Abstract
Heme-Nitric oxide and/or OXygen binding (H-NOX) proteins are a family of diatomic gas binding hemoproteins that have attracted intense research interest. Here we employ X-ray absorption near-edge structure (XANES) spectroscopy to study the nitric oxide (NO) binding site of H-NOX. This is the first time this technique has been utilized to examine the NO/H-NOX signaling pathway. XANES spectra of wildtype and a point mutant (proline 115 to alanine, P115A) of the H-NOX domain from Thermoanaerobacter tengcongensis (Tt H-NOX) were obtained and analyzed for ferrous and ferric complexes of the protein. This work provides specific structural characterization of the solution state of several Tt H-NOX ferrous complexes (-unligated, -NO, and -CO) that were previously unavailable. Our iron K-edges indicate effective charge on the iron center in the various complexes and report on the electronic environment of heme iron. We analyzed the ligand field indicator ratio (LFIR), which is extracted from XANES spectra, for each complex, providing an understanding of ligand field strength, spin state of the central iron, movement of the iron atom upon ligation, and ligand binding properties. In particular, our LFIRs indicate that the heme iron is dramatically displaced towards the distal pocket during ligand binding. Based on these results, we propose that iron displacement towards the distal heme pocket is an essential step in signal initiation in H-NOX proteins. This provides a mechanistic link between ligand binding and the changes in heme and protein conformation that have been observed for H-NOX family members during signaling.
Keywords: Heme-nitric oxide/oxygen binding domain, NO/sGC signaling, X-ray absorption near-edge structure spectroscopy, Edge, Ligand field indicator ratio, Iron-heme displacement
1. Introduction
Soluble guanylate cyclase (sGC) is a heterodimeric (α1/β1 or α2/β1) heme sensor responsive to nitric oxide (NO), an important signaling molecule in eukaryotic organisms [1–4]. The heme-binding domain of sGC has been localized to the N-terminal ~200 amino acids of the β1 subunit [5, 6]. This heme domain shares significant homology (15–40% identity) with a recently discovered family of heme proteins in prokaryotes [7–9]. Based on spectroscopic and kinetic characterization of several of these new sGC-like heme proteins [10–13], this family has been named H-NOX, for Heme-Nitric oxide and/or OXygen binding domain. Although sGC and other H-NOX family members have been the subject of intense investigation [1–16], the molecular details of the signaling mechanism are currently unclear.
The structure of an O2-binding H-NOX domain from Thermoanaerobacter tengcongensis (Tt H-NOX) was solved to a resolution of 1.77 Å (Figure 1) [7]. One of the particularly striking features of the H-NOX fold revealed by this structure is the severely distorted structure of the protoporphyrin IX heme group. CHARMM energy minimizations predict proline 115, invariant among H-NOX proteins, to be primarily responsible for forcing the heme into its strained conformation [7]. Indeed, crystallographic characterization of the Tt H-NOX P115A mutant revealed significant heme relaxation in this mutant [15]. Spectroscopic and structural studies of Tt H-NOX, however, indicate that the heme is quite flexible and can likely assume more than one solution conformation in both wildtype and the P115A mutant (see supplementary materials for more information) [7, 15].
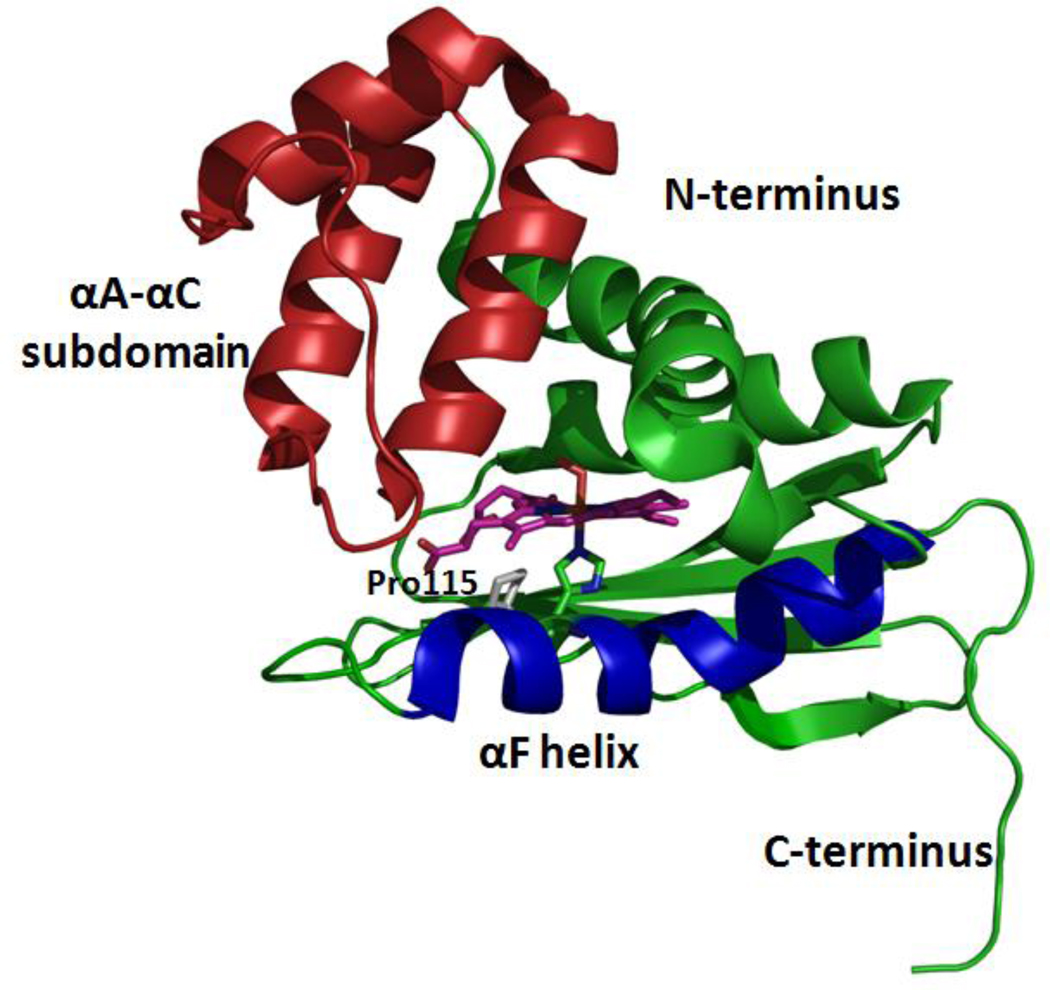
FIG. 1.
Structure of wildtype Tt H-NOX (figure was generated using PDB 1U55: Fe(II)-O2 complex, resolution 1.77 Å) [7]. The heme is shown in purple while proline 115 is shown in gray. The αA–αC subdomain and αF helix are color coded in red and blue, respectively. These two regions are involved in the overall protein conformational change during signal transduction in the H-NOX system.
The results obtained with P115A Tt H-NOX have led to a hypothesis that changes in heme structure are responsible for initiating signaling in the H-NOX family of proteins. However, previous studies have focused on the changes in heme structure that are caused by the P115A mutation, as opposed to changes to heme structure that take place upon ligand binding. Although it is commonly observed that ligand binding causes heme distortion and protein conformational changes in heme proteins [7, 17, 18], the molecular details of how ligand binding, the initial step in signal induction, leads to heme and protein structural changes in H-NOX has not been established. Understanding the molecular impacts of ligand binding is critical to developing a full understanding of signal transduction in this important family of heme sensors.
