Abstract
The role of electrostatics on protein-protein interactions and binding is reviewed in this article. A brief outline of the computational modeling, in the framework of continuum electrostatics, is presented and basic electrostatic effects occurring upon the formation of the complex are discussed. The role of the salt concentration and pH of the water phase on protein-protein binding free energy is demonstrated and indicates that the increase of the salt concentration tends to weaken the binding, an observation that is attributed to the optimization of the charge-charge interactions across the interface. It is pointed out that the pH-optimum (pH of optimal binding affinity) varies among the protein-protein complexes, and perhaps is a result of their adaptation to particular subcellular compartment. At the end, the similarities and differences between hetero- and homo-complexes are outlined and discussed with respect to the binding mode and charge complementarity.
Keywords: electrostatics, protein-protein interactions, pH, salt, nsSNPs, missense mutations
1. Introduction
The living cell is an extremely complicated system and is comprised of hundreds of thousands of types of biological macromolecules, which constantly interact with each other to maintain the function of the cell, reflecting the dynamics of cellular networks [1, 2]. The interactions are very specific and frequently a particular protein macromolecule is able to recognize its partner among hundreds of thousands of candidates [3]. At the same time, the recognition process is fast and thus some protein-protein interactions may be electrostatically guided, perhaps through a long-range force that selects and brings the interacting partners together [4–6]. The best candidate for such a guiding long-range force is the electrostatic force [7–12]. A rough estimate of the electrostatic energy of interaction between two molecules carrying a unit net charge and positioned at a distance 10A away from each other results in almost 1 [KJ/mol], which is much more than any other energy component contributes to the binding at such distances.
Thus, electrostatic forces and energies are essential for the interactions of virtually all biological macromolecules [13–15] (see also excellent review [16]). The central role of electrostatics is due to the fact that most biological macromolecules, especially DNA and RNA, are highly charged. However, they are not easy to calculate because the association occurs in a water phase at specific salt concentration and pH. In this review we first briefly outline the current continuum methods for computing the electrostatic component of the binding free energy. In addition to contributing to the binding free energy, the long-range electrostatic interactions can steer protein molecules toward their pre-binding orientations. However, some complexes are formed of identical macromolecules (homo-complexes), while others involve different entities (hetero-complexes). The main difference between these two cases is the net charge of the monomers, which for homo-dimers is the same for both monomers, while for hetero-complexes the monomers frequently carry opposite net charges. Such a difference is expected to result in different roles of electrostatics on the protein-protein recognition at very large distance, at which the distribution of the charges is not important but the net charge is. As it was mentioned above, charged groups are essential for the interactions of macromolecules, but their probability to be charged (directly connected to their pKa’s) is frequently perturbed in proteins and RNAs due mostly to electrostatic effects. Moreover, the binding itself can induce ionization changes resulting in proton uptake/release. In addition, the ionization phenomena are strongly affected by the pH and ion concentration in the water phase, which are specific for each subcellular compartment. Here we review the progress made in understanding the role of pH and salt on the macromolecular binding and the possibility that the macromolecular association is adapted to the subcellular microenvironment. At the end, we outline the findings related to the effects of single amino acid substitutions (resulting from either disease-causing mutations or non-synonymous single nucleoside polymorphisms, nsSNPs, found in the general population) on the electrostatic component of the binding free energy.
2. Modeling the electrostatic component of binding free energy
To investigate the role of electrostatics on protein-protein association, the electrostatic component of the binding free energy needs to be accurately calculated. However, there are three major obstacles in in silico modeling of the binding free energy of biological macromolecules: (1) the binding occurs in a water phase consisting of millions of water molecules whose positions and orientations vary dynamically [17–19]; (2) the unbound and bound monomers are also dynamical structures existing as an ensemble of structures, and these ensembles could be quite different for unbound versus bound monomers [20–25], and (3) the charged states of the ionizable groups may be non standard in the unbound and bound monomers and even may change due to the binding [26–30]. Taking into account rigorously the above effects in the computational protocol is a challenge.
Computational methods for evaluating the binding free energy can be grouped into two major classes with respect to the treatment of the water phase: explicit and implicit. Explicit models consider the water phase as a sea of explicit water molecules which are allowed to sample different positions and orientation during the calculations of the binding free energy. Typically the corresponding monomers and protein-protein complex are allowed to explore different conformations as well. While such an approach is assumed to better represent the physical reality of protein-protein binding, in this review we will focus on implicit methods of computing the electrostatic component of the binding energy (see Ref. [31–34] for comparison between explicit and implicit treatment). The focus on the implicit model is due to the speed of calculations, the ability to handle large systems (large protein-protein complexes), and the lack of convergence problems [35]. It should be mentioned that hybrid approaches have also been developed that treat bound waters and ions explicitly while the bulk water and ions are modeled as homogenous medium with the corresponding ion concentration [36–39].
In a continuum framework, the electrostatic potential in a system comprised of biological macromolecules (modeled as low dielectric entities) immersed in water (high dielectric medium) and in the presence of mobile ions obeys the Poisson-Boltzmann equation (PBE) [40]. However, because of the irregular shape of the proteins and protein-protein complexes, the PBE must be solved by the means of numerical algorithms [41]. As an alternative to the PBE, the Generalized Born (GB) approach can be applied as well [42–46], although with less impressive accuracy [47]. A typical scheme for computing the electrostatic component of the binding free energy is shown in Fig. 1. For each state (the unbound and bound monomers) the electrostatic energy components are obtained, which in terms of PBE formalism are typically the Coulombic energy of interaction; the interaction energy solvent-water (solvation energy) and the interaction energy of solvent-ions (termed ionic contribution to the energy of the system) [48]. In the case of solving the non linear PBE, the osmotic pressure and electrostatic stress terms must be added to the total electrostatic energy as well [49]. Currently there are many available computer programs designed to deliver these energy terms such as DelPhi [48, 50], Adaptive Poisson-Boltzmann Solver (APBS) and its variants [51–53], and many others [54], including GB based solutions [55–58]. In particular, the DelPhi program provides very convenient output of the abovementioned energies, and thus their individual contributions can be evaluated separately [48, 50] and because of this, DelPhi is extensively used for modeling electrostatic interactions [58–60].
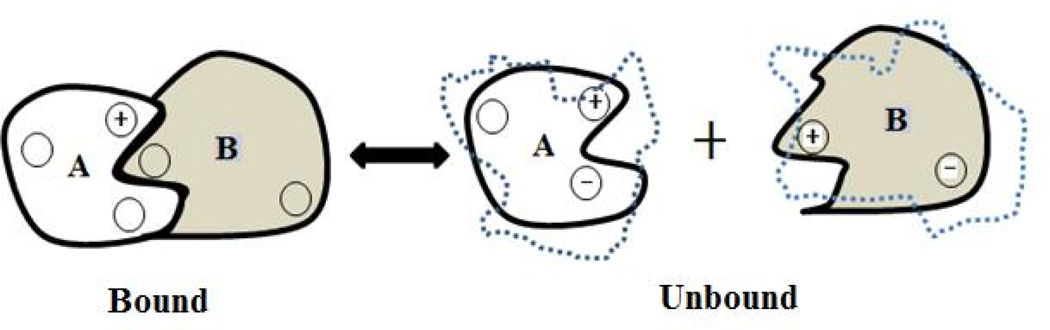
Fig. 1.
Schematic representation of “rigid” body binding concept (solid lines) and “flexible” binding (dash lines).
