Abstract
Rationale: Because acute lung injury is a sporadic disease produced by heterogeneous precipitating factors, previous genetic analyses are mainly limited to candidate gene case-control studies.
Objectives: To develop a genome-wide strategy in which single nucleotide polymorphism associations are assessed for functional consequences to survival during acute lung injury in mice.
Methods: To identify genes associated with acute lung injury, 40 inbred strains were exposed to acrolein and haplotype association mapping, microarray, and DNA-protein binding were assessed.
Measurements and Main Results: The mean survival time varied among mouse strains with polar strains differing approximately 2.5-fold. Associations were identified on chromosomes 1, 2, 4, 11, and 12. Seven genes (Acvr1, Cacnb4, Ccdc148, Galnt13, Rfwd2, Rpap2, and Tgfbr3) had single nucleotide polymorphism (SNP) associations within the gene. Because SNP associations may encompass “blocks” of associated variants, functional assessment was performed in 91 genes within ± 1 Mbp of each SNP association. Using 10% or greater allelic frequency and 10% or greater phenotype explained as threshold criteria, 16 genes were assessed by microarray and reverse real-time polymerase chain reaction. Microarray revealed several enriched pathways including transforming growth factor-β signaling. Transcripts for Acvr1, Arhgap15, Cacybp, Rfwd2, and Tgfbr3 differed between the strains with exposure and contained SNPs that could eliminate putative transcriptional factor recognition sites. Ccdc148, Fancl, and Tnn had sequence differences that could produce an amino acid substitution. Mycn and Mgat4a had a promoter SNP or 3′untranslated region SNPs, respectively. Several genes were related and encoded receptors (ACVR1, TGFBR3), transcription factors (MYCN, possibly CCDC148), and ubiquitin-proteasome (RFWD2, FANCL, CACYBP) proteins that can modulate cell signaling. An Acvr1 SNP eliminated a putative ELK1 binding site and diminished DNA–protein binding.
Conclusions: Assessment of genetic associations can be strengthened using a genetic/genomic approach. This approach identified several candidate genes, including Acvr1, associated with increased susceptibility to acute lung injury in mice.
Keywords: acute respiratory distress syndrome, smoke inhalation, carboxyl stress, transforming growth factor-&beta signaling, ubiquitination
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
A major challenge to critical care is to reliably predict survival of patients with acute lung injury. Individual susceptibility varies greatly (i.e., patients presenting with the same severity score can have markedly different clinical outcomes). For this reason, studies have begun to investigate the role of genetics in determining survival during acute lung injury. However, because acute lung injury is a sporadic disease produced by heterogeneous precipitating factors, previous genetic analyses are mainly limited to case-control studies that evaluate candidate genes associations.
What This Study Adds to the Field
This study presents a strategy in which single nucleotide polymorphism associations are assessed for functional consequences to survival during acute lung injury in mice. This approach identified several candidate genes, and Acvr1 may be particularly worthy of future investigations in acute lung injury.
Acute lung injury is marked by pulmonary edema resulting from increased epithelial and endothelial permeability, decreased clearance of edema fluid, and disruption of surfactant-associated protein function (1–3). This condition can be induced directly (e.g., through smoke inhalation) or indirectly (e.g., through sepsis) (1). In smoke inhalation, the severity of acute lung injury (2) and cardiovascular dysfunction (4) are critical determinants of morbidity and mortality (5–7). Acrolein, a potent irritant (8–10), is the major chemical in smoke responsible for pulmonary edema (11, 12).
A major challenge to critical care is to reliably predict and enhance survival in acute lung injury (7). Individual susceptibility varies greatly (i.e., patients presenting with the same severity score can have markedly different clinical outcomes) (13). For this reason, studies have begun to investigate the role of genetics in determining survival during acute lung injury (14–17). However, because acute lung injury is a sporadic disease produced by heterogeneous precipitating factors, previous genetic analyses are mainly limited to case-control studies that evaluate candidate genes associations.
Constant acquisition of genome-wide information on numerous species, including more than 40 mouse strains (18–21), makes the mouse a useful model to facilitate rapid evaluation of the genetic basis of human physiology and pathophysiology (22, 23). Complementing conventional single gene mapping approaches, genome-wide mapping has mainly used quantitative trait locus (QTL) analysis in mice, which has been a valued tool to identify candidate genes responsible for complex traits underlying human disease (24–26). However, a major obstacle of identifying genes is the difficulty of resolving large QTL regions (10–20 cM) into sufficiently small intervals to make positional cloning practical (27). In this study, haplotype association mapping is used to obtain a dense single-nucleotide polymorphism (SNP) map, which offers finer genetic resolution in the identification of genetic determinants of acute lung injury.
METHODS
Experimental Design
This study was performed in accordance with the Institutional Animal Care and Use Committee of the University of Pittsburgh (Pittsburgh, PA) and mice were housed under specific pathogen-free conditions. Forty inbred strains (n = 507 mice, 6–8 wk old, female; The Jackson Laboratory, Bar Harbor, ME) were used in the study. Mice were exposed to filtered air (control) or acrolein (10 ppm, 24 h) as generated and monitored as described previously (8), and survival time was recorded. Haplotype association analysis was performed using threshold –log(P) value of 6.0 for significant and 4.0 for suggestive linkage as described previously (23). To examine acrolein-induced changes in lung histology, bronchoalveolar lavage, and lung transcripts, 129X1/SvJ and SM/J mice (two strains that represented the opposite ends of the phenotypic spectrum of the analyzed strains) were exposed to filtered air (0 h, control) or acrolein (10 ppm for 6, 12, or 17 h). Microarray analysis and quantitative real time-polymerase chain reaction (qRT-PCR) were used to determine transcript levels of genes previously associated with acute lung injury and to contrast genes identified. Three 5′ untranslated region (UTR) SNPs in Acvr1 were genotyped in 28 strains and the consequences of the rs6406107 variant on DNA–protein binding was assessed by electrophoretic mobility shift assay. Additional details of the methods are presented in the online supplemental.
RESULTS
Survival Time of 40 Mouse Strains during Acute Lung Injury
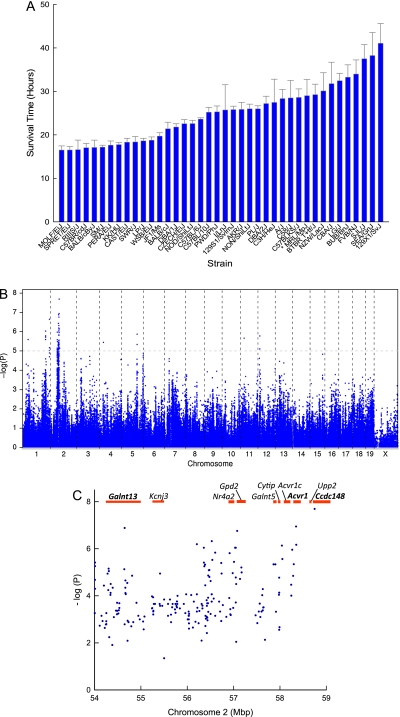
Mice from 40 mouse strains were exposed to acrolein and survival time recorded. The mean survival time varied among mouse strains (Figure 1A). The most polar strains varied approximately 2.5-fold from 16.5 ± 0.9 hours (MOLF/EiJ) to 41.1 ± 4.5 hours (129X1/SvJ). Using the data from all the mice, a haplotype association map was obtained for acrolein-induced acute lung injury (Figure 1B). Significant [−log(P) ≥ 6.0] associations were identified on chromosome 2 with suggestive [−log(P) ≥ 4.2] associations identified on chromosomes 1, 2, 4, 11, and 12. Five genes (Rfwd2, Cacnb4, Galnt13, Acvr1, and Rpap2) had three to four significant or suggestive SNP associations within the gene boundary, whereas two genes (Ccdc148 and Tgfbr3) had one significant or suggestive SNP association within the gene boundary (see Table E1 in the online supplement). Evaluation of SNP associations determined the allelic frequency ranged from 0.20 to 0.54 (or 73 and 197 mice, respectively) among the 40 strains tested for survival (n = 365 mice). Because a sufficient number of mice carried the “minor” allele (≥ 10%), the percentage of phenotype explained by each SNP association was determined using the difference between the mean survival time of mice carrying either allele and dividing it by the total difference between the two polar strains (24.6 h) (Figure E1). The amount of phenotype explained by the SNP associations was substantial (range: 17–34%) (Table E1).
