into the cryo-EM map of the open state. The inner channel delineated by the three ORF16 subunits is similar to that formed by the gp211–552 trimer. However, the sharpened domain of gp211–552 has no counterpart in the ORF16 structure. This is probably due to the supplementary residues present in gp21 (552 for gp211–552 versus 326 for ORF16) that may account for the extra-sharpened domain present in gp21.
To go further into the comparison of SPP1 gp19.1-gp211–552 and p2 ORF15-ORF16, we have addressed the influence of Ca2+ ions on the SPP1 complex structure. Indeed, it has been previously shown that ORF15-ORF16 conformational changes are induced or stabilized by Ca2+ ions (11). We thus investigate the behavior of gp19.1-gp211–552 in the presence of Ca2+ ions by cryo-EM. The reconstruction obtained clearly revealed the opening of gp211–552 trimer upon addition of Ca2+ ions (supplemental Fig. S3).
Full-length gp21 Remains Associated to the SPP1 Tail after DNA Ejection
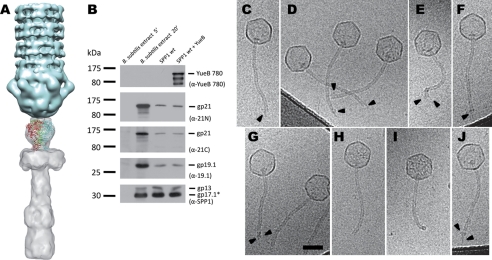
The -fold recognition server Phyre (29) predicted with significant e-value (2e-06 to 1.8e-4) and high precision (95%) that the gp21 C-terminal region (from residue 577 to 1071) is folded either as a globular hydrolase domain followed by a single-stranded β-helix domain or a whole single-stranded β-helix. This is reminiscent of the common single-stranded β-helix or β-helical prism folds of Podoviridae and Myoviridae tail fiber proteins (30–35). Three of the Phyre-predicted structures could be fitted into the proximal part of the tail tip EM map with the β-helix domains pointing toward the saucer-like shaped region (Fig. 4A). This suggests that the gp21 C-terminal region could account for the proximal part of the tail tip. However, the tail tip is not observed in the reconstruction of the SPP1 tail after genome ejection (10). These observations raise the question of the fate of gp21 upon DNA release. To address this question, we investigated the behavior of gp21 in the presence of YueB780, the ectodomain of the B. subtilis protein YueB that is used as receptor by SPP1 for irreversible commitment. Indeed, it was previously shown that YueB780 binds to the SPP1 tail tip and triggers release of phage DNA (36). The presence of gp21 was detected in the virion by Western blot of its structural proteins using antibodies directed either against its N- or C-terminal regions. A single band with the electrophoretic mobility of full-length gp21 (123 kDa) was observed with both sera for intact and YueB780-treated phages (Fig. 4B). Thus gp21 is not cleaved upon DNA ejection, and at least the tip proximal region remains associated to the cap. We then carried out cryo-EM on the extremity of the SPP1 tail after binding to YueB780 (Fig. 4, C–J) using the empty capsids as a testimony of tail tip loss and viral DNA release. The vast majority of tail extremities display heterogeneous profiles where extra densities are observed at the end of tail. In rare cases, tail extremities are well defined (Fig. 4, H and I), whereas others may show a YueB780 fiber bound to the tail (Fig. 4J). This is consistent with the previously description of YueB780 as an elongated dimer in solution forming 36 nm fibers (36). Taken together, these data strongly suggest that gp21 is composed of two structural domains linked by a flexible region and that its C-terminal domain becomes floppy after loss of the tail tip, producing heterogeneous tail end motifs that are smeared out in the map of cap domain symmetry published by Plisson et al. (10).
FIGURE 4.