In this work, X-ray absorption spectroscopy (XAS) is employed, for the first time, to explore the role of ligand binding on heme conformation and electronic structure during signal initiation in the H-NOX family. Although crystal structures are critical for understanding protein structure and function, they do not always accurately represent the solution state structure. XAS reveals the local solution structure of the absorbing atom. Thus it is used to confirm and correct available crystal structures as well as to characterize species for which crystallography data is not yet available. XAS is a powerful method that used to generate unique structural and electronic details of the heme pocket. The XAS spectrum of a metalloprotein provides information about metal oxidation state, metal spin state, the number and type of ligands bound to the metal, and metal-ligand bond lengths; as such, XAS has been widely used to examine the solution-state structure of metal centers in numerous metalloproteins [19–24]. The position of the X-ray absorption iron K-edge, which represents the excitation energy of the core shell electron, is determined by the maximum of the first derivative spectrum. It is directly related to the oxidation state of the iron in the protein and can be used to probe the flow of electron density between the metal center and surrounding ligands. Higher oxidation states have higher X-ray edges, due to an increase in the core shell electron binding energy. For example, met-Mb [Fe(III)] has an X-ray edge that is ~4 eV higher than deoxy-Mb [Fe(II)] [24]. The iron K-edge 1s to 3d pre-edge features have also been shown to vary with oxidation state, spin state and centrosymmetry of the heme iron center [25–27].
Moreover, the X-ray absorption near-edge structure (XANES) region of the XAS spectrum is highly predictive of the local environment of the X-ray absorbing atom. The ligand field indicator ratio (LFIR) is an empirical number that is extracted from the XANES region at about 30–100 eV above the K-edge. LFIR analysis of XANES spectra was developed about 25 years ago using hemoproteins [20]. Here we reintroduce it as a powerful method to analyze changes in heme structure and electronics upon ligation. LFIR is correlated with the magnetic susceptibility, an indicator of the spin state, and with metal-heme displacement [20, 23, 24]. Metal-heme displacement is an important parameter in heme conformation, which has implications in determining the conformational state of the entire protein. LFIR analysis is especially useful for studying changes in metal position among closely related porphyrin species (e.g., various complexes of the same protein or wildtype and mutant proteins), as demonstrated in this work. In addition, the MXAN procedure has been used to model the XANES near-edge features of heme-iron and explore the structure the heme pocket of heme proteins [21, 28, 29].
Here, we present a XANES study on the local electronic and geometric properties of the heme-iron in Tt H-NOX WT and P115A mutant proteins in a variety of Fe(II) and Fe(III) complexes. The solution states of several key Tt H-NOX ferrous compounds (-unligated, -NO, and -CO), which have not been crystallographically studied, were characterized detailing their structural and electronic features. The X-ray absorption edge of each complex is determined, indicating the effective charge on the iron center. We focus on LFIR analysis to obtain electronic and conformational information such as ligand field strength, spin state of the central iron, and movement of the iron upon ligand binding. This information is extrapolated to gain insight into changes in heme distortion and protein conformational change upon heme ligation during a signaling event.
2. Materials and Methods
2.1. Plasmid preparation and protein expression
Cloning of the Tt H-NOX domain into a pET-20b (Novagen) vector has been described previously [10]. For ease of purification, a C-terminal 6×His tag was incorporated to the plasmid ahead of the stop codon. P115A mutagenesis of the 6×His tagged protein construct was carried out using the QuikChange® protocol from Stratagene. Cell culture preparation and expression procedures of H-NOX proteins were carried out as previously described [10], except that upon induction with Isopropyl β-D-1-thiogalactopyranoside (IPTG), overnight protein expression took place at 18 °C.
2.2. Protein purification
Cell lysis and thermal denaturation treatments were carried out as described [10]. The supernatant after thermal denaturation was filtered through a 0.45 µm membrane and the filtrate was applied to a 1 mL HisTrap HP column (GE Life Sciences) pre-equilibrated with buffer A [50 mM sodium phosphate, 300 mM NaCl, pH 7.5]. The column loaded with Tt H-NOX was washed using 30 mL buffer A and then three more washes were performed using 20 mL buffer A with 10 mM imidazole, 20 mL buffer A with 20 mM imidazole, and 10 mL buffer A with 50 mM imidazole. The protein was ultimately eluted using buffer A with 250 mM imidazole. The flow rate was controlled at 1 mL/min. The protein was then exchanged into buffer B [50 mM HEPES, 50 mM NaCl, 5% glycerol and 5 mM DTT, pH 7.5] using a PD-10 column (GE Healthcare), aliquoted, and stored at −80 °C. This procedure (thermal denaturation plus affinity chromatography) resulted in protein that was >95% pure as estimated by SDS-PAGE.
2.3. Electronic spectroscopy
All UV/visible spectra were recorded on a Cary 100 Bio spectrophotometer equipped with a constant temperature bath set to 20 °C. Preparation of ferrous complexes was carried out as previously published [10]. Ferric complexes were prepared as described below.
2.4. XAS sample preparation
Ferrous (-unligated, -O2, -CO, and -NO) complexes of H-NOX were prepared in the glove bag as previously described [10]. The Fe(III)-H2O complex was prepared by oxidizing the protein sample with 10–20 mM potassium ferricyanide followed by desalting with a PD-10 column. Addition of ~50 mM of KCN to the Fe(III)-H2O complex produced the Fe(III)-CN− complex. Protein samples were exchanged into buffer C [50 mM HEPES, 50 mM NaCl, 10% glycerol, pH 7.5] and concentrated to 1–2 mM using spin columns from Millipore [Microcon Ultracel YM-3, 3000 molecular weight cut off (MWCO) and 0.5 mL maximum volume]. The concentrated protein samples were transferred to copper sample holders with Mylar windows (10 mm wide × 5 mm high × 0.5 mm thick), frozen in liquid nitrogen and stored at −80 °C before XAS experiments.
2.5. X-ray absorption spectroscopy
XAS data collection was carried out at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL), on beamline X3B with a sagittally focused Si (111) crystal monochromator. Samples were cooled to at least 40 K before scanning in a closed-cycle liquid helium diplex and kept frozen during data collection. Kα fluorescence was detected with a Canberra 13-element germanium detector. An iron foil was used as a reference to determine the X-ray absorption edge for each spectrum and to account for any shifts in the monochromator. The step size at the edge region was set to 0.2 eV. 4–6 scans for each sample were taken and averaged for data analysis. In all cases, the first and last scans overlap very well, which indicates that X-ray damage was insignificant. Furthermore, as we collected data only in the XANES region, the amount of time the samples were exposed to the X-ray beam was relatively short. XAS data were aligned and merged using Athena. Figure S1 (supplementary materials) illustrates the averaged XANES spectra without further treatment of the data. The K-edge positions were determined from the first derivative of the average XANES spectra after smoothing. The derivative spectra were smoothed using Origin 7.0. In order to extract the ligand field indicator ratios, Origin 7.0 was used to smooth the averaged XANES spectra and OMNIC was applied subsequently to determine the height of the two peaks as described [24].