Figure 1.
Mouse strains vary in sensitivity to acrolein-induced acute lung injury. (A) Acute lung injury survival time of 40 mouse strains. Mice were exposed to 10 ppm acrolein for up to 24 hours and survival time recorded hourly. Values are mean ± SE (n = 6 to 16 mice/strain except MRL/MpJ, n = 3). (B) Haplotype association map for acrolein-induced acute lung injury in mice. The scatter (Manhattan) plot of corresponding −log(P) association probability for single nucleotide polymorphism (SNP) at indicated chromosomal location (ordinate). Transcripts with SNP associations of −log(P) > 4.0 were selected for further analysis. (C) Candidate genes on mouse chromosome 2 associated with acrolein-induced acute lung injury. The Manhattan plot of SNP associations indicates chromosomal location (ordinate) and corresponding –log(P) association probability. Genes with one or more significant SNP (−log[P] > 6.0) included Galnt13 (UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalacto-aminyl transferase 13), Acvr1 (activin A receptor, type 1), and Ccdc148 (coiled-coil domain containing 148). Acvr1c = activin A receptor, type IC; Cytip = cytohesin 1 interacting protein; Galnt5 = UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetyl-galactosaminyl transferase 5; Gpd2 = glycerol phosphate dehydrogenase 2, mitochondrial; Kcnj3 = potassium inwardly-rectifying channel, subfamily J, member 3; Nr4a2 = nuclear receptor subfamily 4, group A, member 2.
Candidate Genes on Chromosome 2 Associated with Acrolein-induced Acute Lung Injury
A number of SNPs with association probabilities −log(P) of 5.0 to 7.0 were identified in a small region on chromosome 2 (54–59 Mbp). Genes in this region with one or more significant SNP associations included Galnt13, Acvr1, and Ccdc148 (Figure 1C). Because any SNP association may encompass blocks of associated variants that reside near the identified SNP (28), functional assessment was performed on SNPs within 91 genes that mapped within 1 Mbp of a significant/suggestive SNP association. Using 10% or greater allelic frequency and 10% or greater phenotype explained as threshold criteria, 16 candidate genes qualified for further analysis.
Histological, Bronchoalveolar Lavage, and Transcriptional Assessment of Acrolein-induced Lung Tissue in SM/J and 129X1/SvJ Mouse Strains
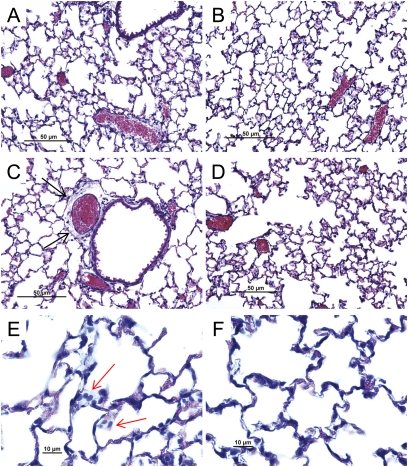
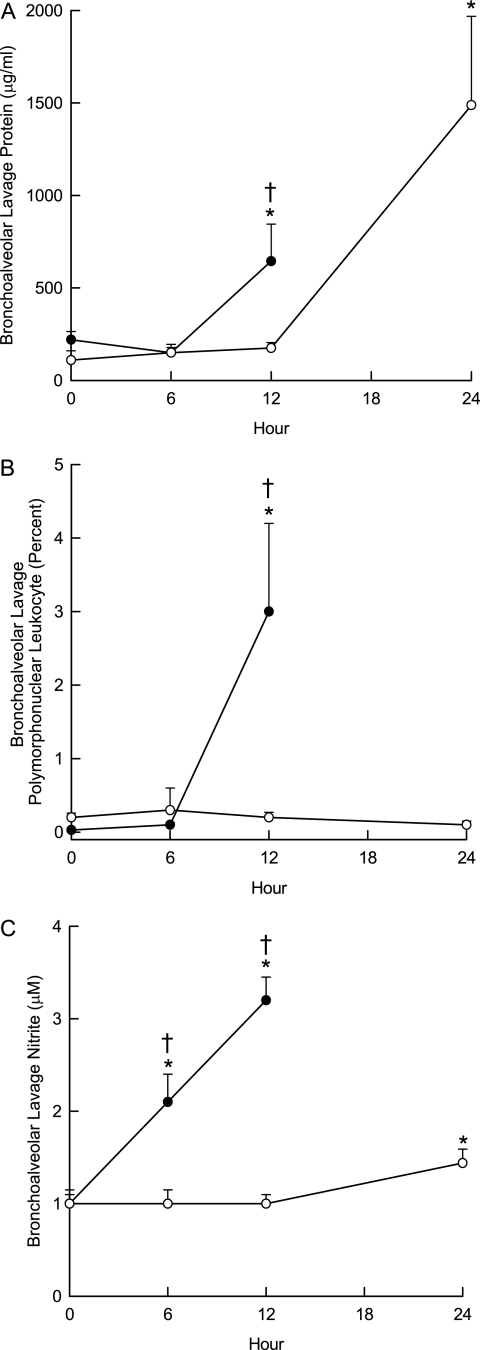
To further assess susceptibility, a sensitive and a resistant mouse strain were contrasted by detailed characterization. For the sensitive strain, we selected SM/J and avoided wild-derived (e.g., MOLF/EiJ, SPRET/EiJ, or PERA/EiJ) or repository (e.g., RIIIS/J) strains. The mean survival time for the SM/J strain (17.2 ± 0.46 h) was not significantly different from that of MOLF/EiJ. The 129X1/SvJ was selected as representative of a resistant strain. Histological analyses of these strains were consistent with acute lung injury. Perivascular enlargement and leukocyte infiltration were more evident in the sensitive SM/J strain than in the resistant 129X1/SvJ strain (Figure 2). Similarly, bronchoalveolar lavage protein concentration (Figure 3A), polymorphonuclear leukocyte percentage (Figure 3B), and nitrite concentration (Figure 3C) increased sooner in the SM/J than in 129X1/SvJ mice.
Figure 2.
Histological assessment of lung tissue from (A) control SM/J mice, (B) control 129X1/SvJ mice, (C, E) acrolein-exposed SM/J mice, or (D, F) acrolein-exposed 129X1/SvJ mice. Consistent with acute lung injury, (C) perivascular enlargement (black arrow) and (E) leukocyte infiltration (red arrow) were more evident in the (C, E) sensitive SM/J strain than in the (D, F) resistant 129X1/SvJ strain. Mice were exposed to filtered air (control) or to acrolein (10 ppm, 17 h) and killed. Lung tissue was obtained, fixed in formaldehyde, and 5-μm sections prepared with hematoxylin and eosin stain. Bars indicate magnification.
Figure 3.
Characterization of acute lung injury by bronchoalveolar lavage. Mice were exposed to 10 ppm acrolein for 0 (filtered air control), 6, or 12 hours, killed, and bronchoalveolar lavage performed with (Ca2+, Mg2+ free) phosphate buffered saline. (A) Bronchoalveolar lavage protein increased sooner in the sensitive (SM/J) than in the resistant (129X1/SvJ) mouse strain. Lavage fluid was centrifuged and total protein in cell-free supernatants was measured using a bicinchoninic acid assay. (B) Bronchoalveolar lavage polymorphonuclear leukocytes increased in the sensitive (SM/J) but not in the resistant (129X1/SvJ) mouse strain. After centrifugation, cell pellet was suspended and an aliquot (200 μl) were cytocentrifuged and the cells were stained with Hemacolor for differential cell analysis according to standard cytological procedures. (C) Bronchoalveolar lavage nitrite concentration increased sooner in the sensitive (SM/J) than in the resistant (129X1/SvJ) mouse strain. Supernatant was analyzed using a fluorometric method in which nitrite reacted with 2,3-diaminonaphthalene. *Significantly different from strain-matched control as determined by analysis of variance with an all pairwise multiple comparison procedure (Holm-Sidak method). †Significantly different between the sensitive SM/J and resistant 129X1/SvJ mouse strain as determined by analysis of variance with an all pairwise multiple comparison procedure (Holm-Sidak method).