In applying such computational protocols, the conformational changes can either be taken into account or not (Fig. 1). In the first case, the method is termed unbound-bound calculations, while if the structures of unbound monomers are considered to be the same as in the bound state, the protocol is called bound-bound calculations. In both cases, however, the 3D structure of the protein-protein complex must be known by either experimental means (X-ray or NMR experiments) or must be predicted in silico [61, 62], however, the last case was not extensively explored in the past due to the significant structural imperfections generated in the models predicted by ab-initio docking methods [63, 64]. As an alternative, accurate models of the 3D structures of protein-protein complexes can be delivered by homology methods using highly homologous templates [65–69]. In the bound-bound approach, sometimes termed “rigid body approach”, the structures of unbound monomers are taken from the protein-protein complex and no conformational changes upon the binding are assumed (solid lines in Fig. 1). The advantage of such an approach is that the internal mechanical energy remains the same for bound and unbound states and needs not to be calculated.
Instead, only the non-bonded energies must be accounted for, the most important one being electrostatics. The disadvantage is that such a model is quite simplified and may not be able to capture all important effects associated with protein-protein association. The bound-unbound approach is much more appropriate (solid lines for the complex and dashed lines for the unbound monomers in Fig. 1), but involves changes of the internal energy of the interacting partners, which is difficult to calculate. However, if we are only interested in the electrostatic component, then the computation is much easier provided that the bound and unbound structures are available. In addition, we are not interested in the absolute energies, but either salt or pH dependence (for most cases). Thus, the change of the internal energy from a bound to unbound state will be a constant contribution that will not affect salt and pH dependence.
The role of protonation states (ionization states) of titratable groups on the calculations of the electrostatic component of the binding energy is obvious. An inappropriate charge assignment to a given ionizable group could affect the calculations dramatically. Therefore, in the best case scenario, pKa calculations must be carried out on the bound and unbound monomers and charges assigned according to the predicted pKa’s. Such predictions are typically not performed prior to the calculations of the electrostatic component of the binding energy because of the assumptions that titratable groups of unbound monomers are charged at neutral pH and do not alter their protonation states upon the binding. Such a protocol can be termed “rigid” charge protocol. While the presumption that all (or most of) titratable groups are ionized in unbound monomers holds in many cases, it frequently may not be valid for the bound state (the protein-protein complex), especially for the titratable groups at the interface of the complex. Thus, the net charge of the complex and unbound monomers may differ resulting in proton uptake/release upon the binding. A protocol that takes into account such a possibility can be termed a “flexible” charge approach (Fig. 1), as for example used in Ref. [26].
3. Role of pH and salt concentration
The role of salt and pH on protein-protein binding was recently reviewed [70]. The importance of proton uptake/release in receptor-ligand interactions is demonstrated by the experimental observation that the vast majority of receptor-ligand interactions are pH-dependent [71–76]. A variation of several pHs can result in binding free energy changes of several KJ/mol [76, 77] or even can change the ligand binding preferences [78]. Even more, different binding interactions can occur at different pHs; for example, as found in the case of beta-lactoglobulin that is a dimer at low pH but forms a tetramer at high pH [79]. Similar phenomena were found in the case of calmodulin, whose domains adopt compact arrangement at low pH while at high pH form “dumbbell” shaped structures [80–82]. From a practical perspective, the ability to re-engineer enzymatic pH-activity profiles is important for the industrial application of enzymes [83]. This possibility has been theoretically and experimentally explored to re-engineer enzymatic pH-activity profiles and pH-dependence of kinetic parameters by changing active site pKa values using point mutations [55, 84–87].
The effect of salt concentration on protein-protein binding free energy and its electrostatic component can also be significant. To the best of our knowledge, the E9 Dnase – Im9 complex exhibits the largest sensitivity to the salt concentration, resulting in δΔΔG(I) /Δln(I) = 8[KJ/mol2] [88, 89] (where ΔΔG(I) is the change of the binding free energy and I is the ionic strength). Thus, a small variation of the salt concentration alters the binding energy by several KJ/mol. Another example is of the complex formed between β-Lactamase (TEM-1) with its protein inhibitor (BLIP), in which the association rate decreases an order of magnitude as salt concentration increases from zero to 0.5M [90]. The list of examples can be extended further to include other complexes with experimentally measured salt dependence of the binding energy [91–94], indicating that most of the protein-protein interactions are affected by the ion concentration in the water phase.
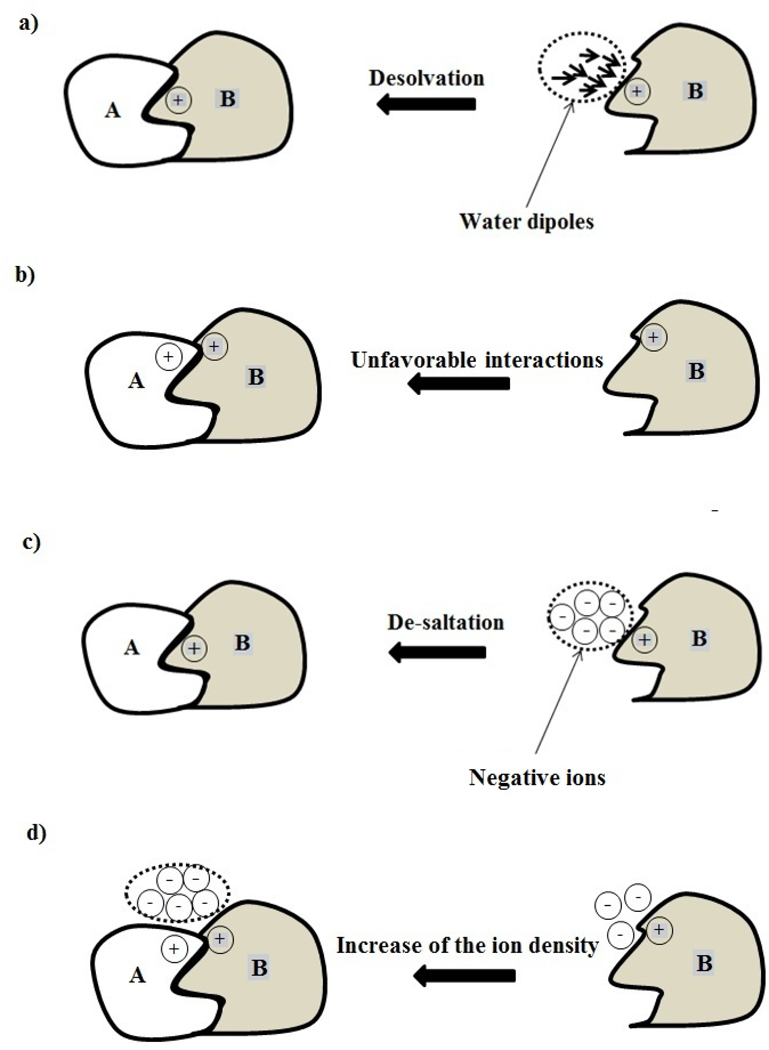
There are significant similarities and at the same time, significant differences of the origin of pH and salt dependence effects (Fig. 2). The overall proton uptake/release induced by protein-ligand association originates from individual pKa shifts of titratable groups induced by the binding. These shifts are typically associated with titratable groups located at the interface of the complex and thus experiencing either significant desolvation energy (Fig. 2a) or being involved in new interactions upon the complex formation (Fig. 2b). In contrast, the salt dependence of the binding originates from the difference of the charges-ions interaction prior to and after binding, i.e. mostly from the difference of the solvent exposure of the charges. This effect is similar to the desolvation effect and thus was termed the de-saltation effect [95] (Fig. 2c). However, the effect depends on charge-charge interactions as well, since new charge interactions can alter the ion distribution in the water phase and thus change the charges-ions interaction (Fig. 2d). Even more complicated cases can occur, when the binding induces ionization change, which in turn affects the salt dependence of the binding affinity.
Fig. 2.
Schematic representation of the major effects associated with the calculation of pH and salt dependence of the electrostatic component of the binding energy.
(a) desolvation effect, which originates from the removal of the charge-water interactions upon binding.
(b) establishment of new interactions upon complex formation.
(c) de-saltation effect, which originates from the removal of favorable charge-ion interactions upon the binding.
(d) change of the ion distribution in the water phase upon complex formation.