2.6. Cyanide association kinetics
All kinetic studies were performed in buffer D [50 mM HEPES, 50 mM NaCl, pH 7.5]. Cyanide (CN−) association to the ferric heme was measured using stopped-flow (for Tt H-NOX WT) or scanning kinetics (for Tt H-NOX P115A) at 20 °C. For Tt H-NOX WT, stopped-flow data were acquired on an Applied Photophysics SX 18MV stopped-flow spectrophotometer equipped with a constant temperature bath set to 20 °C. Difference absorption spectra of Tt H-NOX WT were collected at 424 nm with various cyanide concentrations (0.45–18.16 mM). We collected at least 3 scans for each cyanide concentration. The average difference spectrum was fitted to a single-exponential curve to give observed association rate constant kobs. kobs values were then plotted versus corresponding cyanide concentrations, and the slope of the line revealed the association rate constant (kon) for WT Tt H-NOX.
For Tt H-NOX P115A, scanning kinetics data for various cyanide concentrations (10 mM to 100 mM) were acquired on a Cary 100 Bio spectrophotometer with temperature control set to 20 °C. Each scan started at 480 nm and ended at 370 nm with a 600 nm/min scanning rate and a 1 nm step size. Difference spectra were calculated by subtracting the first scan (t = 0 min) from each subsequent scan. The maximal change of each difference spectrum as a function of time was fitted to a single-exponential curve to generate the kobs, which was subsequently used to calculate kon for Tt H-NOX P115A as was described for wildtype.
3. Results
3.1. UV/visible spectroscopy
UV/visible spectra of each sample were obtained before XAS sample preparation to confirm the ligand binding states (Figure 2). The Soret and α/β bands absorption maxima are listed in Table 1. Electronic spectra of the Tt H-NOX WT and P115A Fe(II) complexes (-unligated, -CO, -O2, and -NO) have been reported previously [10, 14]. Here we also present the spectra for Fe(III) complexes (-H2O and -CN−) of Tt H-NOX WT and the P115A mutant.
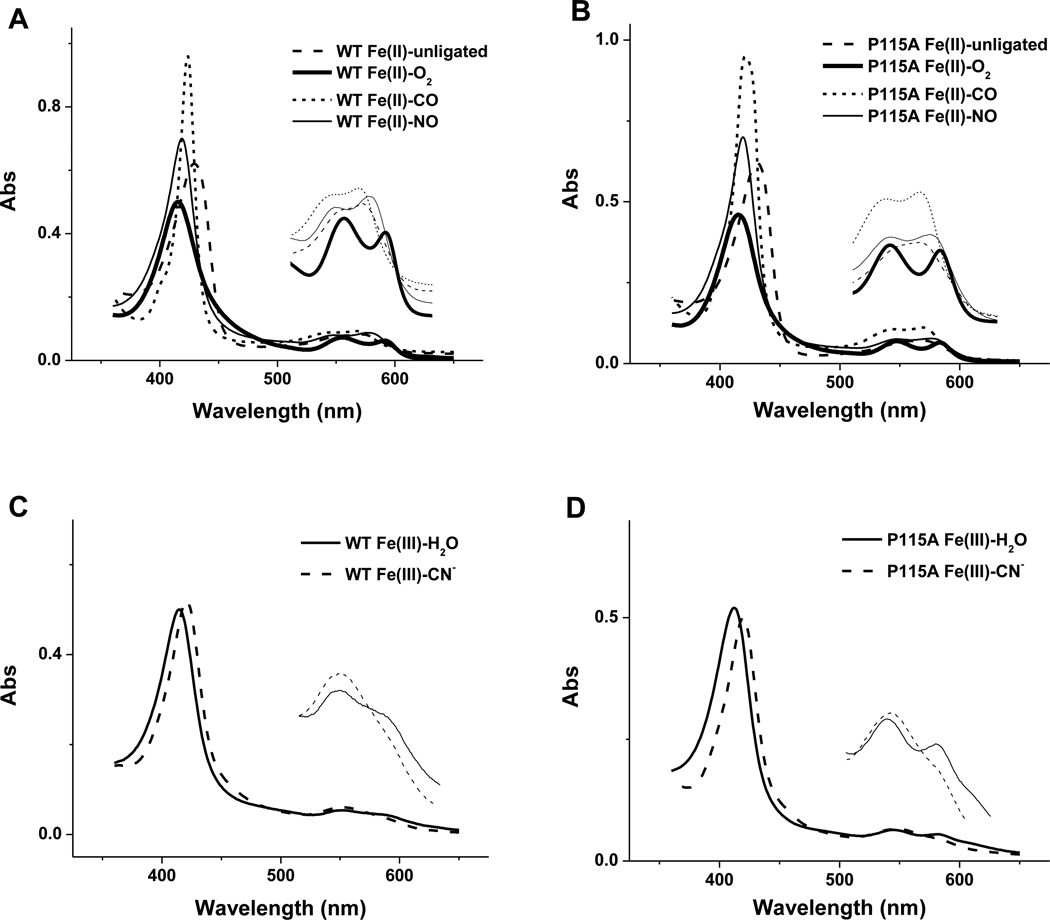
FIG. 2.
UV/visible spectra of WT and P115A Tt H-NOX complexes. Electronic spectra for (A) WT and (B) P115A H-NOX ferrous complexes: Fe(II)-unligated (dash), Fe(II)-O2 (thick solid), Fe(II)-CO (dot), Fe(II)-NO (solid). Spectra for (C) WT and (D) P115A Tt H-NOX ferric complexes: Fe(III)-H2O (solid), Fe(III)-CN− (dash). Insets show enlarged spectra of the α/β bands area.
Table 1.
Soret and α/β band absorption maxima of various Tt H-NOX complexes (unit: nm).
| Soret | β | α | |
|---|---|---|---|
| WT Fe(II)-unligated | 429 | 560 | |
| WT Fe(II)-CO | 424 | 545 | 568 |
| WT Fe(II)-NO | 419 | 547 | 577 |
| WT Fe(II)-O2 | 415 | 555 | 591 |
| WT Fe(III)-H2O | 415 | 552 | 587 |
| WT Fe(III)-CN− | 421 | 550 | - |
| P115A Fe(II)-unligated | 431 | 561 | |
| P115A Fe(II)-CO | 423 | 544 | 568 |
| P115A Fe(II)-NO | 419 | 547 | 577 |
| P115A Fe(II)-O2 | 415 | 547 | 583 |
| P115A Fe(III)-H2O | 405 | 545 | 581 |
| P115A Fe(III)-CN− | 420 | 550 | - |
As shown in Figure 2 and Table 1, our electronic spectra results are the same as those published for the ferrous complexes of Tt H-NOX WT and P115A [10, 14]. In the ferric oxidation state, water-bound [Fe(III)-H2O] Tt H-NOX WT exhibits a Soret band maximum at 415 nm and split α/β bands at 587 nm and 552 nm. Water-bound Tt H-NOX P115A exhibits a Soret band maximum at ~405 nm and split α/β bands at 581 nm and 545 nm, representing a blue shift of ~7 nm from those for WT, an indication of the change in heme structure and electronic environment induced by the mutation. Addition of CN− replaces water from the binding pocket and produces the Fe(III)-CN− complex, giving a Soret band maximum at ~421 nm for both WT and P115A. The α bands for Fe(III)-CN− complexes diminish to a shoulder on the β bands at ~550 nm. Overall, we observe minor changes in the electronic spectra of these ferric complexes upon the P115A mutation, which is consistent with what has been observed for the ferrous complexes [14].