Lung transcript levels were also compared between SM/J and 129X1/SvJ strains using microarray analysis (Table E2). Previously identified transcripts that are altered in mouse lung during acute lung injury include IL-6 (29) and metallothionein 2 (MT2) (30). Microarray results indicated that IL6 transcripts at 6 or 12 hours increased more in SM/J than in 129X1/SvJ lung (Figure E2, top). These differences were confirmed by qRT-PCR (although microarray increases were less than qRT-PCR increases). In contrast, MT2 transcripts increased in both strains when compared with strain-matched controls but were not different between the strains (Figure E2, bottom).
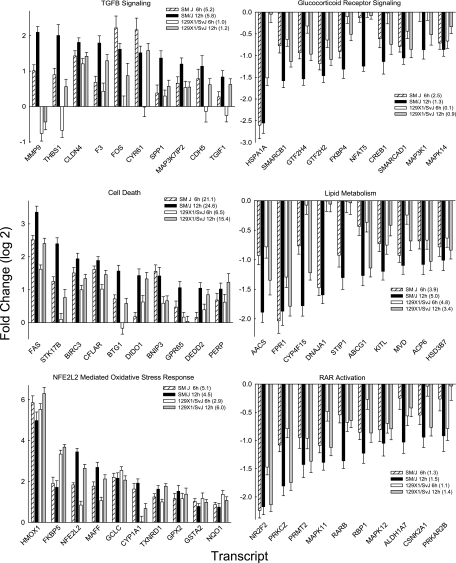
Further evaluation of microarray transcripts that increased with exposure (fold log 2 ≥ 1.0; P < 0.01) revealed enriched pathways including transforming growth factor-β (TGFB) signaling, cell death, and nuclear factor, erythroid derived 2, like 2 (NFE2L2)-mediated oxidative stress response for transcripts that increased with exposure (Figure 4; Table 1). Transcripts in TGFB signaling (e.g., SM/J −log(P) = 5.8 vs. 129X1/SvJ −log(P) = 1.2 at 12 h) or cell death pathway (e.g., SM/J −log(P) = 24.6 vs. 129X1/SvJ –log(P) = 15.4 at 12 h) were increased more in SM/J as compared with 129X1/SvJ mice. In contrast, the response of SM/J and 129X1/SvJ were similar in the NFE2L2-mediated oxidative stress response (e.g., SM/J –log(P) = 5.8 vs. 129X1/SvJ –log(P) = 6.0 at 12 h). The latter is noteworthy in that NFE2L2 activation leads to accumulation of transcripts encoding enzymes that may be viewed as protective (e.g., heme oxygenase [decycling] 1 [HMOX1]) during acute lung injury (31). One transcript in the TGFB signaling pathway, secreted phosphoprotein 1 (SPP1), was also evaluated using qRT-PCR. At 6 or 12 hours, SPP1 transcript increased in SM/J mice, but was not significantly different from control in 129X1/SvJ mice (Figure E3). The increase at 12 hours was significantly greater in the SM/J lung as compared with the 129X1/SvJ lung.
Figure 4.
Pathways enriched in transcripts in sensitive (SM/J) and resistant (129X1/SvJ) mouse lung during acrolein exposure. Mice were exposed to 10 ppm acrolein for 0 (filtered air control), 6, or 12 hours, killed, and lung tissue frozen in liquid nitrogen. Lung transcript levels were then quantified by microarray and compared with filtered air control. Pathway enrichment was determined using significant values (> twofold and P < 0.01 by analysis of variance) analyzed by Ingenuity Pathways Analysis. The top three enriched pathways/lists for “Canonical pathway,” “Biological function,” or “Toxicology list” categories were selected based on the combined 6- and 12-hour −log(P). The strain- and time-specific −log(P) value is presented in parentheses. Left: Increased transcripts were enriched in (top) transforming growth factor-β (TGFB) signaling, (middle) cell death, and (bottom) nuclear factor, erythroid derived 2, like 2 (NFE2L2)-mediated oxidative stress. Increased transcripts in TGFB signaling (e.g., SM/J –log(P) = 5.8 vs. 129X1/SvJ –log(P) = 1.2 at 12 h) and cell death pathway (e.g., SM/J –log(P) = 24.6 vs. 129X1/SvJ –log(P) = 15.4 at 12 h) were increased more in SM/J as compared with 129X1/SvJ mouse strains. In contrast, the response of SM/J and 129X1/SvJ were similar in the NFE2L2-mediated oxidative stress response (e.g., SM/J –log(P) = 5.8 vs. 129X1/SvJ –log(P) = 6.0 at 12 h). Right: Decreased transcripts were enriched in (top) glucocorticoid receptor signaling, (middle) lipid metabolism, and (bottom) retinoic acid receptor-α (RAR) activation. The responses in SM/J and 129X1/SvJ mouse strains were similar (except DnaJ [Hsp40] homolog, subfamily A, member 1 [DNAJA1] and stress-induced phosphoprotein 1 [STIP1]) in these pathways. Bars are mean ± SE (n = 5 mice/strain/time). Additional abbreviations: see Methods section in online supplement.
TABLE 1.
ENRICHED PATHWAYS OF LUNG TRANSCRIPTS IN SENSITIVE (SM/J) AND RESISTANT (129X1/SVJ) MOUSE STRAINS EXPOSE TO ACROLEIN
| SM/J | −log(P) | 129X1/SvJ | −log(P) | ||
|---|---|---|---|---|---|
| Increased | 6 h | 12 h | Increased | 6 h | 12 h |
| Canonical pathway | Canonical pathway | ||||
| TGFB signaling | 5.2 | 5.8 | Phospholipid degradation | 5.2 | 5.0 |
| IL-10 signaling | 4.0 | 5.5 | Sphingosine-1-phosphate signaling | 2.4 | 1.9 |
| mTOR signaling | 2.3 | 4.7 | Protein ubiquitination pathway | 1.8 | 2.6 |
| Biological function | Biological function | ||||
| Cell death | 21.1 | 24.6 | Cell death | 6.5 | 15.4 |
| Immune cell trafficking | 8.5 | 12.9 | Cell movement | 5.2 | 7.4 |
| Inflammatory response | 8.1 | 11.4 | Inflammatory response | 2.0 | 7.2 |
| Toxicology list | Toxicology list | ||||
| NFE2L2-mediated oxidative stress response | 5.1 | 5.8 | NFE2L2-mediated oxidative stress response | 2.9 | 6.0 |
| Oxidative stress | 7.1 | 4.3 | p53 signaling | 1.7 | 4.3 |
| p53 signaling | 4.3 | 3.7 | Antiapoptosis | 3.1 | 2.1 |
| Decreased | Decreased | ||||
| Canonical pathway | Canonical pathway | ||||
| Glucocorticoid receptor signaling | 2.5 | 1.3 | Ephrin receptor signaling | 1.5 | 2.4 |
| Renin-angiotensin signaling | 1.4 | 1.3 | Renin-angiotensin signaling | 1.4 | 0.6 |
| Lysine biosynthesis | 0.1 | 2.0 | Glucocorticoid receptor signaling | 0.1 | 0.9 |
| Biological function | Biological function | ||||
| Lipid metabolism | 3.9 | 4.9 | Lipid metabolism | 3.4 | 4.8 |
| Cardiovascular system development and function | 3.4 | 4.5 | Cardiovascular system development and function | 2.9 | 4.5 |
| Respiratory system development and function | 3.5 | 3.2 | Respiratory system development and function | 3.5 | 3.7 |
| Toxicology list | Toxicology list | ||||
| RAR activation | 1.3 | 1.5 | Cholesterol biosynthesis | 1.8 | 3.5 |
| Cholesterol biosynthesis | 1.2 | 1.3 | RAR activation | 1.1 | 1.4 |
| Cytochrome P450 panel - substrate is an eicosanoid | 1.0 | 0.7 | Cytochrome P450 panel - substrate is an eicosanoid | 0.9 | 0.8 |
Definition of abbreviations: NFE2L2 = nuclear factor, erythroid derived 2, like 2; RAR = retinoic acid receptor.