Salt dependence of protein-protein binding was modeled in the past and compared with experimental data to assess the accuracy of the computational methods. It was shown that Poisson-Boltzmann formalism is capable of capturing non specific salt effects with very good accuracy [95]. The limited cases examined indicated that almost always the increase of the salt concentration makes binding weaker, both experimentally and computationally. Sequential in silico studies on a much larger dataset comprised of 1482 complexes confirmed such a tendency for hereto-complexes, and to a certain extent for homo-complexes as well [96]. The observation that the increase of the salt concentration weakens the protein-protein association in the vast majority of the cases was attributed to the charge-charge optimization at the interface of the interacting partners [97, 98]. Thus, the increase of the salt concentration increases the screening of these favorable interactions and weakens the binding, despite of the net charge of monomers.
The pH-dependence of protein-protein binding is a result of the pKa shifts induced by the binding. Such pKa changes were investigated in silico for the binding of antibiotics to small ribosome units [99], for the enzyme active sites [27], for the kinase-ligand binding [100] and for the protein kinase systems [101]. The pKa changes were calculated to occur upon hirudin-thrombin binding [49] and the binding of small ligands to HIV protease [40, 102]. A recent study on 37 protein-protein complexes showed that in a vast majority of the cases the average pKa shifts for acidic residues induced by the complex formation were negative, indicating that complex formation stabilizes their ionizable states, whereas the histidines were predicted to destabilize the complex [103]. PROPKA method was used on 75 protein-protein complexes, and their corresponding free forms, to model changes in the protonation state of individual residues, and net changes in the protonation state of the complex relative to the unbound proteins [28]. It was concluded that protein-protein binding at neutral pH is often associated with changes in the protonation state of amino acid residues and with changes in the net protonation state of the proteins. Similar observations were made and outlined in a recent review article [104], including the proton uptake/release at pH different from neutral pH. It should be mentioned that proton uptake/release is a pH dependent effect and the pH-dependence profiles are different for each protein-protein complex, perhaps, reflecting the adaptation of each interaction to the environment where the binding is supposed to occur. The possibility of such an adaptation toward the subcellular environment is discussed in the next paragraph.
4. Adaptation of protein-protein interactions to subcellular characteristic pH
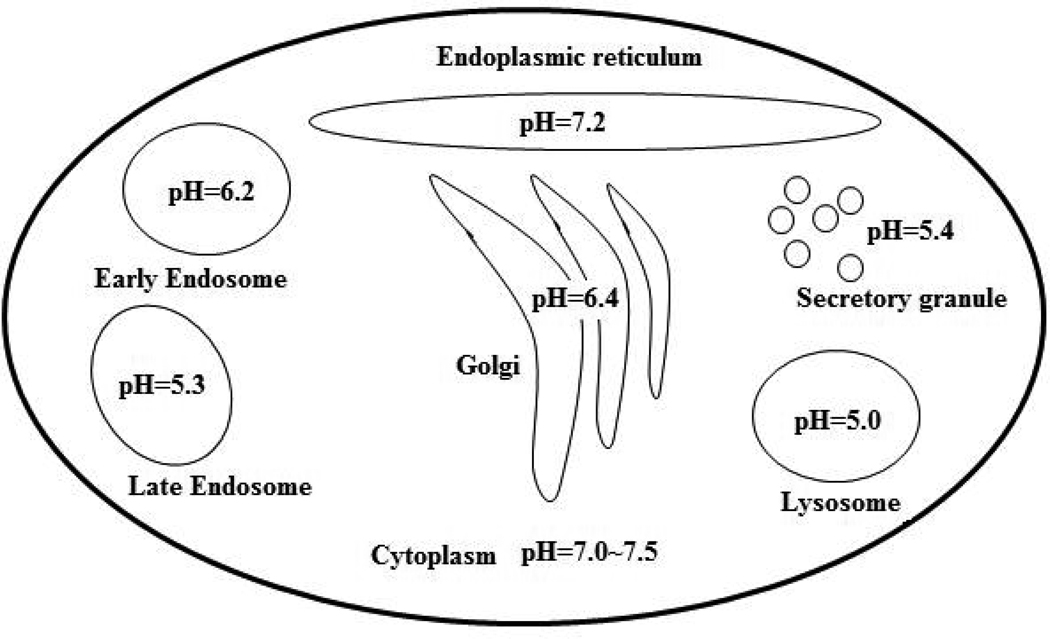
The typical pH-dependence profile of protein-ligand free energy of binding is bell shaped and has an absolute minimum which is termed the pH-optimum of binding. At pH-optimum proton uptake/release is zero and the binding affinity is maximal as indicated by the general formulation of proton binding [70] . To perform their function, protein complexes should be able to tolerate small fluctuations of pH of the environment, which will require that ΔQ be zero or close to zero at the characteristic pH of the subcellular location where the protein complex is supposed to be formed. The characteristic pH varies among cellular organelles as shown in Fig. 3 (a collection of characteristic subcellular pHs can be found in the works of Warwicker and coworkers [105, 106]) and thus the proteins and protein-protein complexes existing in there should have evolved to perform their function at these characteristic pHs (see excellent review [107]). Such a possibility was investigated on a set of 37 proteins, using rigid body approach and it was found that the pH-optimum of binding is correlated with the pH-optimum of stability of individual monomers [103]. Later, similar investigation was carried out, but accounting for the conformational changes induced on the binding and it was confirmed that the correlation does exist between pH-dependent properties of the complex and unbound monomers [108]. It was noticed that the pH-optimum, either of binding or of stability, may not necessarily have to be similar among the binding partners and the complex, but rather they should have similar pH region in which their properties are almost not affected by small fluctuations of the local pH (termed pH-flat). The existence of such a pH-flat region would indicate that the protein-protein complexes are insensitive to small changes of the local characteristic pH, however, there are examples of proteins and protein-protein complexes which function is to sense and respond to the changes in cellular pH [109–111]. Obviously, such protein-protein interactions must not to be independent of pH and should not have pH-flat region, instead their affinity should be strongly affected by the pH changes [109, 112, 113].
Fig. 3.
Schematic representation of a cell with several compartments with the corresponding characteristic pHs.
5. nsSNP modulating of the electrostatic component of the binding free energy
Non-synonymous single nucleoside polymorphism (nsSNP) and in general, any missense mutation results in a change of the amino acid composition of the target protein. Frequently, such changes involve charged or polar amino acids and thus affect the net charge or the dipole moment of the protein. Specifically, if the site of the mutation is located at the protein-protein interface, even a minor change of the charge distribution or the volume of the side chain can induce large effects on the electrostatic component of the binding free energy. Recently an analysis of 264 protein-protein complexes with nsSNPs located at the protein-protein interface was reported [114], with the goal to discriminate the effects caused by mutations found in the general population (non-OMIM) versus mutations known to cause diseases (OMIM). It was found that disease-causing mutations tend to electrostatically destabilize the corresponding protein-protein complexes, while non-OMIM mutations do not exhibit such a trend. An opposite effect was predicted for the HER2 receptor binding to herceptin, where the deleterious nsSNP introduced an additional 2 hydrogen bonds compared with the wild type and thus enhanced electrostatic interactions across the interface of the complex [115]. In contrast, series of charged amino acid missense mutations in the DDR2 gene were found to reduce the ability of the corresponding protein to bind collagen [116]. Since all mutations involve charged residues, the reduction of affinity should be due to change of the electrostatic component of binding free energy. Reduced binding was found to occur also in the case of introducing extra charge on the SHH gene, which in turn affects the corresponding protein affinity to GAS1 [117]. Reduced affinity was found as well in the case of a missense mutation in the BRCA1 gene, which results in an extra positive charge in the binding pocket [118]. Another report on missense mutations in the same gene, the BRCA1 gene, indicated that a charged residues replacement can either reduce or increase binding affinity to p53 [119]. Our recent study on spermidine sythase (SMS) dimerization showed that even a missense mutation introducing polar residue at the interface of the dimer can greatly reduce dimer affinity [120]. Further in silico analysis on the dimerization of the SMS suggested that the effect of the mutation depends not only on the change in charge, but can be amplified by the local electrostatic field created by other, even distant, charges (Zhang et al, PLoS Comp. Biol., submitted). This limited set of examples indicates that the effect of nsSNPs on the electrostatic component of the binding free energy depends on many factors, with the primary being the polarity of the charge, the local potential and the ability of the newly introduced side chain to rearrange and form new interactions. However, in terms of being disease-causing or harmless, any significant deviation of the wild type binding affinity (decrease or increase of the affinity) was found to cause disease.