Structural model of the SSP1 tail Tal extension to the SPP1 tip. A, docking of the gp21 C-terminal domain predicted structure into the SPP1 map of the tip. B, detection of gp19.1 and gp21 in extracts of B. subtilis infected with SPP1 (5 min and 20 min after infection) and in phage particles before and after DNA ejection triggered with YueB780. The amounts of sample applied to the gel lanes of both cellular extracts or of SPP1 particles were identical. DNA ejection from SPP1 phages was triggered by incubation with purified ectodomain receptor YueB780. Proteins were resolved in 10% SDS-PAGE gels and identified by Western blot. Two identical blots were probed sequentially with anti-21N, anti-19.1, and anti-SPP1 or with anti-21C and anti-YueB780 polyclonal antibodies, respectively. Note that YueB is not detected in B. subtilis extracts due to its low level of expression (40). The presence of the major capsid protein (gp13) and the major tail protein (gp17.1*) detected with anti-SPP1 serum is shown at the bottom. The positions of prestained molecular mass markers are indicated on the left (kilodaltons). Polyclonal antibodies used in each case are indicated within parentheses on the right. C–J, cryo-EM images of SPP1 tail ends after DNA ejection. SPP1 phages were incubated with the ectodomain receptor YueB780, inducing tip release and DNA ejection. Most phage capsids are entirely DNA-empty. Arrowheads (C–G) indicate structural heterogeneity of SPP1 tail ends. Sometimes (H and I), it is possible to observe well defined SPP1 tail ends, whereas rarely the YueB780 fiber can be see bound to the tail (J). Scale bar, 50 nm.
DISCUSSION
Present knowledge of the SPP1 tail architecture relies to a large extent on a previous EM study and its structural model derived from volume interpretation of EM densities, structure prediction, and sequence analysis (10). Comparison between tail structures before and after DNA ejection led to the hypothesis that a firing signal generated at the onset of infection is propagated from the distal to the capsid proximal tail extremity as a domino-like cascade along the tail tube (10). The signal targets the head-to-tail connector whose opening leads to DNA release from the viral capsid through the tail tube to exit the phage particle (37). These events are triggered by interaction of the SPP1 tail extremity, composed cap that closes the tail tube and the tip, with the host receptor YueB. Following the recent re-positioning of gp19.1 to the cap structure (22), we have investigated further the organization of this tail region, which appears critical for initiating signaling that is propagated along the MTP helical tube lattice. Bioinformatics showed that gp19.1 and the N terminus of gp21 share sequence homology and that their coding genes follow each other in the phage genome suggesting structural conservation and cross-talk between the two proteins (22). Consequently, we decided to investigate the structural organization of the putative complex formed by gp19.1 and gp21. Considering the high level of sequence conservation observed for Tal N-terminal moieties among Siphoviridae, we recombinantly expressed and purified the N-terminal half of gp21, gp211–552, and demonstrated that it interacts with Dit.
Our light scattering and non-covalent MS results established that one Dit dodecamer was associated with six gp211–552 subunits. Because gp19.1 associates as two back-to-back hexamers in solution, we expected that one gp211–552 trimer would associate with each Dit face. These results were subsequently corroborated by the EM structural study of the gp19.1-gp211–552 complex.
Similarly to some other elongated complexes (38, 39), gp19.1-gp211–552 displays two preferential views (top and side views) that were processed separately. The structural analysis of frozen-hydrated gp19.1-gp211–552 complexes by EM and image processing yielded two major results. Firstly, gp19.1-gp211–552 complexes consist of two structurally independent sub-complexes composed of a gp19.1 hexamer and a gp211–552 trimer. Each sub-complex, which corresponds to the cap structure in SPP1 phage particles (Fig. 2), loosely associates to another sub-complex through interactions between the narrower rings of gp19.1 hexamers (Fig. 1, C and D). Secondly, the structure is asymmetric, a result of the gp211–552 trimers conformation that is open in one sub-complex and closed in the other. The alternating states of gp211–552 trimers in the same complex require a cross-talk mediated through the gp19.1 hexamers, but its structural basis cannot be established at the resolution of our reconstruction. The property of gp211–552 to adopt a closed and an open state are outstandingly reminiscent of the SPP1 tail cap before and after DNA ejection (10) and the p2 ORF16 closed and open structures (11). These data clearly show that the SPP1 tail cap is formed by a gp19.1-gp21 complex that accounts for the transition from 6- to 3-fold symmetry in the tail structure. It also argues for a common structural organization of the cap among Siphoviridae phages. Moreover, the size of the SPP1 full-length gp21 is twice as much as that of the recombinant gp211–552 protein. According to the fit presented in Fig. 4A, the gp21 C-terminal region predicted structure (β-helix) would account for the proximal part of the SPP1 tail tip. This topology of gp21 is consistent with immunoelectron microscopy experiments.4 Otherwise, the SPP1 tip distal EM density remains unassigned. This volume might result from the assembly of several small ORFs following the gp21 tail fiber coding sequence in the SPP1 genome (gp22, gp23, gp23.1, gp24, and gp24.1). However, we previously solved the crystal structures of two of these (gp23.1 and gp22) (41, 42), and the poor quality of their fit in the tip reconstruction suggested that they are not located in this region of the virion.