Mice (n = 5 mice/strain/time) were exposed to 10 ppm acrolein, killed, lung removed, and RNA isolated. Transcript levels were then quantified by microarray and compared to filtered air control. Pathway enrichment was determined using significant values (> twofold and P < 0.01 by analysis of variance) with Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). The top three enriched pathways for “Canonical pathway,” “Biological function,” and “Toxicology list” categories were selected for increased and decreased expression changes, based on the −log(P) for the combined 6- and 12-h data.
For microarray transcripts that decreased with exposure, enriched pathways included glucocorticoid receptor signaling, lipid metabolism, and retinoic acid receptor activation (Figure 4). In general, pathways with decreased transcripts were similar between these strains as determined by the enrichment probability −log(P). Individual transcripts among these pathways were decreased more in the SM/J than in 129X1/SvJ mice, however.
Additional pathways that were enriched in SM/J but were not in 129X1/SvJ mice included (1) increased transcripts: IL-10 signaling, mTOR signaling, and immune cell trafficking; and (2) decreased transcripts: lysine biosynthesis (Table 1). Pathways that were enriched in 129X1/SvJ but not in SM/J mice included (1) increased transcripts: phospholipid degradation, sphingosine-1-phosphate signaling, protein ubiquitination pathway, cell movement, and antiapoptosis; and (2) decreased transcripts: ephrin receptor signaling.
Assessment of Basal Transcript Levels of 16 Candidate Genes for Acrolein-induced Acute Lung Injury in Mice
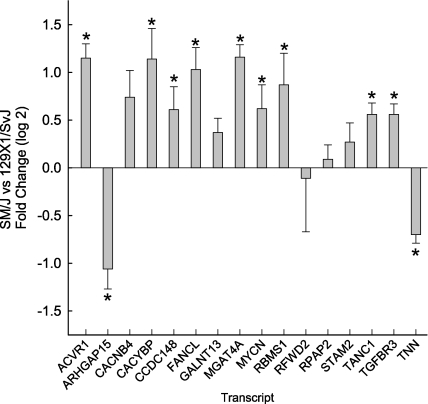
Having found that SM/J and 129X1/J mice differ in the time-course of acrolein-induced lung injury, the identified 16 candidate genes were then evaluated by qRT-PCR. Initially, basal levels were compared and transcript levels of 11 of the 16 genes were significantly different between strains (Figure 5).
Figure 5.
Assessment of basal transcript levels of 16 candidate genes for acrolein-induced acute lung injury in mice. Mice were exposed to filtered air (control) and lung mRNA isolated. Basal transcript levels of SM/J (sensitive) mouse strain were compared with those of 129X1/SvJ (resistant) mouse strain as determined by quantitative real-time polymerase chain reaction. Values are mean ± SE of the transcript level of SM/J (n = 8) as compared with the 129X1/SvJ (n = 8). *Significantly different between the sensitive SM/J and resistant 129X1/SvJ mouse strain as determined by analysis of variance with an all pairwise multiple comparison procedure (Holm-Sidak method). ACVR1 = activin A receptor, type 1; ARHGAP15 = Rho GTPase activating protein 15; CACNB4 = calcium channel, voltage-dependent, β 4 subunit; CACYBP = calcyclin binding protein; CCDC148 = coiled-coil domain containing 148; FANCL = Fanconi anemia, complementation group L; GALNT13 = UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase 13; MGAT4A = mannoside acetylglucosaminyltransferase 4, isoenzyme A; MYCN = v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian); RBMS1 = RNA binding motif, single stranded interacting protein 1; RFWD2 = Ring finger and WD repeat domain 2; RPAP2 = RNA polymerase II associated protein 2; STAM2 = signal transducing adaptor molecule (SH3 domain and ITAM motif) 2; TANC1 = tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1; TGFBR3 = transforming growth factor, β receptor III; TNN = tenascin N.
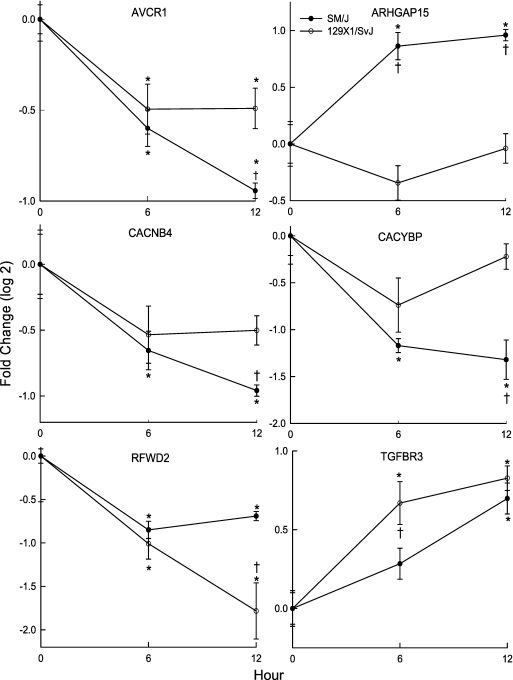
Next, the time course of transcripts of the 16 candidate genes was measured during exposure and compared with the strain-matched control. This analysis identified six transcripts (ACVR1, ARHGAP15, CACNB4, CACYBP, RFWD2, and TGFBR3) that were different between the strains with exposure (Figure 6). For each of these transcripts, the corresponding gene was interrogated to determine whether it contained SNPs that could alter putative transcriptional factor recognition sites (i.e., expression SNP or eSNP). Examining the sequence variability within −2,000 bp in the 5′UTR through intron 1, five genes, Acvr1, Arhgap15, Cacybp, Rfwd2, and Tgfbr3, were identified to have eSNPs that could eliminate ELK1, GAL4, HES-1, YY1, and NF-1 binding motifs as determined by Transcription Element Search Software (32), respectively (Table 2). The survival time associated with either allele was then determined for the mouse population. Mice possessing these eSNPs exhibited mean survival time of 17–30% of the difference between the mean survival times of the most polar strains. The sixth gene, Cacnb4, lacked an eSNP associated with a difference in phenotype in the 5′UTR but contained an SNP that would produce a variant splicing transcript.
Figure 6.
Transcript levels of six candidate genes that differed between the SM/J and 129X1/SvJ mouse strains after acrolein exposure. Mice were exposed to filtered air (control, 0 h) or to acrolein (10 ppm) for 6 or 12 hours, lung mRNA isolated, and transcript expression levels determined by quantitative real-time polymerase chain reaction. Values are mean ± SE of the transcript level of SM/J (n = 8) as compared with the 129X1/SvJ (n = 7–8). *Significantly different from strain-matched control as determined by analysis of variance with an all pairwise multiple comparison procedure (Holm-Sidak method). †Significantly different between the sensitive SM/J and resistant 129X1/SvJ mouse strain as determined by analysis of variance with an all pairwise multiple comparison procedure (Holm-Sidak method). ACVR1 = activin A receptor, type 1; ARHGAP15 = Rho GTPase activating protein 15; CACNB4 = calcium channel, voltage-dependent, β 4 subunit; CACYBP = calcyclin binding protein; RFWD2 = Ring finger and WD repeat domain 2; TGFBR3 = transforming growth factor-β receptor III.
TABLE 2.