6. The role of electrostatics in the cases of hetero- and homo-dimers
The major difference between hetero- and homo-complexes is that hetero-complexes are made of different proteins, which can carry opposite net charge, while homo-complexes being comprised of two identical units can not. Because of this, the role of electrostatics in steering the monomers toward each other is distinctively different for hetero- and homo-complexes. Since in most of the cases the monomers forming a hetero-complex carry opposite net charge, the electrostatics will cause an attraction, while it will be just the opposite for homo-complexes. However, this statement should be clarified since bringing two proteins in close proximity of space does not necessarily help the binding, since binding interfaces may not be positioned properly. Thus, at short distances, the role of electrostatics will depend on the charge distribution, rather on the global net charge of the monomers. Finally, the electrostatic component of the binding free energy will depend on the delicate balance between desolvation energy and pair wise interactions across the interface. A recent study on 260 hetero- and 2148 homo-complexes revealed that electrostatics is predicted to oppose binding in about 80% of the homo-complexes, and this percentage varies from 43% up to 85% for hetero-complexes, depending on the protocol and force field used to make the predictions [121]. The difference is not very significant. However, it should be mentioned that homo-complexes can be formed by two distinctive binding modes: (a) a binding mode that results in the formation of the interface made of identical amino acid interactions across the interface and (b) a binding mode resulting in non-identical interfaces, similar to the interfaces found in hetero-complexes. Much more detailed classification, comprised of (a) cyclic-oligomer, (b) twisted-dimer, (c) dimer-parallel, (d) dimer-perpendicular and (e) dimer-circular was introduced in Ref. [122]. The abovementioned study [121] did not distinguish between these two cases, and perhaps this is the main reason that the predicted effects for hetero- and homo-complexes are not significantly different. In addition, many proteins are know to form both hetero- and homo-complexes at the same time [123–126] and the role of electrostatics will depend on the type of complex formed and the corresponding interface. It was suggested that electrostatics is specifically optimized to avoid homo-complex interactions in the case of LIN-2/7 domains [127]. The role of electrostatics interactions was also demonstrated for homo- and hetero-tetrameric hemoglobin formation [128]. A recent study on 393 non-redundant homo-oligomer interfaces emphasizes the importance of electrostatic complementarity at the interfaces, rather than the global properties on the monomers [122]. This study also delivers electrostatic rules for discriminating the biological-interface from the crystallographic-interface. Apparently the role of electrostatics will depend not only on the type of the complex, hetero- or homo-, but also on the type of binding modes in the case of homo-complex formation (see Ref. [129] for the evolutionary preferences of homo-complexes to form symmetrical and asymmetrical interfaces). It can be expected that in the case of a parallel homo-complex, the electrostatics will always oppose the binding.
5. Conclusions
In this review we outlined the continuum methods for modeling the electrostatic component of the binding free energy and its role on protein interactions and binding. Using a continuum electrostatics framework instead of an explicit water model is typically justified by the need to calculate large number of cases to provide the statistical significance of the obtained results. Even nowadays computing binding free energies with explicit water model for several hundreds or thousand protein-protein complexes is computationally infeasible. Thus, continuum methods must be applied, which despite their macroscopic nature were shown to be quite accurate in describing the electrostatic component of the energy. At the same time, intrinsic limitations of continuum models will result in non accurate energies when explicit waters or ions play significant roles in protein-protein binding. Perhaps, the tradeoff is to use hybrid models [37, 38, 130], which treat the important waters and ions explicitly, while the rest of the water phase is described with the continuum approach.
This review demonstrated that salt- and pH-dependent phenomena, which are mostly electrostatic in origin, can greatly affect protein-protein interactions. Such effects can naturally be modeled with the continuum electrostatics approach as implemented in the PB equation in conjunction with a corresponding pKa algorithm, as with Multi Conformation Continuum Electrostatics (MCCE) [131–133]. As an alternative, the explicit water models, for example constant pH MD methods [134–136], require much more computational resources since the calculations must be repeated at each pH. In addition, the pKa calculations based on the continuum model are still believed to be more accurate than the corresponding algorithms using explicit water representation [137, 138]. However, if necessary to treat some waters or ions explicitly, hybrid approaches do exist, which treat the buried ions and water molecules explicitly within the calculations of the ionizable states [131–133].
Since the characteristic salt concentration and pH vary within subcellular compartments, the pH and salt dependence of protein-protein binding free energy can be associated with a subcellular localization of protein-protein complexes. Protein-protein complexes existing in a particular environment should be able to tolerate its characteristic pH and salt concentration. This review indicates that there is growing evidence that the monomers forming the complex should have similar pH properties and this can be used to transfer subcellular annotations from one partner to another.
The progress in genome sequencing offers vast amounts of data regarding missense mutations, both disease-causing and harmless mutations. It was shown that such mutations may affect the electrostatic component of the binding free energy and thus affect the cellular interaction network. The ability to model such effects will be crucial in understanding the molecular effects associated with disease-causing mutations and in the long run, assist in developing treatments for the corresponding diseases.
This review also outlines the similarities and differences of the electrostatic interactions in hetero- and homo-complexes. While the hetero-complexes are formed by two different molecules, the homo-complexes result from the binding of two identical (on a sequence level) molecules. Frequently, each molecule in the homo-dimer has its own reaction cite, but the biological function occurs only after the dimer is formed. Perhaps the analysis of the electrostatic interactions within the homo-complexes, as contrasted to hetero-complexes, may shed light and may provide some clue for such cases. It may be speculated that the formation of the homo-complex is necessary in order to provide favorable electrostatic environments for the corresponding biological reaction.
Acknowledgements
The work was supported by a grant from the Institute of General Medical Sciences, National Institutes of Health, award number 1R01GM093937-01.
