Abstract
Rationale: Chromosome 12p has been linked to chronic obstructive pulmonary disease (COPD) in the Boston Early-Onset COPD Study (BEOCOPD), but a susceptibility gene in that region has not been identified.
Objectives: We used high-density single-nucleotide polymorphism (SNP) mapping to implicate a COPD susceptibility gene and an animal model to determine the potential role of SOX5 in lung development and COPD.
Methods: On chromosome 12p, we genotyped 1,387 SNPs in 386 COPD cases from the National Emphysema Treatment Trial and 424 control smokers from the Normative Aging Study. SNPs with significant associations were then tested in the BEOCOPD study and the International COPD Genetics Network. Based on the human results, we assessed histology and gene expression in the lungs of Sox5−/− mice.
Measurements and Main Results: In the case-control analysis, 27 SNPs were significant at P ≤ 0.01. The most significant SNP in the BEOCOPD replication was rs11046966 (National Emphysema Treatment Trial–Normative Aging Study P = 6.0 × 10−4, BEOCOPD P = 1.5 × 10−5, combined P = 1.7 × 10−7), located 3′ to the gene SOX5. Association with rs11046966 was not replicated in the International COPD Genetics Network. Sox5−/− mice showed abnormal lung development, with a delay in maturation before the saccular stage, as early as E16.5. Lung pathology in Sox5−/− lungs was associated with a decrease in fibronectin expression, an extracellular matrix component critical for branching morphogenesis.
Conclusions: Genetic variation in the transcription factor SOX5 is associated with COPD susceptibility. A mouse model suggests that the effect may be due, in part, to its effects on lung development and/or repair processes.
Keywords: chronic obstructive pulmonary disease, emphysema, knockout mice, lung development, single nucleotide polymorphism
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
A region on chromosome 12p has been linked to chronic obstructive pulmonary disease (COPD) and related traits, but the specific COPD susceptibility gene in that region has not been identified.
What This Study Adds to the Field
In human genetics studies, we identified a variant in the transcription factor SOX5 that is associated with COPD. In mice, we demonstrate that Sox5 is critical for proper in utero lung morphogenesis. These data suggest SOX5 may be a biologically relevant COPD susceptibility gene.
Genome-wide association studies (GWAS) have recently found several potential candidate genes for chronic obstructive pulmonary disease (COPD) (1–3). A GWAS may identify a specific gene but more commonly points to a multigene locus or an intergenic region, although the chromosomal resolution is much improved over earlier family-based linkage studies. Positional candidate gene studies (4) or fine mapping with a panel of single nucleotide polymorphisms (SNPs) (5) is generally required to localize the specific disease susceptibility gene(s) in a linkage region. Fine mapping may also be required to identify the specific gene within a locus found by GWAS.
Localizing a novel gene or confirming the genetic association of a biologically plausible candidate gene is only the first step in understanding the genetic architecture of COPD or any other common, complex disease. Functional validation using in vitro and in vivo methods is required to determine the specific role of a novel gene in COPD pathogenesis. Genes can influence COPD susceptibility through effects on protease–antiprotease balance, antioxidants, inflammation, innate immunity, cell death, and lung development/repair, among other pathways (6, 7).
In this project, we used a strategy of systematic fine mapping of a chromosomal region previously linked to COPD in the family-based Boston Early-Onset COPD (BEOCOPD) study (8, 9) using a dense set of SNPs to capture the common genetic variation across the region. Using this strategy, we identified a candidate gene for COPD susceptibility, which was then subjected to replication testing in additional human COPD studies. The importance of this gene in lung physiology was assessed in gene-targeted mice. Results of this study have been previously reported as an abstract (10).
METHODS
Detailed methods are available in the online supplement.
Study Subjects
The National Emphysema Treatment Trial (NETT) was a randomized clinical trial of lung volume reduction surgery in patients with emphysema and severe airflow obstruction (11). We compared NETT COPD cases to control smokers from the Normative Aging Study (NAS), a study of aging in initially healthy men (12). From the full set of 389 NETT and 472 NAS subjects (Table 1), 386 NETT and 424 NAS subjects were selected for fine mapping. Subject enrollment and phenotype determination in the family-based BEOCOPD Study have been described previously (13). COPD probands had FEV1 less than 40% predicted at an age less than 53 years. The International COPD Genetics Network (ICGN) was a multicenter family-based study conducted in the United States and Europe (14, 15). Probands were aged 45 to 65 years with FEV1 less than 60% predicted and FEV1/vital capacity less than 90% predicted. Human studies were approved by institutional review boards at participating centers. Subjects provided written informed consent.
TABLE 1.