FUNCTIONAL SINGLE NUCLEOTIDE POLYMORPHISM ASSOCIATIONS WITHIN CANDIDATE GENES
| dbSNP | SNP Type | Phenotype (%) | Symbol | Description | Chr | Start Position |
|---|---|---|---|---|---|---|
| rs32922832 | exon15, 3′UTR | 24.8 | Mgat4a | Mannoside acetylglucosaminyltransferase 4, isoenzyme A | 1 | 37501401 |
| rs31320021 | intron 1 (YY1) | 30.1 | Rfwd2 | Ring finger and WD repeat domain 2 | 1 | 161162457 |
| rs32034023 | Thr1123Met | 25.9 | Tnn | Tenascin N | 1 | 162015167 |
| rs31343079 | 5′UTR (HES-1) | 22.9 | Cacybp | Calcyclin binding protein | 1 | 162133052 |
| rs27124906 | 5′UTR (GAL4) | 26.6 | Arhgap15 | Rho GTPase activating protein 15 | 2 | 43604344 |
| rs27914711 | variant 2 | 18.8 | Cacnb4 | Calcium channel, voltage-dependent, beta 4 subunit | 2 | 52532101 |
| rs6406107 | 5′UTR (ELK1) | 18.9 | Acvr1 | Activin A receptor, type 1 | 2 | 58419239 |
| rs27956803 | Gln81Arg | 25.1 | Ccdc148 | Coiled-coil domain containing 148 (BC062650) | 2 | 58674108 |
| rs51612205 | 5′UTR (NF-1) | 17.4 | Tgfbr3 | Transforming growth factor, beta receptor III | 5 | 107535589 |
| rs4228631 | Asp1Ala | 11.1 | Fancl | Fanconi anemia, complementation group L | 11 | 26287084 |
| rs48672482 | intron 1 (MTF-1) | 28.3 | Mycn | V-myc myelocytomatosis viral related oncogene, neuroblastoma derived | 12 | 12942899 |
dbSNP: single nucleotide polymorphism database identification number. SNP Type: expression SNP (eSNP) includes predicted transcription factor binding site that is lost by polymorphism in parenthesis, and nonsynonymous (cnSNP) includes amino acid substitution with location in gene between amino acid. Phenotype (%): percentage phenotype (mean survival time) difference explained by mice with either allele as a percentage of the difference of two most polar strains. Chr: chromosome location of gene. Start Position: first basepair location of gene.
In addition, four transcripts (CCDC148, FANCL, MYCN, and TNN) were altered by exposure but were not different between the strains (Figure E4). The corresponding genes were then interrogated to determine whether they contained a coding nonsynonymous SNP (i.e., cnSNP) that could produce amino acid changes. Two genes, Ccdc148 and Fancl, had Gln81Arg and Asp1Ala amino acid substitution, respectively (Table 2). Although lacking a cnSNP, Mycn had an eSNP that could eliminate a MTF-1 binding motif. In Tnn, three SNPs (rs4877414, rs46709434, and rs49224805) could produce Ala310Thr, Lys333Gln, and Arg562Cys amino acid substitutions, respectively. Mice having either allele in the identified SNPs exhibited phenotypic differences in survival of 11 to 28%.
The predicted amino acid substitutions (missense mutations) were examined using Prosite (33, 34) to determine whether they are situated in functional domains of the encoded protein. In CCDC148, the rs27956803 Gln81Arg substitution resulted in a gain of a predicted protein kinase C phosphorylation site at amino acid 79 to 81. The Asp1Ala substitution in FANCL was not in a predicted functional domain. In TNN, three amino acid substitutions are located in fibronectin type III domains and the Ala310Thr substitution resulted in a loss of a predicted protein kinase C phosphorylation site.
Last, MGAT4A transcripts did not change significantly with exposure in either strain of mouse tested. As noted above, basal transcript levels were different between the strains. The corresponding sequence was therefore interrogated for eSNP and cnSNP, and none were observed. The 3′UTR was then examined and an SNP was observed in exon 15/3′-UTR, which is in a region that may alter message stability.
Together, the difference in transcript levels combined with the in silico sequence assessment yielded 11 candidates (Mgat4a, Rfwd2, Tnn, Cacybp, Arhgap15, Cacnb4, Acvr1, Ccdc148, Tgfbr3, Fancl, and Mycn) with probable functional associations to diminished survival resulting from acrolein-induced acute lung injury (Table 2). Candidate genes with functional SNP associations were located in eight chromosomal regions (Mbp): 1 (37), 1 (161), 2 (43), 2 (52), 2 (58), 5 (107), 11 (26), and 12 (12) and independently could explain 11 to 30% of the phenotype.
Acvr1 Promoter Analysis
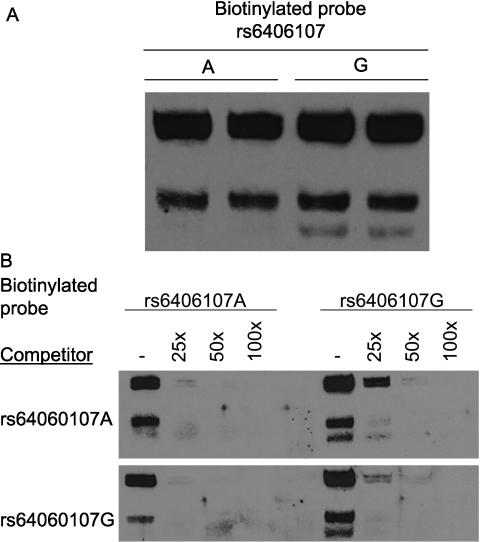
Three SNPs (rs33408603, rs6406068, and rs6406107) were identified in the 5′UTR region of Acvr1. However, few strains had previously been genotyped (129X1/SvJ, C57BL/6J, and A/J for rs33408603, and CZECHII/EiJ and C57BL/6J for rs6406068 or rs6406107). To obtain additional genotype information on these SNPs, 28 strains were genotyped by PCR amplification–DNA sequencing (Table E3). One SNP, rs33408603 (A/G), was observed only in the 129X1/SvJ strain (A-allele). The remaining SNPs exhibited an identical haplotype in all strains typed (i.e., strains with the rs6406068 C-allele had the rs6406107 A-allele). The allele frequency of this SNP haplotype was 12% and could explain 19% of the phenotype. Analysis of the possible consequence of these SNPs indicated that the rs6406107 A-allele would result in the loss of a putative ELK1, member of ETS oncogene family (ELK1) transcription factor binding site. In vitro, an Acvr1 promoter region–derived oligonucleotide containing the rs6406107 G-variant more effectively competed the nuclear protein-binding capacity of a labeled probe than that containing the A-variant (Figure 7).
Figure 7.
Single nucleotide polymorphism (rs6406107) in the 5′ untranslated region of Acvr1 diminishes nuclear protein binding capacity. (A) Oligonucleotide probe containing the G-allele, in contrast to the A-allele, formed a distinct fast-migrating complex (lower band). Electrophoretic mobility shift assay (EMSA) of nuclear protein extract prepared from mouse lung epithelial cells (MLE-15) and biotinylated oligonucleotide probes containing the A- or G-allele. (B) Compared with the A-variant, the G-variant is a more effective competitor of protein-biotinylated DNA complexes formation. Competitive EMSA was performed with excess double-strained oligonucleotides that contained either the A- or G-variant. The biotinylated labeled rs6406107 A-variant oligonucleotide probe is readily competed by nonbiotinylated rs6406107 A- or G-variant, whereas the biotinylated labeled rs6406107 G-variant oligonucleotide probe is more avidly bound, in particular the fast migrating complex (right, lane 2).
DISCUSSION
Past studies using candidate gene approaches have been promising and have provided valuable insights into the genetic architecture of the complex trait of acute lung injury; however, these studies also have had recognized limitations (28, 35–37). For several reasons it has been difficult to assemble a large enough cohort of clinically identical individuals to perform a genome-wide association study in humans. Adding to this barrier are the difficulties in establishing functional significance of genetic associations (38). Thus, using genome-wide association approaches to identification of candidate genes and related pathways (39) in mice could be a valued additional approach (21).
Previously, considerable interstrain differences have been reported in acute lung injury susceptibility (24, 25). In this study with acrolein, we observed an approximately 2.5-fold difference in survival time between the most sensitive and resistant strains, and thus this trait was amenable to mapping. In past decades, QTL mapping has identified chromosomal regions containing genes affecting cancer, diabetes, hypertension, and other disease-related phenotypes. However, a major obstacle of identifying QTL genes is the difficulty of resolving large chromosomal regions (10–20 cM) into sufficiently small intervals to make positional cloning practical. With advances in genome sequencing, dense SNP maps have proven successful in the refinement of previous QTL regions and the identification of new genetic determinants of complex traits (21).
One concern about any SNP association study is that variants identified in genome-wide association study may confer a relatively small increment of risk (explaining 1–3% to the population variance) (28, 35–38). Here, we present a strategy to overcome this limitation in which SNPs within 1 Mbp of an association are assessed for functional consequences to survival in the mice tested. A threshold of 10% or greater survival time between the most sensitive and resistant strains was set for further assessment of candidates. Type I error was diminished by comparing the phenotype (i.e., individual survival time) of every member of the population with the corresponding strain genotype. This approach also limits the associated SNPs to those with feasible functional consequence to expression (eSNP) or amino acid sequence (cnSNP).