3.2. K-edge absorption energy
We obtained XAS spectra for each ferrous (-unligated, -O2, -NO, -CO) and ferric (-H2O, -CN−) complex of both WT and P115A Tt H-NOX. In addition, as controls, we collected spectra for Fe(II) (-unligated, -O2, -CO) and Fe(III)-H2O complexes of Mb (Figure S1).
A summary of iron K-edge absorption energies is presented in Table 2. Determined by the maximum of the derivative XAS spectrum, the X-ray absorption iron K-edge represents the excitation energy of the 1s core shell electron. All edge energies obtained for Mb are within error of those published [24]. Among all Tt H-NOX WT complexes, the Fe(II)-unligated compound, as expected, has the lowest edge energy at 7124.1 eV, since it has the lowest effective charge on heme iron. Binding of CO and NO to the ferrous heme leads to an increase of 1.4 eV in each edge position. When O2 binds, the edge shifts from 7124.1 eV to 7126.5 eV, indicating more extensive electron flow from iron to O2 (or the histidine or heme ligands). Ferric state complexes generally have higher energy edge positions than ferrous species, due to greater charges on the heme iron. The Tt H-NOX WT Fe(III)-CN− complex has a slightly larger edge than WT Fe(III)-H2O. When comparing the edge values of WT and P115A, we can see that Tt H-NOX P115A complexes edge positions are generally similar with those of WT species, and they follow the same trend: Fe(II)-unligated < Fe(II)-CO ~ Fe(II)-NO < Fe(II)-O2 ~ Fe(III)-H2O < Fe(III)-CN−.
Table 2.
X-ray absorption edge, LFIR value, and Fe-Ct displacement for Mb and Tt H-NOX complexes.
| Edge (eV)a | LFIR | Fe-Ct (Å) | ||
|---|---|---|---|---|
| From Crystallography |
From LFIR |
|||
| Mb Fe(II)-unligated | 7123.1 (7123.0)b | |||
| Mb Fe(II)-CO | 7125.0 (7125.5)b | |||
| Mb Fe(II)-O2 | 7126.0 (7126.0)b | |||
| Mb Fe(III)-H2O | 7125.9 | |||
| Tt WT Fe(II)-unligated | 7124.1 | 0.98 | −0.08 | |
| Tt WT Fe(II)-CO | 7125.5 | 0.46 | 0.16 | |
| Tt WT Fe(II)-NO | 7125.5 | 0.94 | −0.06 | |
| Tt WT Fe(II)-O2 | 7126.5 | 0.88 | −0.03c | −0.03 |
| Tt WT Fe(III)-H2O | 7126.5 | 0.94 | −0.06c | −0.06 |
| Tt WT Fe(III)-CN− | 7126.7 | 0.82 | 0.00 | |
| Tt P115A Fe(II)-unligated | 7124.4 | 1.08 | −0.12 | |
| Tt P115A Fe(II)-CO | 7125.7 | 0.43 | 0.18 | |
| Tt P115A Fe(II)-NO | 7125.9 | 0.90 | −0.04 | |
| Tt P115A Fe(II)-O2 | 7126.5 | 0.85 | −0.02c | −0.02 |
| Tt P115A Fe(III)-H2O | 7126.5 | 0.94 | −0.06 | |
| Tt P115A Fe(III)-CN− | 7126.9 | 0.60 | 0.10 | |
X-ray absorption edges were determined assuming the edge for an iron foil is 7112.0eV; errors on edges are ±0.2eV.
Mb X-ray absorption edge values previously determined [24].
Average Fe-Ct distances for all available crystal structures; negative values indicate the iron atom sits in the proximal heme pocket and positive values indicate the iron atom sits in the distal heme pocket.
3.3. Ligand field indicator ratio
Multiple backscattering of atoms neighboring the X-ray absorbing atom (here, Fe) dominates the XANES region of a XAS spectrum. The two peaks in the low energy region used to calculate LFIR (Figure 3A) are primarily due to partially resolved higher shell (second and third shells) ligating atoms [20]. Changes in the LFIR arise from changes in the backscattering of these higher shell ligands. It has been established that LFIR can be used to probe the Fe motion in various heme-complexes and hemoproteins [20, 23, 24].
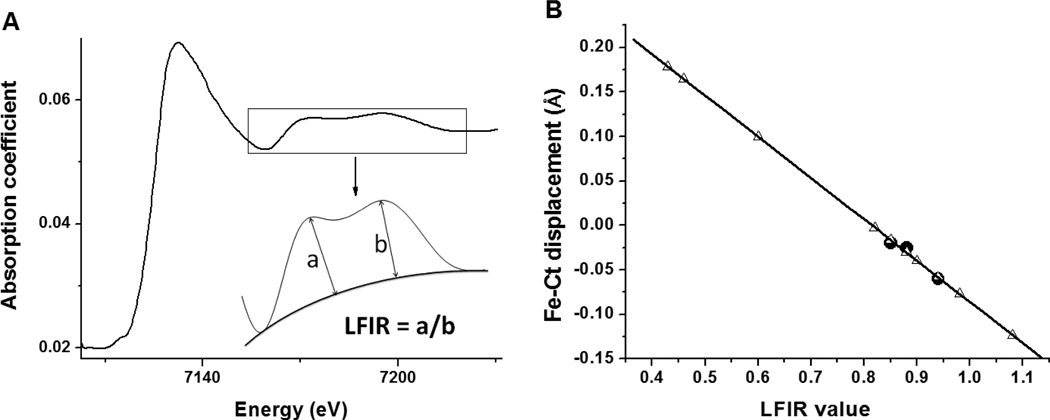
FIG. 3.
(A) Schematic illustration of the extraction of LFIR from a XANES spectrum. (B) Prediction of Fe-Ct displacements from LFIR values for various Tt H-NOX species. The data points in the black circles [WT Fe(II)-O2, WT Fe(III)-H2O and P115A Fe(II)-O2] are based on known crystallographic displacements and were used to determine the correlation line for Tt H-NOX. The LFIR values obtained from XANES spectra are indicated in the white triangles. The estimated error in LFIR ratio values is ±0.05. The equation for the least squares line is: Fe-Ct displacement = −0.46 × LFIR + 0.38 (R2 = 0.95).
To obtain the LFIR, a polynomial baseline is drawn starting from the minimum point preceding the first LFIR peak and ending at the tail of the second LFIR peak. Heights of the two LFIR peaks are measured to the baseline, and the LFIR ratio is the height of the first peak divided by the height of the second peak (Figure 3A). The ligand field indicator ratio yields a good empirical prediction of the spin state of the iron and the iron-heme displacement. For example, Chance, et al. has found that LFIR values linearly correlate to the movement of iron relative to the mean plane of the four porphyrin nitrogens (Fe-Ct displacement) for hemoproteins [23, 24].