CHARACTERISTICS OF SUBJECTS IN THE NATIONAL EMPHYSEMA TREATMENT TRIAL–NORMATIVE AGING STUDY CASE-CONTROL STUDY, THE BOSTON EARLY-ONSET CHRONIC OBSTRUCTIVE PULMONARY DISEASE STUDY, AND THE INTERNATIONAL CHRONIC OBSTRUCTIVE PULMONARY DISEASE GENETICS NETWORK
| NETT |
NAS |
BEOCOPD |
ICGN |
|||
|---|---|---|---|---|---|---|
| Cases | Control Subjects | Probands | Relatives | Probands | Relatives | |
| Subjects | 389 | 472 | 127 | 822 | 1,132 | 1,985 |
| Age, yr | 67.4 (± 5.8) | 69.8 (± 7.5) | 48.1 (± 4.7) | 46.2 (± 18.8) | 58.3 (± 5.4) | 57.9 (± 9.4) |
| Male sex | 250 (64.3) | 472 (100) | 32 (25.2) | 364 (44.3) | 679 (60.0) | 1,025 (51.6) |
| Pack-years of smoking | 66.4 (± 30.4)* | 40.3 (± 27.6) | 38.9 (± 21.9) | 19.0 (± 25.0) | 52.2 (± 29.1) | 39.1 (± 25.0) |
| FEV1 % predicted† | 28.0 (± 7.4) | 100.0 (± 13.1) | 21.9 (± 8.4) | 87.2 (± 20.3) | 36.1 (± 12.9) | 83.1 (± 26.1) |
| FEV1/FVC ratio† | 0.32 (± 0.06) | 0.79 (± 0.05) | 0.31 (± 0.10) | 0.73 (± 0.13) | 0.37 (± 0.12)‡ | 0.64 (± 0.14)‡ |
Definition of abbreviations: BEOCOPD = Boston Early-Onset COPD study; COPD = chronic obstructive pulmonary disease; ICGN = International COPD Genetics Network; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; VC = vital capacity.
Values are presented as mean (± SD) or n (%).
Values for pack-years were missing for three NETT subjects.
Post-bronchodilator values are listed for NETT, BEOCOPD, and ICGN. Prebronchodilator values are listed for NAS.
FEV1/VC was used in ICGN.
SNP Selection and Genotyping
Genome-wide linkage analysis in the BEOCOPD identified a region on chromosome 12p linked to post-bronchodilator FEV1 (16) and moderate airflow obstruction (8). We used a linkage disequilibrium tagging algorithm (r2 > 0.8, minor allele frequency 10%) to select a set of 1,534 SNPs in the region, which ranged from 10.2 to 25.8 Mb on the human genome map (National Center for Biotechnology Information build 36). SNPs were genotyped in the NETT-NAS study using custom-designed Illumina (San Diego, CA) GoldenGate assays. Selected SNPs were genotyped in the BEOCOPD study and the ICGN using Sequenom (San Diego, CA) or TaqMan (Applied Biosystems, Foster City, CA) assays.
Statistical Genetics Analysis
In the NETT-NAS case-control study, SNPs were analyzed under an additive genetic model, without covariate adjustment, using PLINK version 1.0.7 (17). In the BEOCOPD and ICGN family-based studies, SNPs were analyzed for association with COPD status under an additive model, without covariate adjustment, using Golden Helix (Bozeman, MT) PBAT version 6.4.3 (18).
DNA Sequencing
In 23 probands from the BEOCOPD Study and 1 CEPH control subject, we used dye-labeled dideoxy sequencing reactions to sequence the 14 exons and corresponding intron–exon boundaries in SOX5, as well as 10 highly conserved regions in the 3′ end of the gene, due to the 3′ location of rs11046966. SNPs identified through sequencing were genotyped in NETT-NAS and BEOCOPD using Sequenom or TaqMan assays.
Sox5 Null Mouse
All animal experiments were conducted in accordance with the University of Rochester Animal Care and Use Policy and following an approved animal studies protocol. Mice harboring an allele containing a mutated form of the Sox5 gene (19) in a mixed C57BL/6 × 129/SvEv background were bred to generate Sox5 deficient (Sox5−/−), heterozygous (Sox5+/−), and wild-type offspring. Timed matings were performed and offspring were harvested for analysis at 16.5, 17.5, or 18.5 days of embryonic gestation (E) by killing the pregnant dam. Embryos were dissected to isolate the entire thoracic cavity or individual lung lobes and fixed in buffered formalin for histological analysis or snap frozen in liquid nitrogen. RNA was isolated from E17.5 lung tissue for quantitative real-time polymerase chain reaction (qPCR), as previously described (20).
RESULTS
Genetic Association Analysis
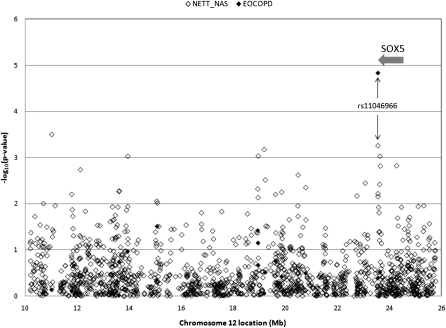
Of the 1,534 SNPs selected in the chromosome 12p linkage region, 1,410 were successfully genotyped in the NETT-NAS case-control study. Twenty-three SNPs showed significant deviation from Hardy-Weinberg Equilibrium (HWE) in the NAS control subjects (P < 0.001) and were removed from further analysis. Of the remaining 1,387 SNPs, 27 SNPs were significantly associated with COPD at P ≤ 0.01 (Table 2). To avoid false-negative results, a P value less than or equal to 0.01 threshold was used to select SNPs for follow-up. Of the 27 SNPs, 26 were successfully genotyped in the BEOCOPD study; SNP rs17381319 failed genotyping. All SNPs were in HWE in the BEOCOPD founders (P > 0.01). Two SNPs were significantly associated with COPD status (Global Initiative for Chronic Obstructive Lung Disease [GOLD] 2 or greater) in the BEOCOPD study, with the same direction of effect as in NETT-NAS (Table 2). The most significant SNP in the BEOCOPD replication was rs11046966 (NETT-NAS P = 6.0 × 10−4, BEOCOPD P = 1.5 × 10−5) (Figure 1). The minor allele led to an increased COPD risk in both study cohorts. The combined P value of 1.7 × 10−7 remained significant after Bonferroni adjustment for the 1,387 SNPs genotyped in NETT-NAS, with P(adjusted) = 2.3 × 10−4. This SNP is located 3′ to the gene SOX5 (sex-determining region Y-box 5). The only other significant SNP on replication in BEOCOPD was rs10744085. The nearest gene is located more than 60 kb away from this SNP, and the combined P = 0.003 was not significant after correction for multiple testing. None of the other follow-up SNPs genotyped in BEOCOPD was significantly associated with COPD in the combined analysis after multiple testing correction. Therefore, we focused on rs11046966 and SOX5 for further analysis.