Of the candidate genes identified, only Mycn has been associated with respiratory failure. Mice with a Mycn mutation (that reduces expression through alternative splicing) die at birth due to respiratory failure (40). However, this effect is probably due to defective lung development (41) and abnormal branching morphogenesis (42), because complete ablation of the gene is embryonic lethal. The other candidate genes identified in this study have not previously been associated with acute lung injury. Nonetheless, these genes are present in the lung and the corresponding proteins have functions related to the cell stress signaling in lung injury.
As with any study, this investigation has several limitations. Although acrolein-induced acute lung injury has relevance to smoke inhalation, numerous other agents (either chemical or infectious) can elicit acute lung injury (43). Until these other forms or phenotypes (e.g., lavage protein) of acute lung injury are evaluated, generalization to other forms of this condition is not warranted. Supportive evidence by another genetic approach (e.g., traditional back-cross or F2 cohort) that likewise identified the candidate genes identified with this haplotype association mapping analysis would strengthen the associations found in this study. In addition, although this approach may improve the assessment of the results obtained from a haplotype mapping analysis, functional assessment of each candidate gene will require further studies (e.g, gene-targeted or transgenic mice). Last, animal models are limited in that species differences in lung structure and function can diminish applicability to humans. Despite these limitations, the identified genes and related pathways may help to direct future human genetic studies that evaluate such pathways using selected tagSNPs.
One of the leading candidate genes was Acvr1. In mice, Acvr1 gene targeting produces a morphological gastrulation defect, which is embryonic lethal (44–46). In humans, mutations in the glycine-serine–rich (GS) activation domain or sites that interact with the GS domain of ACVR1 have been associated with fibrodysplasia ossificans progressiva, which is characterized by progressive heterotopic ossification that can lead to respiratory difficulties (47, 48). The ACVR1 GS domain is involved in phosphorylation and human mutations lead to a gain of function (i.e., augmented signaling) (49). In mice, postnatal expression of mutant ACVR1 can lead to ectopic ossification, but only when combined with infection. Corticosteroid inhibited ossification in mice, suggesting that mutant ACVR1 and inflammation are both required for ectopic ossification (50).
Although named activin A receptor, type 1 (a.k.a. activin receptor-like kinase 2, Alk2), ACVR1 does not bind activins (a.k.a. inhibins), but binds BMP2, 4, 6, and 7, through a heteromeric complex with BMPR2 (51–54) (Figure E5, Table E4). Critical to respiratory organogenesis and development (55, 56), BMPs elicit various effects in adult tissues through type I and II receptors, which in turn phosphorylate receptor-regulated R-SMAD protein (primarily SMAD1, 5, and 8) (57, 58). On activation, R-SMAD proteins associate with the common mediator SMAD4 and translocate to the nucleus, where they act as transcription factors to regulate expression of target genes, including the inhibitory SMADs (I-SMAD), SMAD6, and SMAD7 (59). SMAD6 and SMAD7 inhibit/modulate TGFB/BMP signaling by interfering with the activation of other SMADS (59–61), thereby providing negative feedback control (62).
In this study we found that lung ACVR1 transcripts decreased more in the sensitive SM/J than the resistant 129X1/SvJ mice. In vitro, an Acvr1 promoter region SNP rs6406107 diminished nuclear protein-binding capacity of a labeled probe and was associated with decreased survival in mice. This SNP could lead to diminished ACVR1 transcript levels because the A-allele would eliminate a putative ELK1 binding site. ELK1 can be phosphorylated by a TNF-initiated, JUN-mediated mechanism in pulmonary epithelial cells (63). A possible consequence of decreased ACVR1 transcripts would be augmented TGFB/BMP signaling. This is consistent with increases in transcripts encoded by TGFB target genes noted in the microarray. Previously, TGFB/BMP signaling has been associated with adverse effects during acute lung injury (64–66). Active TGFB1 is increased in edema fluid obtained from patients with acute lung injury (67–70), and TGFB1 decreases pulmonary endothelial (71) or epithelial integrity (72) and diminishes epithelial fluid transport (73, 74). The determination whether ACVR1 may serve to limit TGFB/BMP signaling during lung injury is worthy of future investigation.
Compared with 129X1/SvJ mice, several stress-response transcripts (e.g., CACYBP, DNAJA1, FKBP4, HSPA1A, NFAT5, and STIP1) decreased more in the SM/J lung. Inasmuch as these transcripts encoded chaperon proteins, a decrease could destabilize existing proteins (i.e., aggregation and unfolding of newly translated proteins) and diminish the effectiveness of ubiquitin-proteasome pathway (75, 76). Ubiquitin-proteasome proteins may interact with other candidates identified (77). For example, ACVR1 interacts with FANCL (78) and modulates SMURF2 ubiquitination of various proteins, including SMADs. Two candidate genes, Rfwd2 and Cacybp, encode proteins with ubiquitin-protein ligase activity. Besides limiting TP53 (a.k.a. p53) (79), RFWD2 suppresses JUN (80) and FOXO1 (81) and other transcription factor accumulation and thereby limits cell stress (82). For example, JUN diminishes surfactant-associated protein B synthesis and thereby increases mortality during acute lung injury (3, 83), and FOXO1 is critical to maintaining claudin 5 in acrolein-induced acute lung injury (84).
In summary, haplotype association mapping, microarray/qRT-PCR analyses, and in silico SNP analysis identified 11 candidate genes (Acvr1, Arhgap15, Cacnb4, Cacybp, Ccdc148, Fancl, Mycn, Mgat4a, Rfwd2, Tgfbr3, and Tnn) associated with acrolein-induced acute lung injury in mice. Several genes were related and encoded receptors (ACVR1, TGFBR3), transcription factors (MYCN, possibly CCDC148), and ubiquitin-proteasome (RFWD2, FANCL, CACYBP) proteins that may interact to modulate cell signaling. The Acvr1 SNP rs6406107 eliminates a putative transcription factor binding site and diminished DNA–protein binding making this gene worthy of future investigations in acute lung injury.