We determined the LFIR and Fe-Ct displacement for each complex of WT and P115A Tt H-NOX. Crystal structures of three WT and P115A Tt H-NOX complexes have been solved [WT Fe(II)-O2 (PDB 1U55 and 1U4H), WT Fe(III)-H2O (PDB 1U56) and P115A Fe(II)-O2 (PDB 3EEE)] [7, 15], from which we extracted the crystallographically determined Fe-Ct distances in order to relate them to the measured LFIRs. Figure 3B illustrates the linear relationship between Fe-Ct displacement and LFIR for Tt H-NOX WT and P115A. Based on the least squares fit line determined by the three crystallographically characterized complexes, we predict the iron displacement from heme for all other ligand binding complexes, as shown in Table 2 and Figure 3B. The slope and intercept of the least squares fit line for Tt H-NOX is different than the slope reported for myoglobin, hemoglobin, and other model compounds [23]. The reasons for this are not clear, but apparently Tt H-NOX has different ligand field sensitivity to Fe-Ct displacement. Due to the limiting number of available WT and P115A Tt H-NOX crystal structures, the Fe-Ct displacements used for prediction do not cover the entire range of expected iron motion. However, because we focus on the relative magnitude of the Fe-Ct, and we are comparing various complexes of the same H-NOX domain, we can predict Fe-Ct displacements using measured LFIRs.
Most of the Tt H-NOX WT and P115A complexes have LFIR values between 0.85 and 1.08, which results in negative values for Fe-Ct displacement, indicating the iron is predicted to sit on the proximal side of heme. The CO and CN− complexes, however, have much smaller LFIRs (0.43–0.82) and display positive Fe-Ct displacement values [or no displacement for Tt WT Fe(III)-CN−], indicating that the iron atom moves toward or into the distal heme pocket. The LFIR value and iron displacement towards proximal side for WT and P115A Tt H-NOX follow the same trend: Fe(II)-CO < Fe(III)-CN− < Fe(II)-O2 < Fe(II)-NO ~ Fe(III)-H2O < Fe(II)-unligated.
3.4. Cyanide binding kinetics
Fe-Ct displacement has been correlated with ligand binding affinity in hemoproteins [23, 30]. Since Fe-Ct displacement is obtained from LFIR, ligand binding properties can therefore be predicted from LFIR data. Comparing the LFIR values of WT with P115A complexes (Table 2), we found that they are generally similar except for the Fe(III)-CN− compounds (0.82 for WT versus 0.60 for P115A). Hence, we hypothesized that there would be large differences in CN− binding properties to the ferric complexes of WT and P115A Tt H-NOX.
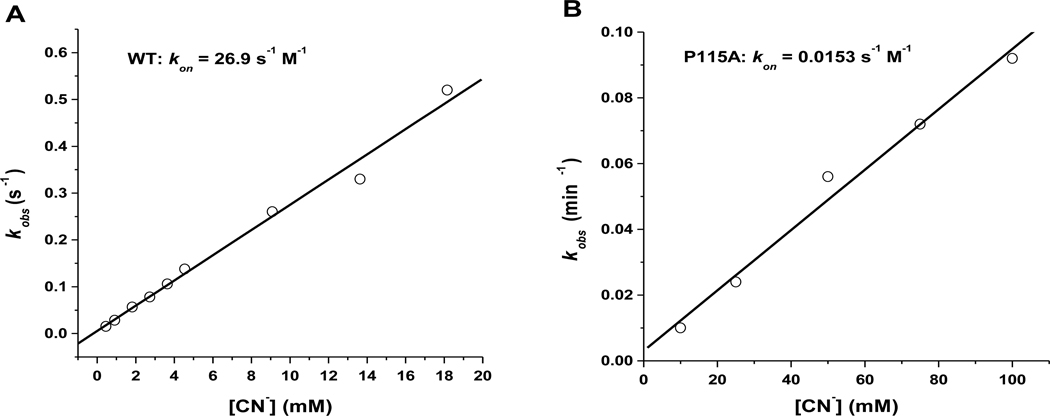
To test this prediction, we performed kinetic and equilibrium studies on Fe(III)-CN− binding properties for both WT and P115A Tt H-NOX. The observed association rate constant (kobs) values were obtained at various cyanide concentrations for both WT and P115A Tt H-NOX and were used to determine the association rate constant (kon) (Figure 4). We found that cyanide binds to WT Fe(III) (kon = 26.9 s−1 M−1) almost 2000 times faster than to P115A Fe(III) (kon = 0.0153 s−1 M−1) (Table 3).
FIG. 4.
Determination of association rate constants for cyanide binding to ferric WT (A) and P115A (B) Tt H-NOX. We found that cyanide binds to ferric Tt H-NOX WT much faster than to P115A.
Table 3.
Rate constants and equilibrium constants of cyanide binding to WT and P115A Tt H-NOX.
| Species | kon (s−1 M−1) | koff (s−1) | KD (µM) |
|---|---|---|---|
| Tt WT | 26.9 ± 1.1 | 2.19 ± 0.02×10−6 | 0.0813 ± 0.0203 |
| Tt P115A | 0.0153± 0.0011 | 4.44 ± 0.12×10−9 | 0.290 ± 0.111 |
We have previously measured the equilibrium binding constant of CN− for both ferric proteins; values of KD for WT and P115A Fe(III)-CN− were determined to be 81.3 nM and 290 nM, respectively [31]. Based on the measured KD and kon values, we calculated the dissociation rate constants koff as 2.19×10−6 s−1 and 4.44×10−9 s−1 for WT and P115A Tt H-NOX, respectively. All the rate and equilibrium constants for cyanide binding to ferric Tt H-NOX are summarized in Table 3.
4. Discussion
The H-NOX family of proteins use a histidine-ligated protoporphyrin IX to bind low concentrations of NO and other diatomic gas molecules. Once the ligand is bound to H-NOX, signal transduction commences. In eukaryotes, this signal transduction results in activation of the enzymatic domain of sGC [1, 2], while in prokaryotes, ligand binding is thought to regulate the activity of histidine kinases and di-guanylate cyclases [9, 32, 33]. In each case, however, the binding of a diatomic molecule to the heme center must be communicated to a different polypeptide domain for signal transduction to occur. The mechanism for this information transfer has not been determined. In this report, we have examined the local structural changes that take place at the heme iron during the first stages of signal transduction in order to shed light on the signaling mechanism of H-NOX proteins.
4.1. Insight into the H-NOX heme pocket and signaling properties based on K-edge absorption energies
Looking at the K-edge data broadly, we observe that the Fe(II) species generally have lower K-edge values than the Fe(III) species of Tt H-NOX. The only exception to this is the Fe(II)-O2 complex, whose edge is about the same as the Fe(III)-H2O complex. However, because of the strong electronegativity of oxygen, it is likely that the Fe(II)-O2 complex has significant contribution from a Fe(III)-O2 − resonance form. Therefore, the central iron in the Tt H-NOX oxy-complex may actually be electronically very similar with ferric complexes and thus this result is not unexpected. Furthermore, we found that the edge positions for WT and P115A complexes are generally similar, and that they follow the same trend. This confirms previous resonance Raman studies that indicate the P115A mutation results in a relatively minor perturbation to heme pocket, considering the dynamic nature of the heme structure in H-NOX (see supplementary materials) [14].