TABLE 2.
RESULTS FOR SINGLE NUCLEOTIDE POLYMORPHISMS WITH P < 0.01 FOR ASSOCIATION WITH CHRONIC OBSTRUCTIVE PULMONARY DISEASE IN THE NATIONAL EMPHYSEMA TREATMENT TRIAL–NORMATIVE AGING STUDY CASE-CONTROL STUDY, THAT WERE GENOTYPED IN THE BOSTON EARLY-ONSET CHRONIC OBSTRUCTIVE PULMONARY DISEASE STUDY
| SNP | Chrom 12 Location* | Nearest Gene | Minor Allele | NETT MAF | NAS MAF | Odds Ratio (95% CI) | NETT-NAS P Value | BEOCOPD P Value | Effect on COPD Risk | Consistent Direction† | Combined P Value‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RS7968536 | 10709511 | STYK1 | G | 0.20 | 0.26 | 0.74 (0.58, 0.93) | 0.010 | 0.87 | Decrease | Y | 0.051 |
| RS1376251 | 11030119 | TAS2R50 | T | 0.30 | 0.38 | 0.68 (0.55, 0.84) | 0.00034 | 0.74 | Increase | N | NA |
| RS2051526 | 11798395 | ETV6 | T | 0.12 | 0.08 | 1.59 (1.14, 2.22) | 0.0067 | 0.78 | Decrease | N | NA |
| RS10845472 | 12129422 | BCL2L14 | C | 0.34 | 0.26 | 1.40 (1.13, 1.73) | 0.0019 | 0.87 | Increase | Y | 0.012 |
| RS1806195 | 13614659 | GRIN2B | G | 0.43 | 0.50 | 0.75 (0.61, 0.92) | 0.0056 | 0.99 | Increase | N | NA |
| RS731086 | 13624996 | GRIN2B | G | 0.46 | 0.39 | 1.33 (1.09, 1.63) | 0.0054 | 0.18 | Decrease | N | NA |
| RS11055681 | 13952836 | GRIN2B | T | 0.22 | 0.16 | 1.56 (1.20, 2.04) | 0.0010 | 0.11 | Decrease | N | NA |
| RS2445402 | 15065856 | C | 0.14 | 0.19 | 0.70 (0.53, 0.91) | 0.0091 | 0.45 | Decrease | Y | 0.027 | |
| RS10744085 | 15086271 | A | 0.12 | 0.16 | 0.69 (0.52, 0.91) | 0.010 | 0.031 | Decrease | Y | 0.0028 | |
| RS10743288 | 18950221 | G | 0.13 | 0.19 | 0.68 (0.52, 0.89) | 0.0051 | 0.038 | Increase | N | NA | |
| RS12810703 | 18961960 | T | 0.13 | 0.17 | 0.69 (0.52, 0.90) | 0.0077 | 0.071 | Increase | N | NA | |
| RS1994641 | 18964611 | G | 0.09 | 0.15 | 0.60 (0.44, 0.82) | 0.0011 | 0.22 | Increase | N | NA | |
| RS1514831 | 19195039 | PLEKHA5 | T | 0.15 | 0.22 | 0.64 (0.49, 0.83) | 0.00074 | 0.89 | Decrease | Y | 0.0054 |
| RS10770448 | 19235485 | PLEKHA5 | G | 0.23 | 0.30 | 0.70 (0.56, 0.89) | 0.0032 | 0.31 | Increase | N | NA |
| RS7960653 | 19633322 | T | 0.41 | 0.34 | 1.32 (1.08, 1.62) | 0.0064 | 0.18 | Increase | Y | 0.0088 | |
| RS11044889 | 19867409 | C | 0.39 | 0.32 | 1.34 (1.09, 1.65) | 0.0058 | 0.38 | Increase | Y | 0.016 | |
| RS11045262 | 20503025 | PDE3A | T | 0.13 | 0.19 | 0.65 (0.49, 0.86) | 0.0026 | 0.65 | Increase | N | NA |
| RS972505 | 20794502 | SLCO1C1 | C | 0.42 | 0.49 | 0.75 (0.61, 0.91) | 0.0046 | 0.32 | Decrease | Y | 0.011 |
| RS261926 | 22761409 | A | 0.43 | 0.50 | 0.76 (0.62, 0.93) | 0.0069 | 0.63 | Decrease | Y | 0.028 | |
| RS10842101 | 23080263 | G | 0.47 | 0.40 | 1.35 (1.10, 1.66) | 0.0037 | 0.86 | Decrease | N | NA | |
| RS10842178 | 23568624 | SOX5 | C | 0.28 | 0.35 | 0.72 (0.57, 0.91) | 0.0063 | 0.41 | Decrease | Y | 0.018 |
| RS11046966 | 23568959 | SOX5 | C | 0.34 | 0.26 | 1.48 (1.18, 1.84) | 0.00060 | 1.45E-05 | Increase | Y | 1.69E-07 |
| RS17381319 | 23570340 | SOX5 | C | 0.19 | 0.14 | 1.46 (1.11, 1.92) | 0.0073 | § | NA | ||
| RS11046991 | 23628593 | SOX5 | T | 0.26 | 0.32 | 0.72 (0.58, 0.90) | 0.0040 | 0.99 | Decrease | Y | 0.026 |
| RS10771006 | 23645476 | SOX5 | C | 0.17 | 0.11 | 1.64 (1.22, 2.20) | 0.0010 | 0.31 | Decrease | N | NA |
| RS7311001 | 23654449 | SOX5 | T | 0.27 | 0.34 | 0.71 (0.57, 0.88) | 0.0016 | 0.90 | Decrease | Y | 0.011 |
| RS2030510 | 24286469 | SOX5 | T | 0.45 | 0.53 | 0.73 (0.59, 0.89) | 0.0016 | 0.68 | Decrease | Y | 0.0084 |
Definition of abbreviations: BEOCOPD = Boston Early-Onset COPD study; CI = confidence interval; COPD = chronic obstructive pulmonary disease; ICGN = International COPD Genetics Network; MAF = minor allele frequency; NAS = Normative Aging Study; NETT = National Emphysema Treatment Trial; SNP = single-nucleotide polymorphism; VC = vital capacity.