Supplementary Material
Originally Published in Press as DOI: 10.1164/rccm.201006-0912OC on February 25, 2011
Supported by National Institutes of Health grants ES015675, HL077763, and HL085655 (G.D.L); HL091938 (Y.P.P.D.), HG003749 and LM009662 (M.M.); HL084932 and HL095397 (N.K.); AT003203, AT005522, CA113793, and CA134433 (M.Y.); and HL075562 (D.R.P.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author Disclosure: G.D.L. is employed by the University of Pittsburgh. V.J.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.S.J. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.A.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.D. is employed by the University of Cincinnati. H.P.-V. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.A.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Y.P.P.D. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Q.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.J.V. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.M. is employed by the University of Cincinnati. N.K. was a consultant for Sanofi-Aventis and Stromedix. He received lecture fees form Medimmune and owns a patent on use of microRNAs to diagnose and treat IPF Us or peripheral blood biomarkers to diagnose and predict outcome in IPF. M.Y. is employed by the Medical College of Wisconsin. D.R.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 2.Rehberg S, Maybauer MO, Enkhbaatar P, Maybauer DM, Yamamoto Y, Traber DL. Pathophysiology, management and treatment of smoke inhalation injury. Expert Rev Respir Med 2009;3:283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bein K, Wesselkamper SC, Liu X, Dietsch M, Majumder N, Concel VJ, Medvedovic M, Sartor MA, Henning LN, Venditto C, et al. Surfactant-associated protein B is critical to survival in nickel-induced injury in mice. Am J Respir Cell Mol Biol 2009;41:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traber DL, Hawkins HK, Enkhbaatar P, Cox RA, Schmalstieg FC, Zwischenberger JB, Traber LD. The role of the bronchial circulation in the acute lung injury resulting from burn and smoke inhalation. Pulm Pharmacol Ther 2007;20:163–166. [DOI] [PubMed] [Google Scholar]
- 5.Erickson SE, Martin GS, Davis JL, Matthay MA, Eisner MD; NIH NHLBI ARDS Network. Recent trends in acute lung injury mortality: 1996–2005. Crit Care Med 2009;37:1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsushima K, King LS, Aggarwal NR, De Gorordo A, D'Alessio FR, Kubo K. Acute lung injury review. Intern Med 2009;48:621–630. [DOI] [PubMed] [Google Scholar]
- 7.Ware LB, Koyama T, Billheimer DD, Wu W, Bernard GR, Thompson BT, Brower RG, Standiford TJ, Martin TR, Matthay MA; NHLBI ARDS Clinical Trials Network. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest 2010;137:288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leikauf GD, Leming LM, O'Donnell JR, Doupnik CA. Bronchial responsiveness and inflammation in guinea pigs exposed to acrolein. J Appl Physiol 1989;66:171–178. [DOI] [PubMed] [Google Scholar]
- 9.Deshmukh HS, Shaver C, Case LM, Dietsch M, Wesselkamper SC, Hardie WD, Korfhagen TR, Corradi M, Nadel JA, Borchers MT, et al. Acrolein-activated matrix metalloproteinase 9 contributes to persistent mucin production. Am J Respir Cell Mol Biol 2008;38:446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshmukh HS, McLachlan A, Atkinson JJ, Hardie WD, Korfhagen TR, Dietsch M, Liu Y, Di PY, Wesselkamper SC, Borchers MT, et al. Matrix metalloproteinase-14 mediates a phenotypic shift in the airways to increase mucin production. Am J Respir Crit Care Med 2009;180:834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hales CA, Barkin PW, Jung W, Trautman E, Lamborghini D, Herrig N, Burke J. Synthetic smoke with acrolein but not HCl produces pulmonary edema. J Appl Physiol 1988;64:1121–1133. [DOI] [PubMed] [Google Scholar]
- 12.Hales CA, Musto SW, Janssens S, Jung W, Quinn DA, Witten M. Smoke aldehyde component influences pulmonary edema. J Appl Physiol 1992;72:555–561. [DOI] [PubMed] [Google Scholar]
- 13.Siempos II, Vardakas KZ, Kyriakopoulos CE, Ntaidou TK, Falagas ME. Predictors of mortality in adult patients with ventilator-associated pneumonia: a meta-analysis. Shock 2010;33:590–601. [DOI] [PubMed] [Google Scholar]
- 14.Flores C, Pino-Yanes Mdel M, Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care 2008;12:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy AJ, Kleeberger SR. Genetic polymorphisms associated with acute lung injury. Pharmacogenomics 2009;10:1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao L, Barnes KC. Recent advances in genetic predisposition to clinical acute lung injury. Am J Physiol Lung Cell Mol Physiol 2009;296:L713–L725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheu CC, Zhai R, Wang Z, Gong MN, Tejera P, Chen F, Su L, Thompson BT, Christiani DC. Heme oxygenase-1 microsatellite polymorphism and haplotypes are associated with the development of acute respiratory distress syndrome. Intensive Care Med 2009;35:1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002;420:520–562. [DOI] [PubMed] [Google Scholar]
- 19.Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2004;2:e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 2006;38:888–895. [DOI] [PubMed] [Google Scholar]
- 21.Burgess-Herbert SL, Tsaih SW, Stylianou IM, Walsh K, Cox AJ, Paigen B. An experimental assessment of in silico haplotype association mapping in laboratory mice. BMC Genet 2009;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, et al. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science 2004;306:690–695. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE 2007;2:e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prows DR, Shertzer HG, Daly MJ, Sidman CL, Leikauf GD. Genetic analysis of ozone-induced acute lung injury in sensitive and resistant strains of mice. Nat Genet 1997;17:471–474. [DOI] [PubMed] [Google Scholar]
- 25.Wesselkamper SC, Prows DR, Biswas P, Willeke K, Bingham E, Leikauf GD. Genetic susceptibility to irritant-induced acute lung injury in mice. Am J Physiol Lung Cell Mol Physiol 2000;279:L575–L582. [DOI] [PubMed] [Google Scholar]
- 26.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ Jr, Wesselkamper SC, Singer JB, Hill AE, Nadeau JH, Leikauf GD. Reciprocal congenic lines of mice capture the aliq1 effect on acute lung injury survival time. Am J Respir Cell Mol Biol 2008;38:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variant create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell SA, Gammon K, Bachurski CJ, Wiest JS, Leikauf JE, Prows DR, Leikauf GD. Differential gene expression in the initiation and progression of nickel-induced acute lung injury. Am J Respir Cell Mol Biol 2000;23:466–474. [DOI] [PubMed] [Google Scholar]
- 30.Wesselkamper SC, McDowell SA, Medvedovic M, Dalton TP, Deshmukh HS, Sartor MA, Case LM, Henning LN, Borchers MT, Tomlinson CR, et al. The role of metallothionein in the pathogenesis of acute lung injury. Am J Respir Cell Mol Biol 2006;34:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter SW, Choi AM. Heme oxygenase-1/carbon monoxide: novel therapeutic strategies in critical care medicine. Curr Drug Targets 2010;11:1485–1494. [DOI] [PubMed] [Google Scholar]
- 32.Schug J. Chapter 2.6. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequence. In: Current protocols in bioinformatics. Baxevanis AD, editors. New York, NY: J. Wiley and Sons; 2003. [DOI] [PubMed]
- 33.Sigrist CJA, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform 2002;3:265–274. [DOI] [PubMed] [Google Scholar]
- 34.Ganguly K, Stoeger T, Wesselkamper SC, Reinhart C, Sartor MA, Medvedovic M, Tomlinson CR, Bolle I, Mason JM, Leikauf GD, et al. Candidate genes controlling pulmonary function in mice: transcript profiling and predicted protein structure. Physiol Genomics 2007;31:410–421. [DOI] [PubMed] [Google Scholar]
- 35.Goldstein DB. Common genetic variation and human traits. N Engl J Med 2009;360:1696–1698. [DOI] [PubMed] [Google Scholar]
- 36.Kraft P, Hunter DJ. Genetic risk prediction–are we there yet? N Engl J Med 2009;360:1701–1703. [DOI] [PubMed] [Google Scholar]
- 37.Dermitzakis ET, Clark AG. Genetics. Life after GWA studies. Science 2009;326:239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirschhorn JN. Genomewide association studies–illuminating biologic pathways. N Engl J Med 2009;360:1699–1701. [DOI] [PubMed] [Google Scholar]
- 40.Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev 1992;6:691–704. [DOI] [PubMed] [Google Scholar]
- 41.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev 1992;6:2235–2247. [DOI] [PubMed] [Google Scholar]
- 42.Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol 2005;283:226–239. [DOI] [PubMed] [Google Scholar]
- 43.Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol 2008;295:L379–L399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishina Y, Crombie R, Bradley A, Behringer RR. Multiple roles for activin-like kinase-2 signaling during mouse embryogenesis. Dev Biol 1999;213:314–326. [DOI] [PubMed] [Google Scholar]