References
- 1.Przytycka TM, Singh M, Slonim DK. Toward the dynamic interactome: it's about time. Brief Bioinform. 2010;11(1):15–29. doi: 10.1093/bib/bbp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger-Wolf TY, Przytycka TM, Singh M, Slonim DK. DYNAMICS OF BIOLOGICAL NETWORKS - Session Introduction. Pac Symp Biocomput. 2010;15:120–122. [Google Scholar]
- 3.Carbonell P, Nussinov R, del Sol A. Energetic determinants of protein binding specificity: insights into protein interaction networks. Proteomics. 2009;9(7):1744–1753. doi: 10.1002/pmic.200800425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elcock AH, Gabdoulline RR, Wade RC, McCammon JA. Computer simulation of protein-protein association kinetics: acetylcholinesterase-fasciculin. J Mol Biol. 1999;291(1):149–162. doi: 10.1006/jmbi.1999.2919. [DOI] [PubMed] [Google Scholar]
- 5.Sept D, Elcock AH, McCammon JA. Computer simulations of actin polymerization can explain the barbed-pointed end asymmetry. J Mol Biol. 1999;294(5):1181–1189. doi: 10.1006/jmbi.1999.3332. [DOI] [PubMed] [Google Scholar]
- 6.Wlodek ST, Shen T, McCammon JA. Electrostatic steering of substrate to acetylcholinesterase: analysis of field fluctuations. Biopolymers. 2000;53(3):265–271. doi: 10.1002/(SICI)1097-0282(200003)53:3<265::AID-BIP6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y, Holst MJ, McCammon JA. Finite element analysis of drug electrostatic diffusion: inhibition rate studies in N1 neuraminidase. Pac Symp Biocomput. 2009:281–292. doi: 10.1142/9789812836939_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persson BA, Lund M. Association and electrostatic steering of alpha-lactalbumin-lysozyme heterodimers. Phys Chem Chem Phys. 2009;11(39):8879–8885. doi: 10.1039/b909179c. [DOI] [PubMed] [Google Scholar]
- 9.Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. J Biol Chem. 2010;285(25):19637–19646. doi: 10.1074/jbc.M110.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persson BA, Jonsson B, Lund M. Enhanced protein steering: cooperative electrostatic and van der Waals forces in antigen-antibody complexes. J Phys Chem B. 2009;113(30):10459–10464. doi: 10.1021/jp904541g. [DOI] [PubMed] [Google Scholar]
- 11.Meltzer RH, Thompson E, Soman KV, Song XZ, Ebalunode JO, Wensel TG, Briggs JM, Pedersen SE. Electrostatic steering at acetylcholine binding sites. Biophys J. 2006;91(4):1302–1314. doi: 10.1529/biophysj.106.081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemsath L, Dvorsky R, Fiegen D, Carlier MF, Ahmadian MR. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol Cell. 2005;20(2):313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 13.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 14.Wong GC, Pollack L. Electrostatics of strongly charged biological polymers: ion-mediated interactions and self-organization in nucleic acids and proteins. Annu Rev Phys Chem. 2010;61:171–189. doi: 10.1146/annurev.physchem.58.032806.104436. [DOI] [PubMed] [Google Scholar]
- 15.McCammon JA. Darwinian biophysics: electrostatics and evolution in the kinetics of molecular binding. Proc Natl Acad Sci U S A. 2009;106(19):7683–7684. doi: 10.1073/pnas.0902767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukic P, Nielsen JE. Electrostatics in proteins and protein-ligand complexes. Future Medicinal Chemistry. 2010;2:647–666. doi: 10.4155/fmc.10.6. [DOI] [PubMed] [Google Scholar]
- 17.Mitomo D, Fukunishi Y, Higo J, Nakamura H. Calculation of protein-ligand binding free energy using smooth reaction path generation (SRPG) method: a comparison of the explicit water model, gb/sa model and docking score function. Genome Inform. 2009;23(1):85–97. [PubMed] [Google Scholar]
- 18.Yamane T, Okamura H, Ikeguchi M, Nishimura Y, Kidera A. Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment. Proteins. 2008;71(4):1970–1983. doi: 10.1002/prot.21874. [DOI] [PubMed] [Google Scholar]
- 19.Frembgen-Kesner T, Elcock AH. Computational sampling of a cryptic drug binding site in a protein receptor: explicit solvent molecular dynamics and inhibitor docking to p38 MAP kinase. J Mol Biol. 2006;359(1):202–214. doi: 10.1016/j.jmb.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omori S, Fuchigami S, Ikeguchi M, Kidera A. Linear response theory in dihedral angle space for protein structural change upon ligand binding. J Comput Chem. 2009;30(16):2602–2608. doi: 10.1002/jcc.21269. [DOI] [PubMed] [Google Scholar]
- 23.Gunasekaran K, Nussinov R. How different are structurally flexible and rigid binding sites? Sequence and structural features discriminating proteins that do and do not undergo conformational change upon ligand binding. J Mol Biol. 2007;365(1):257–273. doi: 10.1016/j.jmb.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 24.Ikeguchi M, Ueno J, Sato M, Kidera A. Protein structural change upon ligand binding: linear response theory. Phys Rev Lett. 2005;94(7) doi: 10.1103/PhysRevLett.94.078102. 078102. [DOI] [PubMed] [Google Scholar]
- 25.McCammon MG, Robinson CV. Structural change in response to ligand binding. Curr Opin Chem Biol. 2004;8(1):60–65. doi: 10.1016/j.cbpa.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Alexov E. Calculating proton uptake/release and binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association: application to plasmepsin, cathepsin D and endothiapepsin-pepstatin complexes. Proteins. 2004;56(3):572–584. doi: 10.1002/prot.20107. [DOI] [PubMed] [Google Scholar]
- 27.Lund H, Christensen BP, Nielsen AD, Westh P. Proton exchange coupled to the specific binding of alkylsulfonates to serum albumins. Biochim Biophys Acta. 2006;1764(7):1243–1251. doi: 10.1016/j.bbapap.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Mason AC, Jensen JH. Protein-protein binding is often associated with changes in protonation state. Proteins. 2008;71(1):81–91. doi: 10.1002/prot.21657. [DOI] [PubMed] [Google Scholar]
- 29.Shan Y, Seeliger MA, Eastwood MP, Frank F, Xu H, Jensen MO, Dror RO, Kuriyan J, Shaw DE. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106(1):139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilat Z, Antosiewicz JM. Multiple protonation equilibria in electrostatics of protein-protein binding. J Phys Chem B. 2008;112(47):15074–15085. doi: 10.1021/jp8029659. [DOI] [PubMed] [Google Scholar]
- 31.Tan C, Yang L, Luo R. How well does Poisson-Boltzmann implicit solvent agree with explicit solvent? A quantitative analysis. J Phys Chem B. 2006;110(37):18680–18687. doi: 10.1021/jp063479b. [DOI] [PubMed] [Google Scholar]
- 32.Wong SE, Bernacki K, Jacobson M. Competition between intramolecular hydrogen bonds and solvation in phosphorylated peptides: simulations with explicit and implicit solvent. J Phys Chem B. 2005;109(11):5249–5258. doi: 10.1021/jp046333q. [DOI] [PubMed] [Google Scholar]
- 33.Ma B, Nussinov R. Explicit and implicit water simulations of a beta-hairpin peptide. Proteins. 1999;37(1):73–87. doi: 10.1002/(sici)1097-0134(19991001)37:1<73::aid-prot8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Olson MA. Modeling loop reorganization free energies of acetylcholinesterase: a comparison of explicit and implicit solvent models. Proteins. 2004;57(4):645–650. doi: 10.1002/prot.20294. [DOI] [PubMed] [Google Scholar]
- 35.Wagoner J, Baker NA. Solvation forces on biomolecular structures: a comparison of explicit solvent and Poisson-Boltzmann models. J Comput Chem. 2004;25(13):1623–1629. doi: 10.1002/jcc.20089. [DOI] [PubMed] [Google Scholar]
- 36.Brancato G, Rega N, Barone V. A hybrid explicit/implicit solvation method for first-principle molecular dynamics simulations. J Chem Phys. 2008;128(14):144501. doi: 10.1063/1.2897759. [DOI] [PubMed] [Google Scholar]
- 37.Lee MS, Olson MA. Evaluation of Poisson solvation models using a hybrid explicit/implicit solvent method. J Phys Chem B. 2005;109(11):5223–5236. doi: 10.1021/jp046377z. [DOI] [PubMed] [Google Scholar]