Chromosomal locations based on National Center for Biotechnology Information build 36.
Consistent direction of effect on COPD in the NETT-NAS and BEOCOPD studies.
Combined P values calculated using Fisher method (47). P values were not combined for SNPs with inconsistent direction of effect in the NETT-NAS and BEOCOPD studies.
SNP RS17381319 failed genotyping in BEOCOPD.
Figure 1.
Genetic associations with chronic obstructive pulmonary disease (COPD) on chromosome 12p. P values for 1,387 single-nucleotide polymorphisms (SNPs) in the National Emphysema Treatment Trial–Normative Aging Study case-control study and 26 SNPs followed up in the Boston Early-Onset COPD Study are shown. Chromosomal locations are based on National Center for Biotechnology Information build 36.
The association between rs11046966 and case status in NETT-NAS remained significant in a logistic regression model adjusted for age and pack-years of smoking (odds ratio [OR], 1.49; P = 0.0017). The association with COPD remained significant in BEOCOPD as well, after adjusting for age, sex, and pack-years of smoking (P = 7.0 × 10−5). In BEOCOPD, the minor allele of rs11046966 was significantly associated with an increased risk of severe COPD, defined as GOLD 3 or greater (unadjusted P = 2.1 × 10−5; adjusted for age, sex, and pack-years, P = 1.2 × 10−4); this phenotype definition is more comparable with the COPD severity seen in NETT. In BEOCOPD, rs11046966 was associated with reduced values of post-bronchodilator FEV1 as a continuous trait (P = 0.0043, adjusted for age, sex, pack-years of smoking, and height). In NETT, rs11046966 was not associated with post-bronchodilator FEV1 or emphysema (−950 Hounsfield units) on chest computed tomography scans, likely due to the narrow ranges of these variables in NETT cases. In analyses restricted to current and former smokers only in the BEOCOPD study, rs11046966 remained significantly associated with COPD status (unadjusted P = 1.4 × 10−5, adjusted P = 3.9 × 10−5) and post-bronchodilator FEV1 (P = 2.5 × 10−4).
In the family-based ICGN study, rs11046966 was not associated with COPD (GOLD 2 or greater), severe COPD (GOLD 3 or greater), or post-bronchodilator values for FEV1. There was a trend for increased CT emphysema with increasing copies of the minor allele (P = 0.16), but no association with airway wall thickness (P = 0.37). The five other SOX5 SNPs from Table 2 were also not significantly associated with COPD in the ICGN. To identify associations with early-onset COPD, we restricted the ICGN analysis to families of probands under age 53 years, the age cut-off in the BEOCOPD study. Adjusted for age, sex, and pack-years, rs11046966 trended toward association with COPD (P = 0.11). In the online data from the British 1958 Birth Cohort (http://www.b58cgene.sgul.ac.uk/, accessed October 21, 2009), the minor allele of rs11046966 showed a trend toward reduced values of FEV1 (genotype model P = 0.097, allele model P = 0.055).
DNA Sequencing
Sequencing identified 30 SNPs, 13 of which were not found in the dbSNP database, including one synonymous exonic SNP (see Table E1 in the online supplement). Of the 17 known SNPs, 4 had been previously genotyped in NETT-NAS and 1 in BEOCOPD. Twenty-one SNPs were present in more than one chromosome (minor allele frequency > 2%). These 21 SNPs plus the synonymous exonic SNP were genotyped in NETT-NAS and BEOCOPD. One SNP, rs7970223, failed genotyping. The other SNPs were in HWE in NAS control subjects and BEOCOPD founders. In NETT-NAS, only one SNP identified through sequencing was significantly associated with COPD: rs7485662, located in an intron (unadjusted OR = 1.25, P = 0.021; covariate-adjusted OR = 1.34, P = 0.0093). However, this SNP was not significantly associated with COPD or post-bronchodilator FEV1 in the BEOCOPD study.