This structural organization raised the questions: 1) the possible fate of gp21 during DNA ejection, because the tip is not observed anymore upon interaction with YueB (10), and 2) notably, why the SPP1 cap structure after DNA ejection (10) did not reveal the presence of additional densities corresponding to the second half of gp21 sequence. The two possible explanations are that either gp21 is cleaved in the virion and the C-terminal domain is released with the tail tip upon receptor binding, or the second domain of gp21 is linked to the N terminus domain by a flexible hinge and becomes disorganized upon SPP1 interaction with YueB, and consequently its density is averaged out in the map of SPP1 tail cap.
To address the hypothesis of a putative gp21 cleavage, as is the case in Tuc2009 and TP901-1 Tal proteins (19, 43, 44), we carried out Western blots of SPP1 particles before and after challenge with the receptor ectodomain YueB780. Detection with antibodies raised either against the gp21 N or C termini demonstrated that the full-length protein remains intact after DNA ejection. In addition, the cryo-EM images of SPP1 phages incubated with YueB780 show irregular SPP1 tail ends whose appearance varies between different viral particles. The extra densities observed are attributed to the disorganized gp21 C terminus, YueB780, and eventually to other tail tip components. The resulting model is that the gp21 N terminus forms the cap bottom structure that is connected by a gp21 flexible hinge to the C terminus that accounts for the tail tip proximal region.
Therefore, interaction with YueB leads to opening of the gp21 N terminus region and concomitant disassembly of the C terminus organization. The challenge is now to understand how the sturdy β-helix fold proposed for the gp21 terminus is disassembled during this process.
The conserved sequences of Tal-like proteins, including T4 gp27 (PDB ID 1K28), Mu gp44 (PDB ID 1WRU), p2 ORF16 (PDB ID 2WZP and 2X53), Shewanella oneidensis prophage MuSO2 Q8EDP4 (PDB ID 3CDD), L. monocytogenes prophage EGD-e gp18 (PDB entry 3GS9), and the Escherichia coli c3393 Type 6 secretion system protein (2P5Z) concern the first 400 residues. Among them, it was shown that ORF16, the Tal protein of phage p2, could adopt either a closed or open conformation (11). The docking of the p2 ORF15-ORF16 complex into our cryo-EM map of the gp19.1-gp211–552 open and closed states showed that the opening of Tal proteins is a common feature in siphophages p2 and SPP1 leading to formation of a channel for DNA release. In the case of the p2 baseplate, the ORF16 open conformation was triggered by addition of Ca2+ ions that is also correlated with a 200° rotation downward of the six receptor-binding protein (RBP) that are anchored in ORF15. Interestingly, Ca2+ ions stabilize also the gp211–552 open conformation (supplemental Fig. S3). Millimolar amounts of Ca2+ favor SPP1 infection in vivo (45) but are not necessary to trigger DNA ejection in vitro (36) rendering it difficult to assess their biological importance for gp21 biological function at present.
In an evolutionary context, several SPP1 tail components show significant sequence and/or structural similarities to equivalent proteins from lactococcal phages Tuc2009, TP901-1, and p2. Hence, we recently showed that the central part of the TP901-1 baseplate is formed by Dit and Tal proteins (18). It becomes now apparent that Siphoviridae phages share a common core at the tail distal extremity composed of MTP, Dit, and Tal proteins. However, Siphoviridae exhibit different types of distal tail architectures that seem to be related to the nature of the host recognition process and affinity between partners. Thus, phages TP901-1 or p2, which have a low avidity toward saccharidic receptors, have evolved by developing a strategy related to the presence of a high number of RBPs, 18 and 54 for phage p2 and TP901-1, respectively. On the contrary, SPP1 phage, which recognizes a proteinaceous receptor with a high affinity, displays an elongated tail tip (36). In phage p2, the large baseplate conformational changes observed might be transmitted to the first MTP ring, and, by a domino-like concerted effect, to all other MTP rings along the tail, finally resulting in connector opening and DNA release. However, in the case of TP901, RBPs are immediately available to bind to host receptors (18), and large conformational changes do not appear to be involved for initiating infection. Similarly, the SPP1 elongated tail tips that do not possess RBPs do not seem to be subjected to large conformational changes induced by receptor binding. Thus, the initiation of signal transmission to the first MTP ring might be more subtle than the direct consequence of RBPs displacement.