- 38.Lee MS, Salsbury FR, Jr, Olson MA. An efficient hybrid explicit/implicit solvent method for biomolecular simulations. J Comput Chem. 2004;25(16):1967–1978. doi: 10.1002/jcc.20119. [DOI] [PubMed] [Google Scholar]
- 39.Prabhu NV, Panda M, Yang Q, Sharp KA. Explicit ion, implicit water solvation for molecular dynamics of nucleic acids and highly charged molecules. J Comput Chem. 2008;29(7):1113–1130. doi: 10.1002/jcc.20874. [DOI] [PubMed] [Google Scholar]
- 40.Grochowski P, Trylska J. Continuum molecular electrostatics, salt effects, and counterion binding--a review of the Poisson-Boltzmann theory and its modifications. Biopolymers. 2008;89(2):93–113. doi: 10.1002/bip.20877. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA. Poisson-Boltzmann methods for biomolecular electrostatics. Methods Enzymol. 2004;383:94–118. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 42.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Alexov E, Honig B. Comparative study of generalized born models: Born radii and peptide folding. J Phys Chem B. 2005;109(7):3008–3022. doi: 10.1021/jp046307s. [DOI] [PubMed] [Google Scholar]
- 44.Sigalov G, Fenley A, Onufriev A. Analytical electrostatics for biomolecules: beyond the generalized Born approximation. J Chem Phys. 2006;124(12):124902. doi: 10.1063/1.2177251. [DOI] [PubMed] [Google Scholar]
- 45.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: application to the implicit modeling of biological membranes. J Chem Phys. 2005;122(12):124706. doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 46.Fan H, Mark AE, Zhu J, Honig B. Comparative study of generalized Born models: protein dynamics. Proc Natl Acad Sci U S A. 2005;102(19):6760–6764. doi: 10.1073/pnas.0408857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrascal N, Green DF. Energetic decomposition with the generalized-born and Poisson-Boltzmann solvent models: lessons from association of G-protein components. J Phys Chem B. 2010;114(15):5096–5116. doi: 10.1021/jp910540z. [DOI] [PubMed] [Google Scholar]
- 48.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent ions. Journal of Physical Chemistry B. 2001;105(28):6507–6514. [Google Scholar]
- 49.Sharp KA, Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 50.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu B, Cheng X, Huang J, McCammon JA. AFMPB: An Adaptive Fast Multipole Poisson-Boltzmann Solver for Calculating Electrostatics in Biomolecular Systems. Comput Phys Commun. 2010;181(6):1150–1160. doi: 10.1016/j.cpc.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu B, Cheng X, Huang J, McCammon JA. An Adaptive Fast Multipole Boundary Element Method for Poisson-Boltzmann Electrostatics. J Chem Theory Comput. 2009;5(6):1692–1699. doi: 10.1021/ct900083k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altman MD, Bardhan JP, White JK, Tidor B. Accurate solution of multi-region continuum biomolecule electrostatic problems using the linearized Poisson-Boltzmann equation with curved boundary elements. J Comput Chem. 2009;30(1):132–153. doi: 10.1002/jcc.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu HY, Grinter SZ, Zou X. Multiscale generalized born modeling of ligand binding energies for virtual database screening. J Phys Chem B. 2009;113(35):11793–11799. doi: 10.1021/jp901212t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu HY, Zou X. Electrostatics of ligand binding: parametrization of the generalized Born model and comparison with the Poisson-Boltzmann approach. J Phys Chem B. 2006;110(18):9304–9313. doi: 10.1021/jp060334w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang X, Shafer RH, Kuntz ID. Calculation of ligand-nucleic acid binding free energies with the generalized-born model in DOCK. Biopolymers. 2004;73(2):192–204. doi: 10.1002/bip.10541. [DOI] [PubMed] [Google Scholar]
- 58.Macchiarulo A, Costantino G, Sbaglia R, Aiello S, Meniconi M, Pellicciari R. The role of electrostatic interaction in the molecular recognition of selective agonists to metabotropic glutamate receptors. Proteins. 2003;50(4):609–619. doi: 10.1002/prot.10301. [DOI] [PubMed] [Google Scholar]
- 59.Oron A, Wolfson H, Gunasekaran K, Nussinov R. Using DelPhi to compute electrostatic potentials and assess their contribution to interactions. Curr Protoc Bioinformatics. 2003;Chapter 8 doi: 10.1002/0471250953.bi0804s02. Unit 8 4. [DOI] [PubMed] [Google Scholar]
- 60.Selzer T, Schreiber G. Predicting the rate enhancement of protein complex formation from the electrostatic energy of interaction. J Mol Biol. 1999;287(2):409–419. doi: 10.1006/jmbi.1999.2615. [DOI] [PubMed] [Google Scholar]
- 61.Mashiach E, Schneidman-Duhovny D, Peri A, Shavit Y, Nussinov R, Wolfson HJ. An integrated suite of fast docking algorithms. Proteins. 2010;78(15):3197–3204. doi: 10.1002/prot.22790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein-protein docking. Proteins. 2008;73(2):271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiehe K, Peterson MW, Pierce B, Mintseris J, Weng Z. Protein-protein docking: overview and performance analysis. Methods Mol Biol. 2008;413:283–314. doi: 10.1007/978-1-59745-574-9_11. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Z, Felts AK, Friesner RA, Levy RM. Comparative performance of several flexible docking programs and scoring functions: enrichment studies for a diverse set of pharmaceutically relevant targets. J Chem Inf Model. 2007;47(4):1599–1608. doi: 10.1021/ci7000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kundrotas PJ, Lensink MF, Alexov E. Homology-based modeling of 3D structures of protein-protein complexes using alignments of modified sequence profiles. Int J Biol Macromol. 2008;43(2):198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Kundrotas P, Georgieva P, Shosheva A, Christova P, Alexov E. Assessing the quality of the homology-modeled 3D structures from electrostatic standpoint: test on bacterial nucleoside monophosphate kinase families. J Bioinform Comput Biol. 2007;5(3):693–715. doi: 10.1142/s0219720007002709. [DOI] [PubMed] [Google Scholar]
- 67.Kundrotas PJ, Alexov E. Predicting 3D structures of transient protein-protein complexes by homology. Biochim Biophys Acta. 2006;1764(9):1498–1511. doi: 10.1016/j.bbapap.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Korkin D, Davis FP, Alber F, Luong T, Shen MY, Lucic V, Kennedy MB, Sali A. Structural modeling of protein interactions by analogy: application to PSD-95. PLoS Comput Biol. 2006;2(11):e153. doi: 10.1371/journal.pcbi.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis FP, Braberg H, Shen MY, Pieper U, Sali A, Madhusudhan MS. Protein complex compositions predicted by structural similarity. Nucleic Acids Res. 2006;34(10):2943–2952. doi: 10.1093/nar/gkl353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen JH. Calculating pH and salt dependence of protein-protein binding. Curr Pharm Biotechnol. 2008;9(2):96–102. doi: 10.2174/138920108783955146. [DOI] [PubMed] [Google Scholar]
- 71.Blundell CD, Mahoney DJ, Cordell MR, Almond A, Kahmann JD, Perczel A, Taylor JD, Campbell ID, Day AJ. Determining the molecular basis for the pH-dependent interaction between the link module of human TSG-6 and hyaluronan. J Biol Chem. 2007;282(17):12976–12988. doi: 10.1074/jbc.M611713200. [DOI] [PubMed] [Google Scholar]
- 72.MacKnight ML, Gillard JM, Tollin G. Flavine-protein interactions in flavoenzymes. pH dependence of the binding of flavine mononucleotide and riboflavine to Azotobacter flavodoxin. Biochemistry. 1973;12(21):4200–4206. doi: 10.1021/bi00745a025. [DOI] [PubMed] [Google Scholar]