Looking at the data more closely, however, we can understand details of the electronic structure of H-NOX upon ligand binding. In hemoproteins, the iron edge position reports on the iron-ligand electron density distribution. As discussed, we found the 5-coordinate Fe(II)-unligated complexes have the lowest absorption edges of all the H-NOX complexes. Qualitatively, upon binding of the sixth ligand in the distal pocket of Tt H-NOX, there is σ electron donation from the ligand to central iron that should act to reduce the effective charge of iron. However, as the sixth ligand pulls valence electrons away from the iron cation due to stronger electronegativity, the electrons around the central metal (in lower energy orbitals) are pulled closer to the nucleus to increase shielding of the existing nuclear charge, and hence increase the core shell electron excitation energy [27]. Apparently, the latter effect overwhelms the former as evidenced by the increasing K-edge values of CO-bound, NO-bound and especially the O2-bound complexes. The K-edge value for the NO-bound complex is slightly larger than that for CO-complex, however it is not as much larger as would be expected based on the electronegativity difference between nitrogen and carbon. This is because of π-back bonding, in which iron donates its 3d electrons back to the π* anti-bonding orbitals of CO. Also as a result of the π-back bond donating electrons from iron to CN− in the ferric states, the cyanide-bound complexes have slightly larger K-edges than water-bound complexes for both Tt H-NOX WT and P115A.
Because the edge position is a good indication of iron-ligand electron density distribution, it is related to electronic properties of the heme, such as iron reduction potential [34–37]. The heme iron in P115A has a lower reduction potential (−3.8 mV versus SHE) than that of WT Tt H-NOX (167.0 mV versus SHE) [15]. Indeed, we see this difference reflected in the K-edges of the Fe(II)-unligated complexes of WT (7124.1 eV) and P115A (7124.4 eV). This difference (0.3 eV) is close to the limit of energy resolution in our data collection. However, we have repeated these scans several times with different samples and we always see a 0.3 eV difference, thus we believe this is reflective of a real difference in edge energy. The Fe(III)-H2O K-edges are comparable, both at 7126.5 eV. Furthermore, iron porphyrins with more severe saddling are known to have more positive reduction potentials, which serves to link heme conformation with redox properties [38]. Therefore we attribute the difference in ferrous K-edge energies to electronic structure changes that arise from a combination of changes in heme structure as well as overall changes in electrostatic interactions in the heme pocket, introduced upon mutation, that alter the dielectric constant and consequently lead to a different iron reduction potential [39, 40].
The significance of these results lies in the fact that we are able to address the re-distribution of electron density upon the binding of various ligands to H-NOX proteins, which is important in small molecule sensing processes. Our data predict that the heme structure and environment are more similar in the ferric state than the ferrous state. As only the ferrous oxidation state of H-NOX is expected to be physiologically relevant, this may indicate that heme structure is especially important for priming the H-NOX unligated protein for ligand (NO, CO or O2) binding.
4.2. Insight into the H-NOX heme pocket based on ligand field indicator ratios
Crystallographically determined displacements of hemoglobin as well as many artificial porphyrin macrocycles, have established that the LFIR correlates with spin state and iron-heme displacement [20]. Miller et al. also found that there is a linear relationship between Fe-Ct displacement and LFIR in myoglobin [24]; a reduction in Fe-Ct displacement towards the distal side is always accompanied by an increase in LFIR. Here we have predicted the Fe-Ct for each complex of the Tt H-NOX domain using our LFIR data. Notably, the trend in Fe-Ct displacements implied by the LFIR data agrees with available crystallography data (Table 2 and Figure 3B).
As indicated before, we see a trend in the movement of iron towards the proximal pocket for both WT and P115A Tt H-NOX as a function of ligand binding: Fe(II)-CO < Fe(III)-CN− < Fe(II)-O2 < Fe(II)-NO ~ Fe(III)-H2O < Fe(II)-unligated. This trend is easily explained with the spectrochemical series and indicates ligand field strength. These data suggest that the strongest ligands studied here, CO and CN−, which have significant π-back bonding interaction with the heme iron, “pull” the iron back to the distal side (where the sixth ligand locates), resulting in smaller iron movements to the proximal heme pocket in these complexes. For the same reason, in a strong ligand field, Fe uses 3d orbitals instead of 4d orbitals for hybridization, which also helps to reduce the Fe-ligand bond length and pull the iron away from the proximal side. Indeed, Fe-Ct displacements determined from recent crystal structures of Ns H-NOX support this interpretation. The Ns H-NOX Fe-Ct displacement changes from −0.31 Å (proximal side) for Fe(II)-unligated to +0.02 Å (distal side) for Fe(II)-CO complex [11], indicating a substantial movement of iron towards the distal pocket after binding of a strong ligand. Conversely, since there is no or only a weak ligand for Fe(II)-unligated (5-coordinate) and Fe(III)-H2O, these complexes of WT and P115A Tt H-NOX have the largest LFIR values and the largest displacement towards the proximal heme pocket (towards the proximal histidine ligand).
LFIR ratios also correlate with the magnetic moment of the heme. Chance et al. have obtained a good linear fit with a positive slope when plotting the LFIR values of various hemoproteins to their magnetic moments; lower LFIR values yield smaller magnetic moments [20]. The magnetic moments of Tt H-NOX complexes have not been independently measured. However, based on ligand strength and our LFIR values, we can predict their relative magnitude. For the high spin Fe(II)-unligated and Fe(III)-H2O complexes, the 3d electrons are trying to occupy different d orbitals, since in this case the electron pairing energy is larger than the crystal field splitting energy, which results in more unpaired electrons and larger magnetic moments. However, the strong ligands CO and CN− impose strong ligand fields to the central iron and lead to low-spin iron with fewer unpaired electrons and a smaller magnetic moment. Therefore, we expect the magnetic moment for ferrous complexes in the Tt H-NOX system follow the trend: Fe(II)-CO < Fe(II)-O2 < Fe(II)-NO < Fe(II)-unligated, and ferric states to follow this trend: Fe(III)-CN− < Fe(III)-H2O.
The spin state of iron is key to the local geometry of its complexes. Low spin iron porphyrin complexes tends to have less iron displacement towards the proximal histidine [11, 30, 41]. This is consistent with our observations that CO and CN− complexes have the least iron displacement toward the proximal heme pocket. Therefore, the two seemingly separate parameters - the magnetic moment and Fe-Ct displacement - can both be predicted by our LFIR data and explained by the strength of the binding ligand.
4.3. Insight into H-NOX ligand binding properties based on ligand field indicator ratios
Fe-Ct displacement is also related to ligand binding affinity in hemoproteins [23]. We found large differences in the LFIR-predicted Fe-Ct values for the Fe(III)-CN− complexes of WT and P115A H-NOX. Hence we predicted this was indicative of differences in the CN− binding properties of the two proteins. Indeed, large differences in the cyanide binding properties of ferric WT and P115A Tt H-NOX were observed (Figure 4 and Table 3). This can be attributed to increased steric crowding and a larger ligand binding activation energy barrier in P115A, which result from a larger Fe-Ct displacement towards the distal pocket in this mutant protein.
As shown in Table 2, during CN− binding to the ferric heme, the central iron in P115A Tt H-NOX experiences a much larger shift (0.16 Å) than that for WT (0.06 Å). In this case, the iron in the WT Tt H-NOX Fe(III)-CN− complex is located almost exactly at the center of the porphyrin nitrogens, while the iron in the P115A Tt H-NOX Fe(III)-CN− complex moves dramatically into the distal pocket where cyanide locates; this results in a more crowded distal heme pocket, and hence more steric crowding in P115A. Consequently, Fe-Ct displacement into the distal pocket leads to a larger energy barrier for both CN− association with and dissociation from P115A Fe(III). This is evidenced by kon and koff values for P115A that are much smaller than those measured for WT. Also due to the larger steric strain, the Fe(III)-CN− complex for P115A is not as stable as that for WT Tt H-NOX, as indicated by the equilibrium dissociation constants [31].