The emerging picture is that the Siphoviridae tail end is built around a conserved core of Dit and Tal that corresponds to the SPP1 cap structure. This platform serves to dock RBPs, which can be assembled in baseplate structures gathering multiple high avidity saccharidic receptor binders (e.g. p2 and TP901-1) and/or single tips that bind a proteinaceous receptor with high affinity (e.g. SPP1). The platform provides also the attachment site for the MTP and the tape measure protein that builds the tail tube. We propose that interaction of RBPs with host receptors triggers a signal converging to the Dit platform. The signal switches Tal to its open conformation and initiates the domino-like cascade in the first ring of MTP hexamers that propagates along the helical tail to cause DNA ejection (10).
Accession Codes
The EM map has been deposited at EMDB (http://www.emdatabank.org), with accession code EMD-1900.
Supplementary Material
Acknowledgments
We gratefully thank Prof. Elena Orlova for providing us with the EM maps of SPP1 cap, Prof. Alan Davidson for fruitful discussion, Dr. Arie Geerlof who kindly provided the GatewayTM plasmid pETG-20A, Sandrine Brasilès for purified YueB780, and Isabelle Petitpas for anti-gp21 and anti-gp19.1 polyclonal sera. Visualization of cryo-EM maps and molecular graphic images were produced using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
This work was supported by ANR BLAN07-1_191968 Structural insights on Siphoviridae Bacteriophages infection, Conseil Régional Languedoc-Roussillon, Montpellier I University and INSERM.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S3.
I. Vinga, C. Baptista, I. Auzat, I. Petipas, R. Lurz, P. Tavares, M. A. Santos, and C. Sao-Jose, submitted for publication.
- MTP
- major tail protein
- Tal
- protein gp21
- Dit
- protein gp19.1
- SEC
- size-exclusion chromatography
- RBP
- receptor-binding protein.
REFERENCES
- 1. Wommack K. E., Colwell R. R. (2000) Microbiol. Mol. Biol. Rev. 64, 69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ackermann H. W. (2007) Arch. Virol. 152, 227–243 [DOI] [PubMed] [Google Scholar]
- 3. Ackermann H. W. (2003) Res. Microbiol. 154, 245–251 [DOI] [PubMed] [Google Scholar]
- 4. Casjens S., Henrix R. (1988) Control Mechanisms in dsDNA Bacteriophage Assembly, Plenum Press, New York [Google Scholar]
- 5. Molineux I. J. (2006) Virology 344, 221–229 [DOI] [PubMed] [Google Scholar]
- 6. Effantin G., Boulanger P., Neumann E., Letellier L., Conway J. F. (2006) J. Mol. Biol. 361, 993–1002 [DOI] [PubMed] [Google Scholar]
- 7. Böhm J., Lambert O., Frangakis A. S., Letellier L., Baumeister W., Rigaud J. L. (2001) Curr. Biol. 11, 1168–1175 [DOI] [PubMed] [Google Scholar]
- 8. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pell L. G., Liu A., Edmonds L., Donaldson L. W., Howell P. L., Davidson A. R. (2009) J. Mol. Biol. 389, 938–951 [DOI] [PubMed] [Google Scholar]
- 10. Plisson C., White H. E., Auzat I., Zafarani A., São-José C., Lhuillier S., Tavares P., Orlova E. V. (2007) EMBO J. 26, 3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sciara G., Bebeacua C., Bron P., Tremblay D., Ortiz-Lombardia M., Lichière J., van Heel M., Campanacci V., Moineau S., Cambillau C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J., Hofnung M., Charbit A. (2000) J. Bacteriol. 182, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baptista C., Santos M. A., São-José C. (2008) J. Bacteriol. 190, 4989–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monteville M. R., Ardestani B., Geller B. L. (1994) Appl. Environ. Microbiol. 60, 3204–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katsura I., Hendrix R. W. (1984) Cell 39, 691–698 [DOI] [PubMed] [Google Scholar]
- 16. Johnsen M. G., Neve H., Vogensen F. K., Hammer K. (1995) Virology 212, 595–606 [DOI] [PubMed] [Google Scholar]