SOX5 Expression in COPD
We have previously described analysis of gene expression in two independent microarray data sets derived from the lung tissue of subjects with COPD. In a data set from case subjects with severe COPD (21), we observed a significant decrease in SOX5 expression in COPD cases as compared with control subjects (207336_at; mean signal intensity 50.5 vs. 18.8 in cases vs. control subjects; P = 0.016). In this population, SOX5 expression was also significantly correlated with lung function. SOX5 displayed a strong positive correlation with prebronchodilator FEV1 (r = 0.435, P = 0.034) and diffusing capacity (r = 0.507, P = 0.004) and a strong negative correlation with total lung capacity (r = −0.451, P = 0.011). These data are consistent with a decrease in SOX5 expression in COPD. No significant differences in SOX5 gene expression were observed in a second microarray data set derived from a more heterogeneous COPD population (22).
Animal Model
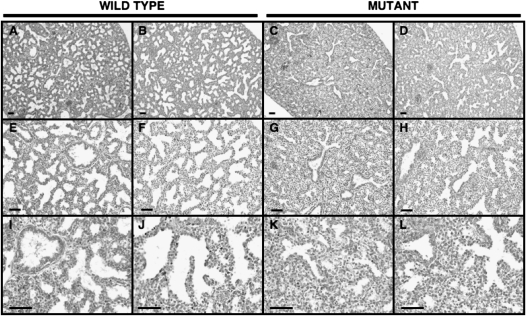
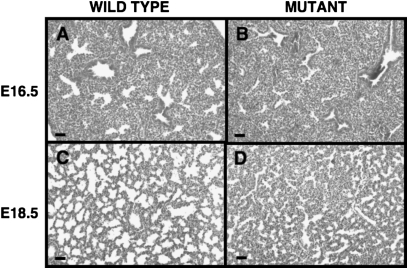
Respiratory distress and neonatal death have previously been reported in Sox5−/− and Sox5−/−; Sox6−/− double-mutant mice (19). However, no studies of the pulmonary manifestations or description of lung histology in these mice are available. We assessed gross lung histology in Sox5−/−, Sox5+/−, and wild-type littermate mice at 16.5, 17.5, and 18.5 days of embryonic gestation (E). No differences in embryo size, weight, or lung size were apparent. Abnormalities in the structure of Sox5−/− lungs were most evident at the end of the canalicular stage of development (Figure 2). In wild-type E17.5 lungs, the initiation of distal respiratory structures, including reduced cellular density and the morphological appearance of respiratory epithelial cells, was noted. However, E17.5 Sox5−/− lungs displayed histology similar to E16.5, with increased cellularity and decreased airspace complexity. At high magnification, alterations in tissue organization, with a reduction in the emergence of discrete distal airspaces lined by squamous (respiratory) epithelium, was observed in Sox5−/− lungs. At the transition from the pseudoglandular to canalicular stage of development (E16.5), a similar reduction in the complexity of the lung was noted in the Sox5−/− mouse (Figure 3). Developmental delays in the lungs of Sox5−/− mice persisted at E18.5, where developing saccules were substantially less organized than in their wild-type counterparts (Figure 3). These abnormalities in Sox5−/− lung structure were obvious in all litters analyzed (N > 6), whether lung tissue was isolated or the entire thoracic cavity was fixed in situ to minimize manipulation-related artifacts in histology. Embryonic lungs from Sox5+/− heterozygotes displayed a range of intermediate phenotypes, ranging from mildly abnormal to substantially affected.
Figure 2.
Abnormalities in the structure of Sox5−/− embryonic lungs. Lung structure was assessed in (A, B, E, F, I, J) wild-type and (C, D, G, H, K, L) and Sox5−/− mouse lungs at 17.5 days of embryonic gestation by hematoxylin and eosin staining of formalin-fixed paraffin-embedded tissues. (A–D) Low magnification images demonstrate structural abnormalities, including increased cellular density and reduced airspace complexity, at the end of the canalicular stage in Sox5−/− lungs. (E–K) Higher magnification images reveal abnormalities in organization of distal airspaces. Representative images are shown from a total of more than 12 mice of each genotype from more than 6 litters. Bar indicates 50 μm.
Figure 3.
Age-related abnormalities in the structure of Sox5−/− embryonic lungs. Lung structure was assessed in (B, D) Sox5−/− and (A, C) wild-type littermates at 16.5 and 18.5 days of embryonic gestation (E) by hematoxylin and eosin staining of formalin-fixed paraffin-embedded tissues. Sox5−/− lungs appear immature with increased cellularity at all ages. Representative images are shown from a total of more than 12 mice of each genotype from more than 6 litters. Bar indicates 50 μm.