- 45.Gu Z, Reynolds EM, Song J, Lei H, Feijen A, Yu L, He W, MacLaughlin DT, van den Eijnden-van Raaij J, Donahoe PK, et al. The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development 1999;126:2551–2561. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu Y, Scott G, Nagy A, Kaartinen V, Mishina Y. BMP type I receptor ALK2 is essential for proper patterning at late gastrulation during mouse embryogenesis. Dev Dyn 2007;236:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, Connor JM, Delai P, Glaser DL, LeMerrer M, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet 2006;38:525–527. [DOI] [PubMed] [Google Scholar]
- 48.Kaplan FS, Xu M, Seemann P, Connor JM, Glaser DL, Carroll L, Delai P, Fastnacht-Urban E, Forman SJ, Gillessen-Kaesbach G, et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum Mutat 2009;30:379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song GA, Kim HJ, Woo KM, Baek JH, Kim GS, Choi JY, Ryoo HM. Molecular consequences of the ACVR1(R206H) mutation of fibrodysplasia ossificans progressiva. J Biol Chem 2010;285:22542–22553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein M L, Hong DW, McManus PM, Katagiri T, Sachidanandan C, et al. BMP type I receptor inhibition reduces heterotropic ossification. Nat Med 2008;14:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koenig BB, Cook JS, Wolsing DH, Ting J, Tiesman JP, Correa PE, Olson CA, Pecquet AL, Ventura F, Grant RA, et al. Characterization and cloning of a receptor for BMP-2 and BMP-4 from NIH 3T3 cells. Mol Cell Biol 1994;14:5961–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003;425:577–584. [DOI] [PubMed] [Google Scholar]
- 53.Renlund N, O'Neill FH, Zhang L, Sidis Y, Teixeira J. Activin receptor-like kinase-2 inhibits activin signaling by blocking the binding of activin to its type II receptor. J Endocrinol 2007;195:95–103. [DOI] [PubMed] [Google Scholar]
- 54.Kevenaar ME, Themmen AP, van Kerkwijk AJ, Valkenburg O, Uitterlinden AG, de Jong FH, Laven JS, Visser JA. Variants in the ACVR1 gene are associated with AMH levels in women with polycystic ovary syndrome. Hum Reprod 2009;24:241–249. [DOI] [PubMed] [Google Scholar]
- 55.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 1996;10:1580–1594. [DOI] [PubMed] [Google Scholar]
- 56.Warburton D, El-Hashash A, Carraro G, Tiozzo C, Sala F, Rogers O, De Langhe S, Kemp PJ, Riccardi D, Torday J, et al. Lung organogenesis. Curr Top Dev Biol 2010;90:73–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishidou Y, Kitajima I, Obama H, Maruyama I, Murata F, Imamura T, Yamada N, ten Dijke P, Miyazono K, Sakou T. Enhanced expression of type I receptors for bone morphogenetic proteins during bone formation. J Bone Miner Res 1995;10:1651–1659. [DOI] [PubMed] [Google Scholar]
- 58.Ebendal T, Bengtsson H, Söderström S. Bone morphogenetic proteins and their receptors: potential functions in the brain. J Neurosci Res 1998;51:139–146. [DOI] [PubMed] [Google Scholar]
- 59.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature 1997;389:622–666. [DOI] [PubMed] [Google Scholar]
- 60.Topper JN, Cai J, Qiu Y, Anderson KR, Xu YY, Deeds JD, Feeley R, Gimeno CJ, Woolf EA, Tayber O, et al. Vascular MADs: two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci USA 1997;94:9314–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao J, Shi W, Chen H, Warburton D. Smad7 and Smad6 differentially modulate transforming growth factor beta-induced inhibition of embryonic lung morphogenesis. J Biol Chem 2000;275:23992–23997. [DOI] [PubMed] [Google Scholar]
- 62.Afrakhte M, Morén A, Jossan S, Itoh S, Sampath K, Westermark B, Heldin CH, Heldin NE, ten Dijke P. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-beta family members. Biochem Biophys Res Commun 1998;249:505–511. [DOI] [PubMed] [Google Scholar]
- 63.Adiseshaiah P, Kalvakolanu DV, Reddy SP. A JNK-independent signaling pathway regulates TNF alpha-stimulated, c-Jun-driven FRA-1 protooncogene transcription in pulmonary epithelial cells. J Immunol 2006;177:7193–7202. [DOI] [PubMed] [Google Scholar]
- 64.Pittet JF, Griffiths MJ, Geiser T, Kaminski N, Dalton SL, Huang X, Brown LA, Gotwals PJ, Koteliansky VE, Matthay MA, et al. TGF-beta is a critical mediator of acute lung injury. J Clin Invest 2001;107:1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhainaut JF, Charpentier J, Chiche JD. Transforming growth factor-beta: a mediator of cell regulation in acute respiratory distress syndrome. Crit Care Med 2003;31:S258–S264. [DOI] [PubMed] [Google Scholar]
- 66.Wesselkamper SC, Case LM, Henning LN, Borchers MT, Tichelaar JW, Mason JM, Dragin N, Medvedovic M, Sartor MA, Tomlinson CR, et al. Gene expression changes during the development of acute lung injury: role of transforming growth factor beta. Am J Respir Crit Care Med 2005;172:1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamacher J, Lucas R, Lijnen HR, Buschke S, Dunant Y, Wendel A, Grau GE, Suter PM, Ricou B. Tumor necrosis factor-alpha and angiostatin are mediators of endothelial cytotoxicity in bronchoalveolar lavages of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med 2002;1660:651–656. [DOI] [PubMed] [Google Scholar]
- 68.Fahy RJ, Lichtenberger F, McKeegan CB, Nuovo GJ, Marsh CB, Wewers MD. The acute respiratory distress syndrome: a role for transforming growth factor-beta 1. Am J Respir Cell Mol Biol 2003;28:499–503. [DOI] [PubMed] [Google Scholar]
- 69.Budinger GR, Chandel NS, Donnelly HK, Eisenbart J, Oberoi M, Jain M. Active transforming growth factor-beta1 activates the procollagen I promoter in patients with acute lung injury. Intensive Care Med 2005;31:121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Synenki L, Chandel NS, Budinger GR, Donnelly HK, Topin J, Eisenbart J, Jovanovic B, Jain M. Bronchoalveolar lavage fluid from patients with acute lung injury/acute respiratory distress syndrome induces myofibroblast differentiation. Crit Care Med 2007;35:842–848. [DOI] [PubMed] [Google Scholar]
- 71.Hurst V IV, Goldberg PL, Minnear FL, Heimark RL, Vincent PA. Rearrangement of adherens junctions by transforming growth factor-beta1: role of contraction. Am J Physiol 1999;276:L582–L595. [DOI] [PubMed] [Google Scholar]
- 72.Ganter MT, Roux J, Miyazawa B, Howard M, Frank JA, Su G, Sheppard D, Violette SM, Weinreb PH, Horan GS, et al. Interleukin-1beta causes acute lung injury via alphavbeta5 and alphavbeta6 integrin-dependent mechanisms. Circ Res 2008;102:804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frank J, Roux J, Kawakatsu H, Su G, Dagenais A, Berthiaume Y, Howard M, Canessa CM, Fang X, Sheppard D, et al. Transforming growth factor-beta1 decreases expression of the epithelial sodium channel alphaENaC and alveolar epithelial vectorial sodium and fluid transport via an ERK1/2-dependent mechanism. J Biol Chem 2003;278:43939–43950. [DOI] [PubMed] [Google Scholar]
- 74.Roux J, Carles M, Koh H, Goolaerts A, Ganter MT, Chesebro BB, Howard M, Houseman BT, Finkbeiner W, Shokat KM, et al. Transforming growth factor beta1 inhibits cystic fibrosis transmembrane conductance regulator-dependent cAMP-stimulated alveolar epithelial fluid transport via a phosphatidylinositol 3-kinase-dependent mechanism. J Biol Chem 2010;285:4278–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J Biol Chem 2005;280:34178–34185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 2008;47:7001–7011. [DOI] [PubMed] [Google Scholar]
- 77.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci 2008;99:2107–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 2005;307:1621–1625. [DOI] [PubMed] [Google Scholar]
- 79.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004;429:86–92. [DOI] [PubMed] [Google Scholar]
- 80.Bianchi E, Denti S, Catena R, Rossetti G, Polo S, Gasparian S, Putignano S, Rogge L, Pardi R. Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J Biol Chem 2003;278:19682–19690.12615916 [Google Scholar]
- 81.Kato S, Ding J, Pisck E, Jhala US, Du K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem 2008;283:35464–35473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yi C, Li S, Chen X, Wiemer EA, Wang J, Wei N, Deng XW. Major vault protein, in concert with constitutively photomorphogenic 1, negatively regulates c-Jun-mediated activator protein 1 transcription in mammalian cells. Cancer Res 2005;65:5835–5840. [DOI] [PubMed] [Google Scholar]
- 83.Bein K, Leight H, Leikauf GD. JUN-CCAAT/enhancer binding protein (C/EBP) complexes inhibit surfactant associated protein B promoter activity. Am J Respir Cell Mol Biol. 2010; Dec 10 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 84.Jang AS, Concel VJ, Bein K, Brant KA, Liu S, Pope-Varsalona H, Dopico Jr RA, Di YP, Knoell DL, Barchowsky A, Leikauf GD. Endothelial dysfunction and Claudin 5 regulation during acrolein-induced lung injury. Am J Respir Cell Mol Biol 2011;44:483–490. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.