- 17. McGrath S., Neve H., Seegers J. F., Eijlander R., Vegge C. S., Brøndsted L., Heller K. J., Fitzgerald G. F., Vogensen F. K., van Sinderen D. (2006) J. Bacteriol. 188, 3972–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bebeacua C., Bron P., Lai L., Vegge C. S., Brøndsted L., Spinelli S., Campanacci V., Veesler D., van Heel M., Cambillau C. (2010) J. Biol. Chem. 285, 39079–39086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vegge C. S., Brøndsted L., Neve H., McGrath S., van Sinderen D., Vogensen F. K. (2005) J. Bacteriol. 187, 4187–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campanacci V., Veesler D., Lichière J., Blangy S., Sciara G., Moineau S., van Sinderen D., Bron P., Cambillau C. (2010) J. Struct. Biol. 172, 75–84 [DOI] [PubMed] [Google Scholar]
- 21. Katsura I., Kühl P. W. (1975) J. Mol. Biol. 91, 257–273 [DOI] [PubMed] [Google Scholar]
- 22. Veesler D., Robin G., Lichière J., Auzat I., Tavares P., Bron P., Campanacci V., Cambillau C. (2010) J. Biol. Chem. 285, 36666–36673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 24. Mindell J. A., Grigorieff N. (2003) J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 25. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 26. van Heel M. (1986) Optik 73, 119–122 [Google Scholar]
- 27. Kanamaru S., Kondabagil K., Rossmann M. G., Rao V. B. (2004) J. Biol. Chem. 279, 40795–40801 [DOI] [PubMed] [Google Scholar]
- 28. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 29. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 30. Barbirz S., Müller J. J., Uetrecht C., Clark A. J., Heinemann U., Seckler R. (2008) Mol. Microbiol. 69, 303–316 [DOI] [PubMed] [Google Scholar]
- 31. Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. (2002) Nature 415, 553–557 [DOI] [PubMed] [Google Scholar]
- 32. Müller J. J., Barbirz S., Heinle K., Freiberg A., Seckler R., Heinemann U. (2008) Structure 16, 766–775 [DOI] [PubMed] [Google Scholar]
- 33. Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. (1994) Science 265, 383–386 [DOI] [PubMed] [Google Scholar]
- 34. Thomassen E., Gielen G., Schütz M., Schoehn G., Abrahams J. P., Miller S., van Raaij M. J. (2003) J. Mol. Biol. 331, 361–373 [DOI] [PubMed] [Google Scholar]
- 35. van Raaij M. J., Schoehn G., Burda M. R., Miller S. (2001) J. Mol. Biol. 314, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 36. São-José C., Lhuillier S., Lurz R., Melki R., Lepault J., Santos M. A., Tavares P. (2006) J. Biol. Chem. 281, 11464–11470 [DOI] [PubMed] [Google Scholar]
- 37. Lhuillier S., Gallopin M., Gilquin B., Brasilès S., Lancelot N., Letellier G., Gilles M., Dethan G., Orlova E. V., Couprie J., Tavares P., Zinn-Justin S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8507–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen S., Roseman A. M., Hunter A. S., Wood S. P., Burston S. G., Ranson N. A., Clarke A. R., Saibil H. R. (1994) Nature 371, 261–264 [DOI] [PubMed] [Google Scholar]
- 39. Harris J. R., Plückthun A., Zahn R. (1994) J. Struct. Biol. 112, 216–230 [DOI] [PubMed] [Google Scholar]
- 40. São-José C., Baptista C., Santos M. A. (2004) J. Bacteriol. 186, 8337–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veesler D., Blangy S., Lichière J., Ortiz-Lombardía M., Tavares P., Campanacci V., Cambillau C. (2010) Protein Sci. 19, 1812–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Veesler D., Blangy S., Spinelli S., Tavares P., Campanacci V., Cambillau C. (2010) Protein Sci. 19, 1439–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kenny J. G., McGrath S., Fitzgerald G. F., van Sinderen D. (2004) J. Bacteriol. 186, 3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brøndsted L., Pedersen M., Hammer K. (2001) Appl. Environ. Microbiol. 67, 5626–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santos M. A., De Lencastre H., Archer L. J. (1984) J. Gen. Virol. 65, 2067–2072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.