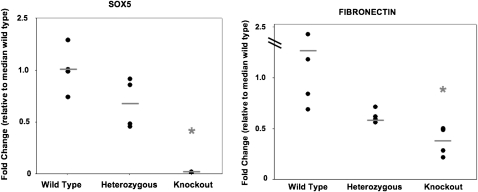
We studied gene expression in Sox5−/−, Sox5+/−, and wild-type (n = 4 each genotype) lungs at E17.5 by qPCR. As expected, we observed a significant reduction in Sox5 expression in Sox5−/− lungs to less than 2% (P < 0.01) of the level of expression observed in the lungs of wild-type animals (Figure 4). Sox5+/− lungs displayed close to 50% of wild-type expression levels. We screened for the expression of a panel of lung development–related genes in the Sox5−/− lungs by qPCR. No significant differences were noted for the expression of Sftpc, Scgb1a1, Titf1, Acta2, Ki67, Shh, Bmp4, or Aqp5. However, we observed a significant reduction (55% reduction; P < 0.05) in the expression of fibronectin (Fn1) in Sox5−/− lungs (Figure 4). As Fn1 is necessary for proper branching morphogenesis in the lung (23), by providing clefts to regulate bud branching and elongation, the reduction in Fn1 gene expression may partly explain the observed phenotype in Sox5−/− lungs. Interestingly, Sox5+/− lungs displayed intermediate levels of Fn1 expression (30% reduction), consistent with their intermediate morphological abnormalities.
Figure 4.
Decreased fibronectin expression in Sox5−/− lungs. Whole lung tissue was dissected from Sox5−/− (knockout), Sox5+/− (heterozygous), and wild-type embryos at E17.5. Quantitative real-time reverse transcriptase–polymerase chain reaction demonstrated a significant reduction in Sox5 (< 2% wild type) and fibronectin (< 50% wild type) steady state mRNA levels. *P < 0.05
DISCUSSION
Using a dense set of SNPs to fine map a region on chromosome 12p linked to COPD, we identified SOX5 as a candidate gene for COPD. The association was discovered in the NETT-NAS case-control study and replicated in the family-based BEOCOPD study. This combined analysis remained significant with a strict multiple testing correction. The SNP association could not be confirmed in a third study, the ICGN. However, there was a trend for association with earlier-onset COPD in the ICGN. SOX5 gene expression was reduced, and significantly correlated with lung function, in the lung tissue of some subjects with COPD. A Sox5 null mouse model demonstrated abnormalities in embryonic lung development and reduced expression of fibronectin, an extracellular matrix component previously shown to be critical for lung morphogenesis. Lungs of Sox5+/− heterozygous mice showed less severe developmental abnormalities, which may be more comparable to lung disease in humans carrying SOX5 variants than is the extreme phenotype of Sox5−/− mice.
There are no previous animal or human reports on the role of SOX5 in lung disease. There are two major isoforms of SOX5 (24). The short isoform is expressed primarily in testis, whereas the long isoform is expressed in many tissues, including lung (24, 25). In mouse models, Sox5 has been primarily studied in relation to cartilage (19) and nervous system development (26). Smits and colleagues observed that Sox5−/− mice died at birth from respiratory distress, with a cleft palate and abnormalities of the bony thoracic cage (19). Developmental abnormalities in the lungs of Sox5−/− mice had not been previously described; thus, it was unknown if lethality was due to defects in lung development. The lungs of newborn Sox5−/− mice appeared severely affected, demonstrating a premature (canalicular rather than saccular) structure (data not shown). To determine if these abnormalities contributed to neonatal death, we studied embryonic lung development in these mice. We observed a profound and consistent deficiency in lung development in embryonic Sox5−/− mutant lungs. It is possible that abnormalities in rib cage and/or palate formation contributed to the developmental lung defects in these mice. Prior animal models have demonstrated associations between cartilage/bone formation and lung development (27, 28).
We studied molecular changes in the lungs of Sox5−/− mice, but found no significant differences in the expression of markers for cell proliferation or cell lineage at the steady-state mRNA level. Sox5 is known to regulate the expression of many extracellular matrix genes. Such a process could contribute, at least in part, to the defects in lung development observed in mutant mice. We found a significant reduction in the expression of fibronectin, but not other extracellular matrix molecules. This is of particular interest as fibronectin is specifically necessary for branching morphogenesis. Interestingly, fibronectin expression was intermediately affected in Sox5+/− heterozygous lungs consistent with the mild to moderate histological abnormalities in the developing lungs of these mice. Further in vivo and in vitro studies will be required to define whether changes in fibronectin expression explain the phenotype in these mice and to more completely determine the specific contributions of Sox5 to mammalian lung development.
The animal model demonstrated the importance of Sox5 in normal mouse lung development. It is plausible that events in early human lung development may eventually predispose to COPD (29, 30). Abnormalities in lung development may reduce the maximally attained level of lung function, predisposing to reduced lung function in adulthood, according to the classic Fletcher and Peto diagram (31) and supported by more recent longitudinal studies (32, 33). Additionally, lungs with subtle developmental abnormalities, which may not be clinically relevant in childhood, may have an increased susceptibility to the damaging effects of exposures such as cigarette smoke (30). Similarly, functional variants of developmentally important genes may be less capable of contributing to injury-repair processes necessary for lung maintenance. Several genes have been identified that may have effects on lung development as well as COPD susceptibility, including members of the TGF-β signaling pathway (34–37). For example, mice genetically deficient in latent TGF-β binding protein-4 (ltpb4) have emphysematous changes present at birth (38); our group has shown that genetic variants in LTBP4 may influence COPD-related phenotypes (39).
Recently, variants in SOX5 have been associated with a variety of human traits through GWAS. None of these studies has focused on musculoskeletal or lung diseases. In GWAS, SNPs in SOX5 have been associated with rapid progression of AIDS (40), electrocardiographic P-R interval (41), triglyceride levels in subjects taking statins (42), and metabolic side effects of antipsychotic drugs, specifically high-density lipoprotein levels in patients taking perphenazine (43). Most of these associations have been confined to the 5′ end of the gene, whereas the SNP we identified to be associated with COPD was located 3′ to the SOX5 transcript. SOX5 SNPs have also been associated with pharmacogenetic effects of bupropion for smoking cessation, a potentially relevant phenotype for COPD (44). However, the specific associated SNPs were not detailed.