- 73.Gramberg T, Soilleux E, Fisch T, Lalor PF, Hofmann H, Wheeldon S, Cotterill A, Wegele A, Winkler T, Adams DH, et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology. 2008;373(1):189–201. doi: 10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bauman AT, Jaron S, Yukl ET, Burchfiel JR, Blackburn NJ. pH Dependence of peptidylglycine monooxygenase. Mechanistic implications of Cu-methionine binding dynamics. Biochemistry. 2006;45(37):11140–11150. doi: 10.1021/bi060905a. [DOI] [PubMed] [Google Scholar]
- 75.Syme CD, Nadal RC, Rigby SE, Viles JH. Copper binding to the amyloid-beta (Abeta) peptide associated with Alzheimer's disease: folding, coordination geometry, pH dependence, stoichiometry, and affinity of Abeta-(1–28): insights from a range of complementary spectroscopic techniques. J Biol Chem. 2004;279(18):18169–18177. doi: 10.1074/jbc.M313572200. [DOI] [PubMed] [Google Scholar]
- 76.Sprague ER, Martin WL, Bjorkman PJ. pH dependence and stoichiometry of binding to the Fc region of IgG by the herpes simplex virus Fc receptor gE-gI. J Biol Chem. 2004;279(14):14184–14193. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- 77.Bidwai AK, Ok EY, Erman JE. pH dependence of cyanide binding to the ferric heme domain of the direct oxygen sensor from Escherichia coli and the effect of alkaline denaturation. Biochemistry. 2008;47(39):10458–10470. doi: 10.1021/bi800872d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gramberg T, Soilleux E, Fisch T, Lalor PF, Hofmann H, Wheeldon S, Cotterill A, Wegele A, Winkler T, Adams DH, et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: Differential pH dependence, internalization and virion binding. Virology. 2008;373(1):189–201. doi: 10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakurai K, Oobatake M, Goto Y. Salt-dependent monomer-dimer equilibrium of bovine beta-lactoglobulin at pH 3. Protein Sci. 2001;10(11):2325–2335. doi: 10.1110/ps.17001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fallon J, Quiocho F. A Closed Compact Structure of Native Ca2+Calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Slaughter B, Unruh J, Allen M, Bieber R, Johnson C. Conformational substates of calmodulin revealed by single-pair fluorescence resonance energy transfer: influence of solution conditions and oxidative modification. Biochemistry. 2005;44:3694–3707. doi: 10.1021/bi048595o. [DOI] [PubMed] [Google Scholar]
- 82.Isvoran A, Craescu CT, Alexov E. Electrostatic control of the overall shape of calmodulin: numerical calculations. Eur Biophys J. 2007;36(3):225–237. doi: 10.1007/s00249-006-0123-1. [DOI] [PubMed] [Google Scholar]
- 83.Konvalinka J, Horejsi M, Andreansky M, Novek P, Pichova I, Blaha I, Fabry M, Sedlacek J, Foundling S, Strop P. An engineered retroviral proteinase from myeloblastosis associated virus acquires pH dependence and substrate specificity of the HIV-1 proteinase. Embo J. 1992;11(3):1141–1144. doi: 10.1002/j.1460-2075.1992.tb05154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tynan-Connolly BM, Nielsen JE. Redesigning protein pKa values. Protein Sci. 2007;16(2):239–249. doi: 10.1110/ps.062538707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Labrou NE, Rigden DJ, Clonis YD. Engineering the pH-dependence of kinetic parameters of maize glutathione S-transferase I by site-directed mutagenesis. Biomol Eng. 2004;21(2):61–66. doi: 10.1016/j.bioeng.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 86.Kusano M, Yasukawa K, Hashida Y, Inouye K. Engineering of the pH-dependence of thermolysin activity as examined by site-directed mutagenesis of Asn112 located at the active site of thermolysin. J Biochem. 2006;139(6):1017–1023. doi: 10.1093/jb/mvj112. [DOI] [PubMed] [Google Scholar]
- 87.Neves-Petersen M, Petersen E, Fojan P, Noronha M, Madsen R, Petersen S. Engineering the pH-optimum of triglyceride lipase: from predictions based on electrostatic computations to experimental results. J Biotechnol. 2001;87:225–254. doi: 10.1016/s0168-1656(01)00240-1. [DOI] [PubMed] [Google Scholar]
- 88.Wallis R, Moore GR, James R, Kleanthous C. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry. 1995;34(42):13743–13750. doi: 10.1021/bi00042a004. [DOI] [PubMed] [Google Scholar]
- 89.Wallis R, Leung KY, Pommer AJ, Videler H, Moore GR, James R, Kleanthous C. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry. 1995;34(42):13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- 90.Albeck S, Schreiber G. Biophysical characterization of the interaction of the beta-lactamase TEM-1 with its protein inhibitor BLIP. Biochemistry. 1999;38(1):11–21. doi: 10.1021/bi981772z. [DOI] [PubMed] [Google Scholar]
- 91.Ahl IM, Jonsson BH, Tibell LA. Thermodynamic characterization of the interaction between the C-terminal domain of extracellular superoxide dismutase and heparin by isothermal titration calorimetry. Biochemistry. 2009;48(41):9932–9940. doi: 10.1021/bi900981k. [DOI] [PubMed] [Google Scholar]
- 92.Glaser BT, Bergendahl V, Anthony LC, Olson B, Burgess RR. Studying the salt dependence of the binding of sigma70 and sigma32 to core RNA polymerase using luminescence resonance energy transfer. PLoS One. 2009;4(8):e6490. doi: 10.1371/journal.pone.0006490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao H, Naganathan S, Beckett D. Thermodynamic and structural investigation of bispecificity in protein-protein interactions. J Mol Biol. 2009;389(2):336–348. doi: 10.1016/j.jmb.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ozkirimli E, Yadav SS, Miller WT, Post CB. An electrostatic network and long-range regulation of Src kinases. Protein Sci. 2008;17(11):1871–1880. doi: 10.1110/ps.037457.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertonati C, Honig B, Alexov E. Poisson-Boltzmann calculations of nonspecific salt effects on protein-protein binding free energies. Biophys J. 2007;92(6):1891–1899. doi: 10.1529/biophysj.106.092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Talley K, Kundrotas P, Alexov E. Modeling salt dependence of protein-protein association: Linear vs non-linear Poisson-Boltzmann equation. Communications in Computational Physics. 2008;3(5):1071–1086. [Google Scholar]
- 97.Brock K, Talley K, Coley K, Kundrotas P, Alexov E. Optimization of electrostatic interactions in protein-protein complexes. Biophys J. 2007;93(10):3340–3352. doi: 10.1529/biophysj.107.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Das M, Basu G. Coulomb energies of protein-protein complexes with monopole-free charge distributions. J Mol Graph Model. 2009;27(7):846–851. doi: 10.1016/j.jmgm.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 99.Tandori J, Maroti P, Alexov E, Sebban P, Baciou L. Key role of proline L209 in connecting the distant quinone pockets in the reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 2002;99(10):6702–6706. doi: 10.1073/pnas.092327799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sims PA, Wong CF, McCammon JA. Charge optimization of the interface between protein kinases and their ligands. J Comput Chem. 2004;25(11):1416–1429. doi: 10.1002/jcc.20067. [DOI] [PubMed] [Google Scholar]
- 101.Sims PA, Wong CF, Vuga D, McCammon JA, Sefton BM. Relative contributions of desolvation, inter- and intramolecular interactions to binding affinity in protein kinase systems. J Comput Chem. 2005;26(7):668–681. doi: 10.1002/jcc.20207. [DOI] [PubMed] [Google Scholar]
- 102.Mardis K, Luo R, Gilson M. Interpreting Trends in the Binding of Cyclic Ureas to HIV-1 Protease. J Mol Biol. 2001;309:507–517. doi: 10.1006/jmbi.2001.4668. [DOI] [PubMed] [Google Scholar]
- 103.Kundrotas PJ, Alexov E. Electrostatic properties of protein-protein complexes. Biophys J. 2006;91(5):1724–1736. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mitra R, Shyam R, Mitra I, Miteva MA, Alexov E. Calculation of the protonation states of proteins and small molecules: Implications to ligand-receptor interactions. Current computer-aided drug design. 2008;4:169–179. [Google Scholar]