Furthermore, differences in the LFIR-predicted Fe-Ct displacements also correlate with the ligand binding properties for NO, CO, and O2 in reaction with ferrous WT and P115A Tt H-NOX (see supplementary materials for further discussion). In addition to the crystallographically measured displacements discussed above, the correlation we observe between ligand binding affinity and LFIR-predicted Fe-Cts supports our assignment of Fe-Cts in this system.
4.4. Insight into the H-NOX involved signaling pathway based on LFIR predicted iron-heme displacement
The current view on signal transduction in H-NOX proteins is that upon ligand binding, the proximal histidine protein residue dissociates, leading to protein conformational changes [2, 42, 43]. New data has also suggested a role for heme distortion in triggering protein conformational changes [11, 17]. Structural data indicate that upon ligation, the heme flattens and the αF and N-terminal αA–αC helices shift [11].
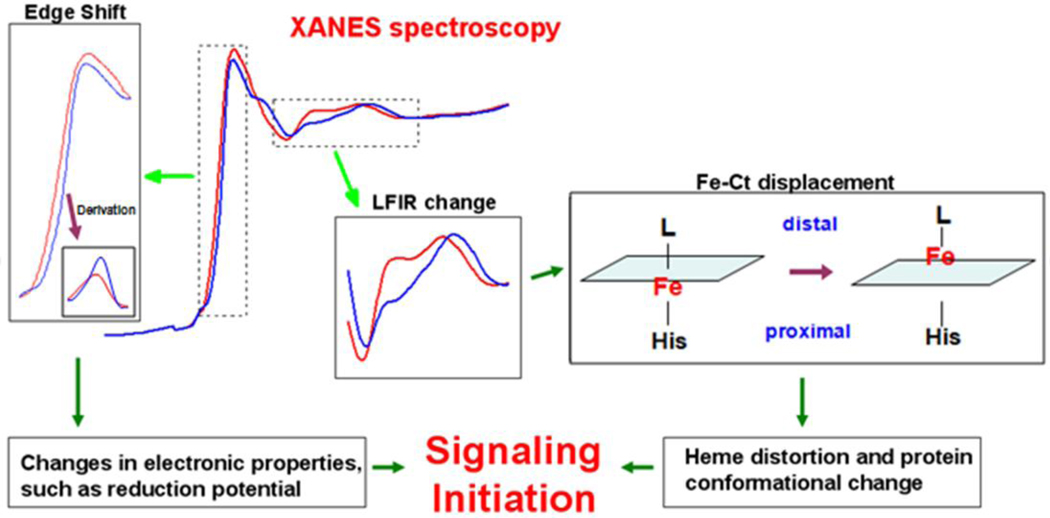
The data presented here provide a description of the changes in the electronic structure and position of the heme iron that occur upon ligand binding. We suggest movement of the heme iron is the initial step in signaling that gives rise to subsequent conformational changes in the histidine ligand, heme conformation, and protein conformation. Our XAS data indicate that diatomic gas binding to the H-NOX heme alters the position of the central iron, moving it towards or into the distal heme pocket. Because the heme is in contact with a large number of atoms in the protein matrix of H-NOX, porphyrin iron displacement alone could trigger changes in the tertiary structure needed for signal transduction. Iron movement into the H-NOX distal pocket can cause protein tertiary structural changes by one or both of two mechanisms: through lengthening or breaking the Fe-histidine (proximal) bond and/or through distal pocket rearrangements brought about by changes in heme structure. Iron moving toward the distal side of the heme pocket would lengthen and/or break the bond with the proximal histidine ligand. This would then lead to a shift in the position of histidine residue and relax the αF helix where the histidine resides, causing the αF helix to move further from the heme pocket (Figure 1). In addition, ligand binding in the distal pocket, accompanied by iron movement into the distal pocket, would result in a crowded distal pocket with substantial steric strain, as evidenced by our cyanide binding data. Hence, a rearrangement of this pocket, involving the N-terminal αA–αC helices, would be required to release the strain.
These alterations in tertiary structure would provide for direct communication between ligand binding and downstream signaling events (Figure 5). Our hypothesis is supported by recent subpicosecond time-resolved resonance Raman and femtosecond transient absorption spectroscopy results which emphasize the importance of heme iron motion for heme and protein conformational changes in hemoproteins [44].
FIG. 5.
Insight into the molecular basis of signal initiation of H-NOX domains based on XANES spectroscopy. The synergism between local structural rearrangement and electronic environment change leads to the signaling initiation step of the NO/sGC signaling pathway.
Our data indicate that CO binding, as well as NO binding, causes the iron to move substantially into the distal pocket, thus this mechanism does not discriminate against CO as an activating molecule for H-NOX proteins. Indeed, in combination with an allosteric activator such as YC-1, CO fully activates sGC [45]. Recent resonance Raman experiments have shown that during activation of CO-bound sGC by YC-1, the iron moves so far into the distal pocket that there is evidence that the Fe-His bond breaks, creating a population of 5-coordinated sGC-CO complex [18]. It is proposed that this population of 5-coordinate sGC, in which Fe has substantially moved towards the distal pocket, is the active state in both NO and CO/YC-1 bound sGC. These data further emphasize the importance of iron-heme displacement in the H-NOX signaling cascade.
Our K-edge and LFIR data indicate that the greatest differences in the electronic and geometric properties of the heme pocket between WT and P115A occur in the Fe(II)-unligated and Fe(III)-CN− complexes. As H-NOX is expected to remain in the ferrous oxidation states throughout its physiological function, this may indicate that H-NOX heme structure has been optimized to produce the largest changes in the H-NOX heme environment upon ligation by O2, CO, or NO, the likely functional ligands in this family.
5. Conclusion
Understanding the molecular details of how sGC and other H-NOX proteins sense small signaling molecules is still under investigation. We employed X-ray absorption spectroscopy to study the local electronic and geometric properties of the iron center in the heme pocket of WT and P115A Tt H-NOX, a prokaryotic analog of sGC. Our results shed light on electronic and conformational changes that occur in the heme pocket of H-NOX proteins after ligation and during initiation of signaling, which confirms that XAS is a promising approach in studying the NO/sGC signaling pathway.
The K-edge absorption energies measured here yield the effective charges on the iron center as well as the distribution of electron density for various complexes. Based on our XAS data, we evaluated the spin states of iron and predicted the Fe-Ct displacement for Tt H-NOX WT and P115A mutant complexes. Our XANES predictions of Fe-Ct were confirmed by known crystallographic data and ligand binding properties. We propose that this iron displacement upon ligand binding is the driving force leading to protein conformation changes in the αF helix and the N-terminal αA–αC helices, which subsequently contribute to signal initiation in the H-NOX family.