In the present study, the most strongly associated SOX5 SNP in NETT-NAS (rs11046966) was also strongly associated with COPD in the BEOCOPD study. We were unable to replicate the associations in a third population, the ICGN. The genetic associations in the first two populations and the animal model both point to the potential importance of SOX5 in COPD, yet rs11046966 may not be the functional variant. SNP rs11046966 is located more than 7 kb downstream from the 3′ UTR of SOX5, so it is unlikely to affect an miRNA binding site. Despite sequencing the exons of SOX5 and highly conserved regions in the 3′ end of the gene, we were unable to identify a putative functional variant. SNP rs11046966 is located in a conserved region that extends from the 3′ end of SOX5. According to MAPPER analysis (45), rs11046966 is located within a predicted binding sequence for interferon response factor. Based on SNP data in whites from phase 3 of the International HapMap project (46), rs11046966 is in moderate linkage disequilibrium (r2 > 0.66) with additional SNPs 3′ to the transcript and with intronic SNPs (Table E3), but the roles of these SNPs in COPD are unknown as well. However, these SNPs do point to SOX5 as the relevant gene in the linkage region, justifying the studies in the Sox5 null mouse model. Subtle differences in the linkage disequilibrium patterns between rs11046966 and the functional variant comparing the two U.S. populations (NETT-NAS and BEOCOPD) to the combined North American and European subjects in the ICGN may account for the lack of association in the latter study. Differences in COPD severity across the study populations may also explain the nonreplication in ICGN (Figure E1).
In summary, using a systematic fine mapping approach, we identified genetic associations between a variant near SOX5 and COPD in two of the three populations tested. SOX5 gene expression was reduced in lung tissue from patients with COPD in one of two populations tested. A Sox5 null mouse demonstrated abnormal lung development, and heterozygous animals showed an intermediate severity phenotype. However, the specific role of SOX5 as it relates to human COPD is not clear. For example, we do not know whether its role in human COPD is confined to lung development or whether it plays a role in lung repair. Future research in the mouse model, such as determining the effects of cigarette smoke exposure in the heterozygous model, and future studies in emphysematous human lung tissue will be required to answer these questions about the role of SOX5 in COPD.
Supplementary Material
Acknowledgments
The authors thank Scott Weiss, Frank Speizer, Jeffrey Drazen, Hal Chapman, Leo Ginns and Steven Mentzer for their roles in developing the Boston Early-Onset COPD Study. Coinvestigators in the NETT Genetics Ancillary Study include Joshua Benditt, Gerard Criner, Malcolm DeCamp, Philip Diaz, Mark Ginsburg, Larry Kaiser, Marcia Katz, Mark Krasna, Neil MacIntyre, Barry Make, Rob McKenna, Fernando Martinez, Zab Mosenifar, Andrew Ries, Paul Scanlon, Frank Sciurba, and James Utz. International COPD Genetics Network (ICGN) Investigators (in addition to Drs. Lomas and Silverman): Alvar Agusti, Son Dureta Hospital and Fundación Caubet-Cimera, Palma de Mallorca, Spain; Peter M. A. Calverley, University of Liverpool, Liverpool, UK; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, Canada; Barry J. Make, National Jewish Medical and Research Centre, Denver, Colorado; Peter D. Paré, University of British Columbia, Vancouver, Canada; Stephen I. Rennard, University of Nebraska, Omaha, Nebraska; Jørgen Vestbo, Department of Cardiology and Respiratory Medicine, Hvidovre Hospital, Copenhagen, Denmark; Emiel F. M. Wouters, University Hospital Maastricht, The Netherlands.
Supported by National Institutes of Health grants HL080242, HL094635, HL071885, HL075478, P01 HL083069, HL72303, and a grant from the Alpha-1 Foundation. The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute grants N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119; the Centers for Medicare and Medicaid Services; and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), Boston, MA. The International COPD Genetics Network was supported by GlaxoSmithKline.