- 105.Chan P, Lovric J, Warwicker J. Subcellular pH and predicted pH-dependent features of proteins. Proteomics. 2006;6(12):3494–3501. doi: 10.1002/pmic.200500534. [DOI] [PubMed] [Google Scholar]
- 106.Chan P, Warwicker J. Evidence for the adaptation of protein pH-dependence to subcellular pH. BMC Biol. 2009;7:69. doi: 10.1186/1741-7007-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garcia-Moreno B. Adaptations of proteins to cellular and subcellular pH. J Biol. 2009;8(11):98. doi: 10.1186/jbiol199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitra R, Zhang Z, Alexov E. In silico modeling of pH-optimum of protein-protein binding. Proteins. 2010 doi: 10.1002/prot.22931. in press and available online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sachleben JR, McElroy CA, Gollnick P, Foster MP. Mechanism for pH-dependent gene regulation by amino-terminus-mediated homooligomerization of Bacillus subtilis anti-trp RNA-binding attenuation protein. Proc Natl Acad Sci U S A. 2010;107(35):15385–15390. doi: 10.1073/pnas.1004981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O'Callaghan KM, Ayllon V, O'Keeffe J, Wang Y, Cox OT, Loughran G, Forgac M, O'Connor R. Heme-binding protein HRG-1 is induced by insulin-like growth factor I and associates with the vacuolar H+ATPase to control endosomal pH and receptor trafficking. J Biol Chem. 2010;285(1):381–391. doi: 10.1074/jbc.M109.063248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kawai C, Pessoto FS, Rodrigues T, Mugnol KC, Tortora V, Castro L, Milicchio VA, Tersariol IL, Di Mascio P, Radi R, et al. pH-sensitive binding of cytochrome c to the inner mitochondrial membrane. Implications for the participation of the protein in cell respiration and apoptosis. Biochemistry. 2009;48(35):8335–8342. doi: 10.1021/bi9006463. [DOI] [PubMed] [Google Scholar]
- 112.Pei Z, Anderson H, Myrskog A, Duner G, Ingemarsson B, Aastrup T. Optimizing immobilization on two-dimensional carboxyl surface: pH dependence of antibody orientation and antigen binding capacity. Anal Biochem. 2010;398(2):161–168. doi: 10.1016/j.ab.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 113.Heng BC, Gribbon PM, Day AJ, Hardingham TE. Hyaluronan binding to link module of TSG-6 and to G1 domain of aggrecan is differently regulated by pH. J Biol Chem. 2008;283(47):32294–32301. doi: 10.1074/jbc.M804155200. [DOI] [PubMed] [Google Scholar]
- 114.Teng S, Madej T, Panchenko A, Alexov E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys J. 2009;96(6):2178–2188. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rajasekaran R, George Priya Doss C, Sudandiradoss C, Ramanathan K, Purohit R, Sethumadhavan R. Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. C R Biol. 2008;331(6):409–417. doi: 10.1016/j.crvi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 116.Ali BR, Xu H, Akawi NA, John A, Karuvantevida NS, Langer R, Al-Gazali L, Leitinger B. Trafficking defects and loss of ligand binding are the underlying causes of all reported DDR2 missense mutations found in SMED-SL patients. Hum Mol Genet. 2010;19(11):2239–2250. doi: 10.1093/hmg/ddq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martinelli DC, Fan CM. A sonic hedgehog missense mutation associated with holoprosencephaly causes defective binding to GAS1. J Biol Chem. 2009;284(29):19169–19172. doi: 10.1074/jbc.C109.011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tischkowitz M, Hamel N, Carvalho MA, Birrane G, Soni A, van Beers EH, Joosse SA, Wong N, Novak D, Quenneville LA, et al. Pathogenicity of the BRCA1 missense variant M1775K is determined by the disruption of the BRCT phosphopeptide-binding pocket: a multi-modal approach. Eur J Hum Genet. 2008;16(7):820–832. doi: 10.1038/ejhg.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quaresima B, Faniello MC, Baudi F, Crugliano T, Di Sanzo M, Cuda G, Costanzo F, Venuta S. Missense mutations of BRCA1 gene affect the binding with p53 both in vitro and in vivo. Oncol Rep. 2006;16(4):811–815. [PubMed] [Google Scholar]
- 120.Zhang Z, Teng S, Wang L, Schwartz CE, Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31(9):1043–1049. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Talley K, Ng C, Shoppell M, Kundrotas P, Alexov E. On the electrostatic component of protein-protein binding free energy. PMC Biophys. 2008;1(1):2. doi: 10.1186/1757-5036-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsuchiya Y, Kinoshita K, Nakamura H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Des Sel. 2006;19(9):421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- 123.Liu GH, Qu J, Carmack AE, Kim HB, Chen C, Ren H, Morris AJ, Finck BN, Harris TE. Lipin proteins form homo- and hetero-oligomers. Biochem J. 2010;432(1):65–76. doi: 10.1042/BJ20100584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wyatt CN, Peers C. Hetero or homo, hypoxia has them all. J Physiol. 2009;587(Pt 12):2717–2718. doi: 10.1113/jphysiol.2009.174078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jansma A, Handel TM, Hamel DJ. Chapter 2. Homo- and hetero-oligomerization of chemokines. Methods Enzymol. 2009;461:31–50. doi: 10.1016/S0076-6879(09)05402-0. [DOI] [PubMed] [Google Scholar]
- 126.Zhang F, Fu G, Wang C, Cao L, Yang HY, Wang GY, Chen YZ, He C. Detection of homo- or hetero-association of Doks by fluorescence resonance energy transfer in living cells. Mol Imaging Biol. 2009;11(3):188–194. doi: 10.1007/s11307-008-0189-5. [DOI] [PubMed] [Google Scholar]
- 127.Petrosky KY, Ou HD, Lohr F, Dotsch V, Lim WA. A general model for preferential hetero-oligomerization of LIN-2/7 domains: mechanism underlying directed assembly of supramolecular signaling complexes. J Biol Chem. 2005;280(46):38528–38536. doi: 10.1074/jbc.M506536200. [DOI] [PubMed] [Google Scholar]
- 128.Yamaguchi T, Pang J, Reddy KS, Surrey S, Adachi K. Role of beta112 Cys (G14) in homo- (beta4) and hetero- (alpha2 beta2) tetramer hemoglobin formation. J Biol Chem. 1998;273(23):14179–14185. doi: 10.1074/jbc.273.23.14179. [DOI] [PubMed] [Google Scholar]
- 129.Dayhoff JE, Shoemaker BA, Bryant SH, Panchenko AR. Evolution of protein binding modes in homooligomers. J Mol Biol. 2010;395(4):860–870. doi: 10.1016/j.jmb.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou R. Free energy landscape of protein folding in water: explicit vs. implicit solvent. Proteins. 2003;53(2):148–161. doi: 10.1002/prot.10483. [DOI] [PubMed] [Google Scholar]
- 131.Alexov E. Role of the protein side-chain fluctuations on the strength of pair-wise electrostatic interactions: comparing experimental with computed pK(a)s. Proteins. 2003;50(1):94–103. doi: 10.1002/prot.10265. [DOI] [PubMed] [Google Scholar]
- 132.Alexov EG, Gunner MR. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys J. 1997;72(5):2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83(4):1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mongan J, Case DA. Biomolecular simulations at constant pH. Curr Opin Struct Biol. 2005;15(2):157–163. doi: 10.1016/j.sbi.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 135.Khandogin J, Brooks CL., 3rd Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89(1):141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stern H. Molecular simulation with variable protonation states at constant pH. J Chem Phys. 2007;126(16):164112. doi: 10.1063/1.2731781. [DOI] [PubMed] [Google Scholar]
- 137.Kuhn B, Kollman PA, Stahl M. Prediction of pKa shifts in proteins using a combination of molecular mechanical and continuum solvent calculations. J Comput Chem. 2004;25(15):1865–1872. doi: 10.1002/jcc.20111. [DOI] [PubMed] [Google Scholar]
- 138.Simonson T, Carlsson J, Case DA. Proton binding to proteins: pK(a) calculations with explicit and implicit solvent models. J Am Chem Soc. 2004;126(13):4167–4180. doi: 10.1021/ja039788m. [DOI] [PubMed] [Google Scholar]