Supplementary Material
Acknowledgments
We sincerely thank all X3B beamline staff for technical assistance, and Dr. Lisa Miller for help with data collection, analysis, and critical reading of the manuscript. We also thank Stephanie Georgiou for construction of the Tt H-NOX 6×His plasmid and the Boon group for discussion of this work. This work was supported by New York State Foundation for Science, Technology and Information grant C060030 to E.M.B., NSF grant CHE-0910771 to E.M.B., and Stony Brook University. The X3B beamline at the CWRU Center for Synchrotron Biosciences is supported by the National Institute for Biomedical Imaging and Bioengineering under P30-EB-09998. Use of the NSLS at BNL, is supported by the U.S. DOE, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
Abbreviations
- DTT
dithiothreitol
- Fe-Ct displacement
the movement of iron relative to the mean plane of the four porphyrin nitrogens
- H-NOX
heme-nitric oxide and/or oxygen binding domain
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- LFIR
ligand field indicator ratio
- Mb
myoglobin
- Ns
Nostoc sp
- sGC
soluble guanylate cyclase
- SHE
standard hydrogen electrode
- Tt
Thermoanaerobacter tengcongensis
- WT
wildtype
- XANES
X-ray absorption near-edge structure
- XAS
X-ray absorption spectroscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary Materials
Supplementary materials associated with this article can be found, in the online version, at
References
- 1.Ignarro LJ. Biochem. Pharmacol. 1991;41:485–490. doi: 10.1016/0006-2952(91)90618-f. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Brandish PE, Ballou DP, Marletta MA. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evgenov OV, Pacher P, Schmidt PM, Hasko G, Schmidt HHHW, Stasch J-P. Nat. Rev. Drug Discov. 2006;5:755–768. doi: 10.1038/nrd2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin E, Berka V, Bogatenkova E, Murad F, Tsai A-L. J. Biol. Chem. 2006;281:27836–27845. doi: 10.1074/jbc.M601078200. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Marletta MA. Biochemistry. 1997;36:15959–15964. doi: 10.1021/bi971825x. [DOI] [PubMed] [Google Scholar]
- 6.Namiki S, Hirose K, Iino M. Biochem. Biophys. Res. Commun. 2001;288:798–804. doi: 10.1006/bbrc.2001.5836. [DOI] [PubMed] [Google Scholar]
- 7.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boon EM, Marletta MA. Curr. Opin. Chem. Biol. 2005;9:441–446. doi: 10.1016/j.cbpa.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Boon EM, Marletta MA. J. Inorg. Biochem. 2005;99:892, 902. doi: 10.1016/j.jinorgbio.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, Sayed N, Beuve A, van den Akker F. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, Marletta MA. J. Biol. Chem. 2006;281:21892–21902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- 13.Boon EM, Huang SH, Marletta MA. Nat. Chem. Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- 14.Tran R, Boon EM, Marletta MA, Mathies RA. Biochemistry. 2009 doi: 10.1021/bi900563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. ACS Chem. Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrick D. Biochemistry. 2002;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 17.Capece L, Estrin DA, Marti MA. Biochemistry. 2008;47:9416–9427. doi: 10.1021/bi800682k. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim M, Derbyshire ER, Marletta MA, Spiro TG. Biochemistry. 2010;49:3815–3823. doi: 10.1021/bi902214j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianconi A, Congiu-Castellano A, Dell'Ariccia M, Giovannelli A, Durham PJ, Burattini E, Barteri M. FEBS Lett. 1984;178:165–170. doi: 10.1016/0014-5793(84)81263-6. [DOI] [PubMed] [Google Scholar]
- 20.Chance B, Powers L, Ching Y, Poulos T, Schonbaum GR, Yamazaki I, Paul KG. Arch. Biochem. Biophys. 1984:596–611. doi: 10.1016/0003-9861(84)90234-0. [DOI] [PubMed] [Google Scholar]
- 21.D’Angelo P, Lapi A, Migliorati V, Arcovito A, Benfatto M, Roscioni OM, Meyer-Klaucke W, Della-Longa S. Inorg. Chem. 2008;47:9905–9918. doi: 10.1021/ic800982a. [DOI] [PubMed] [Google Scholar]
- 22.Levina A, Armstrong RS, Lay PA. Coord. Chem. Rev. 2005;249:141–160. [Google Scholar]
- 23.Chance M, Parkhurst L, Powers L, Chance B. J. Biol. Chem. 1986;261:5689–5692. [PubMed] [Google Scholar]
- 24.Miller LM, Chance MR. Biochemistry. 2002;34:10170–10179. doi: 10.1021/bi00032a010. [DOI] [PubMed] [Google Scholar]
- 25.Roe AL, Schneider DJ, Mayer RJ, Pyrz JW, Widom J, Que L. J. Am. Chem. Soc. 1984;106:1676–1681. [Google Scholar]
- 26.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]
- 27.Wirt MD, Sagi I, Chen E, Frisbie SM, Lee R, Chance MR. J. Am. Chem. Soc. 1991;113:5299–5304. [Google Scholar]
- 28.Benfatto M, Della Longa S, Natoli CR. J. Synchrotron Rad. 2003;10:51–57. doi: 10.1107/s0909049502018137. [DOI] [PubMed] [Google Scholar]
- 29.Della Longa S, Pin S, Cortes R, Soldatov AV, Alpert B. Biophys. J. 1998;75:3154–3162. doi: 10.1016/S0006-3495(98)77757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopfield JJ. J. Mol. Biol. 1973;77:207–222. doi: 10.1016/0022-2836(73)90332-x. [DOI] [PubMed] [Google Scholar]
- 31.Dai Z, Boon EM. J. Am. Chem. Soc. 2010;132:11496–11503. doi: 10.1021/ja101674z. [DOI] [PubMed] [Google Scholar]
- 32.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. J. Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erbil WK, Price MS, Wemmer DE, Marletta MA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19753–19760. doi: 10.1073/pnas.0911645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kassner RJ. Proc. Natl. Acad. Sci. U.S.A. 1972;69:2263–2267. doi: 10.1073/pnas.69.8.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy ML, Silchenko S, Houndonougbo Nv, Gibney BR, Dutton PL, Rodgers KR, Benson DR. J. Am. Chem. Soc. 2001;123:4635–4636. doi: 10.1021/ja0156441. [DOI] [PubMed] [Google Scholar]
- 36.Mao J, Hauser K, Gunner MR. Biochemistry. 2003;42:9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 37.Mauk AG, Moore GR. J. Biol. Inorg. Chem. 1997;2:119–125. [Google Scholar]
- 38.Grinstaff M, Hill M, Labinger J, Gray H. Science. 1994;264:1311–1313. doi: 10.1126/science.8191283. [DOI] [PubMed] [Google Scholar]
- 39.Cowley AB, Kennedy ML, Silchenko S, Lukat-Rodgers GS, Rodgers KR, Benson DR. Inorg. Chem. 2006;45:9985–10001. doi: 10.1021/ic052205k. [DOI] [PubMed] [Google Scholar]
- 40.Tezcan FA, Winkler JR, Gray HB. J. Am. Chem. Soc. 1998;120:13383–13388. [Google Scholar]
- 41.Perutz MF. Nature. 1970;228:726–734. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 42.Marti MA, Capece L, Crespo A, Doctorovich F, Estrin DA. J. Am. Chem. Soc. 2005;127:7721–7728. doi: 10.1021/ja042870c. [DOI] [PubMed] [Google Scholar]
- 43.Martí MA, Scherlis DA, Doctorovich FA, Ordejón P, Estrin DA. J. Biol. Inorg. Chem. 2003;8:595–600. doi: 10.1007/s00775-003-0452-9. [DOI] [PubMed] [Google Scholar]
- 44.Kruglik SG, Yoo B-K, Franzen S, Vos MH, Martin J-L, Negrerie M. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13678–13683. doi: 10.1073/pnas.0912938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friebe A, Koesling D. Mol. Pharmacol. 1998;53:123–127. doi: 10.1124/mol.53.1.123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.