Originally Published in Press as DOI: 10.1164/rccm.201010-1751OC on February 17, 2011
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
Author Disclosure: C.P.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.K.S. was a consultant for GlaxoSmithKline (GSK) and AstraZeneca (AZ). He received lecture fees from GSK, AZ, and Bayer and received grant support from GSK. J.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J.K. owns a patent through the Massachusetts Institute of Technology on methods and products related to genotyping and DNA analysis and is employed by Brigham and Women's Hospital. A.A.L. receives royalties from Up-to-Date. V.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.J.R. owns a patent through Brigham and Women's Hospital on software algorithm for CT scan analysis and receives royalties from Up-to-Date. W.H.A. is a full-time employee of, owns stocks or options of, and received travel accommodations from GSK. D.A.L. was a consultant for GSK, Novartis, Genzyme, and Amicus. He was on the Advisory Board for GSK, Talecris, Thorax, and the Medical Research Council . He received lecture fees from GSK, AZ, Boehringer Ingelheim, and London Fire Brigade (LFB). He received grant support from GSK, Merck Sharp & Dohme, the Medical Research Council, the Wellcome Trust, the British Lung Foundation, and the Alpha-1 Foundation. T.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 2009;5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, et al. Transforming growth factor-beta receptor-3 is associated with pulmonary emphysema. Am J Respir Cell Mol Biol 2009;41:324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersh CP, Pillai SG, Zhu G, Lomas DA, Bakke P, Gulsvik A, DeMeo DL, Klanderman BJ, Lazarus R, Litonjua AA, et al. Multistudy fine mapping of chromosome 2q identifies XRCC5 as a chronic obstructive pulmonary disease susceptibility gene. Am J Respir Crit Care Med 2010;182:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacNee W, Tuder RM. New paradigms in the pathogenesis of chronic obstructive pulmonary disease I. Proc Am Thorac Soc 2009;6:527–531. [DOI] [PubMed] [Google Scholar]
- 7.Sethi S, Mallia P, Johnston SL. New paradigms in the pathogenesis of chronic obstructive pulmonary disease II. Proc Am Thorac Soc 2009;6:532–534. [DOI] [PubMed] [Google Scholar]
- 8.Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet 2002;11:623–632. [DOI] [PubMed] [Google Scholar]
- 9.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002;70:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh CP, Mariani TJ, Litonjua AA, Klanderman BJ, Gascon J, Sparrow D, Reilly JJ, Silverman EK. SOX5 is a candidate gene for COPD susceptibility and is necessary for lung development [abstract]. Am J Respir Crit Care Med 2010;181:A4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 12.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Dev 1972;3:5–17. [Google Scholar]
- 13.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 1998;157:1770–1778. [DOI] [PubMed] [Google Scholar]
- 14.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, Donner CF, Make BJ, Muller NL, Rennard SI, Vestbo J, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:500–505. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173. [DOI] [PubMed] [Google Scholar]
- 16.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell 2001;1:277–290. [DOI] [PubMed] [Google Scholar]
- 20.Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, Mariani TJ. Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. Am J Respir Crit Care Med 2010;181:838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol 2004;31:601–610. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya S, Srisuma S, Demeo DL, Shapiro SD, Bueno R, Silverman EK, Reilly JJ, Mariani TJ. Molecular biomarkers for quantitative and discrete COPD phenotypes. Am J Respir Cell Mol Biol 2009;40:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature 2003;423:876–881. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda T, Zhang J, Chano T, Mabuchi A, Fukuda A, Kawaguchi H, Nakamura K, Ikegawa S. Identification and characterization of the human long form of Sox5 (L-SOX5) gene. Gene 2002;298:59–68. [DOI] [PubMed] [Google Scholar]
- 25.Wunderle VM, Critcher R, Ashworth A, Goodfellow PN. Cloning and characterization of SOX5, a new member of the human SOX gene family. Genomics 1996;36:354–358. [DOI] [PubMed] [Google Scholar]
- 26.Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA 2008;105:16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet 1995;11:415–421. [DOI] [PubMed] [Google Scholar]
- 28.Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development 1998;125:3615–3623. [DOI] [PubMed] [Google Scholar]
- 29.Maciewicz RA, Warburton D, Rennard SI. Can increased understanding of the role of lung development and aging drive new advances in chronic obstructive pulmonary disease? Proc Am Thorac Soc 2009;6:614–617. [DOI] [PubMed] [Google Scholar]
- 30.Shi W, Warburton D. Is COPD in adulthood really so far removed from early development? Eur Respir J 2010;35:12–13. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher C, Peto R. The natural history of chronic airflow obstruction. BMJ 1977;1:1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalhan R, Arynchyn A, Colangelo LA, Dransfield MT, Gerald LB, Smith LJ. Lung function in young adults predicts airflow obstruction 20 years later. Am J Med 2010;123:468.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern DA, Morgan WJ, Wright AL, Guerra S, Martinez FD. Poor airway function in early infancy and lung function by age 22 years: a non-selective longitudinal cohort study. Lancet 2007;370:758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-{beta}1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649–1656. [DOI] [PubMed] [Google Scholar]
- 35.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Nolte IM, Boezen HM. Decorin and TGF-beta1 polymorphisms and development of COPD in a general population. Respir Res 2006;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 2004;59:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morty RE, Konigshoff M, Eickelberg O. Transforming growth factor-beta signaling across ages: from distorted lung development to chronic obstructive pulmonary disease. Proc Am Thorac Soc 2009;6:607–613. [DOI] [PubMed] [Google Scholar]
- 38.Sterner-Kock A, Thorey IS, Koli K, Wempe F, Otte J, Bangsow T, Kuhlmeier K, Kirchner T, Jin S, Keski-Oja J, et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev 2002;16:2264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hersh CP, Demeo DL, Lazarus R, Celedon JC, Raby BA, Benditt JO, Criner G, Make B, Martinez FJ, Scanlon PD, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Clerc S, Limou S, Coulonges C, Carpentier W, Dina C, Taing L, Delaneau O, Labib T, Sladek R, Deveau C, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03). J Infect Dis 2009;200:1194–1201. [DOI] [PubMed] [Google Scholar]
- 41.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, et al. Genome-wide association study of PR interval. Nat Genet 2010;42:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE 2010;5:e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adkins DE, Aberg K, McClay JL, Bukszar J, Zhao Z, Jia P, Stroup TS, Perkins D, McEvoy JP, Lieberman JA, et al. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Mol Psychiatry 2011;16:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry 2008;65:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics 2005;6:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 47.Fisher RA. Statistical methods for research workers. London: Oliver and Boyd, Ltd.; 1925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.