Abstract
Autophagy (self-eating) is an evolutionary conserved simple process by which cells target their own cellular organelles and long-lived proteins for degradation. Recently, this simple ancient process has proved to be involved in many biological aspects, including host defense, cell survival and death, innate and adaptive immunity, and cancer. The implications of aberrant regulation of autophagy in human diseases are just beginning to unravel. This is a brief review of recent progress in the association of autophagy with innate and adaptive immunity relevant to lung biology and disease.
Keywords: autophagy, immunity, toll-like receptors, asthma, viruses
THE PROCESS OF CELLULAR AUTOPHAGY
Autophagy is an evolutionary highly conserved process in virtually all eukaryotic cells. The three types of autophagy are macroautophagy, microautophagy, and chaperone-mediated autophagy. In this review, we focus on macroautophagy, hereafter called autophagy, which plays an important role in human health (1). Autophagy involves the sequestration of regions of the cytosol within double-membrane–bound compartments and delivery of the contents to the lysosomes for degradation. Autophagy has been shown to be an important regulator in many critical biological processes, such as cellular response to starvation, cell death, cancer, neurodegenerative diseases, and more recently, host defense. Rapidly accumulating evidence has shown that autophagy is a component of innate immunity and is involved in host defense elimination of pathogens, including viruses. Several pathogens, including common viral agents, have evolved to circumvent the autophagic responses (2).
Although the process of autophagy was described decades ago, its genetic components have been only recently identified through extensive studies using yeast genetics. Substantial progress has been made in understanding genetic factors contributing to the formation of the autophagic vesicles, but the molecular mechanisms and the signaling pathways leading to induction of autophagy in innate immunity are still enigmatic. Because autophagy is a rather new research field being unfolded, there are still many unanswered questions. Future studies are needed to map a more complete signaling pathway for autophagy, determine how the cells use autophagy for cell survival or death and evaluate the physiological role of autophagy in pathogens elimination.
CELLULAR REGULATION OF AUTOPHAGY
Autophagy can be divided into three stages: initiation, execution, and maturation (2). The most studied initiator of cellular autophagy is nutrient starvation. Starvation-induced autophagy is mainly regulated by target of rapamycin (TOR), a kinase that inhibits autophagy. TOR regulates protein translation and amino acid synthesis. Rapamycin and nutrient starvation induce autophagy by inhibiting TOR. Starvation has been suggested to induce autophagy independent of TOR inhibition, possibly by activating trimeric G proteins (2). It has also been shown that class III phosphoinositide 3-kinases (PI3Ks) are essential for starvation-induced autophagosome formation (1, 2). Commonly used inhibitors of autophagy, such as 3-methyladenine (3-MA) and wartmannin, target PI3Ks.
Execution of autophagy is mediated by a ubiquitination-like system that involves two key covalent conjugation pathways (3). In the first pathway, autophagy gene 12 (Atg12) is activated by transient covalent linkage first to Atg7 and then to Atg10 before becoming covalently linked to Atg5. Atg16 binding generates an Atg5-Atg12-Atg16 complex that is present in the autophagy isolation membrane. In the second conjugation pathway, a terminal amino acid at the carboxyl terminus of microtubule-associated protein light chain 3 (LC3, also known as Atg8) is cleaved by the cysteine protease Atg4 to leave an exposed conserved glycine residue that is required for autophagosome formation. Cleaved LC3 is then transiently linked to Atg7, then to Atg3, and then to phosphatidylethanolamine. Lipidation of LC3 is one of the best markers for autophagy because modified LC3 remains associated with autophagosomes until destruction at the autolysosomal stage. Maturation step for autophagosomes occurs by their fusion with endosomal vesicles forming intermediate autophagosomes, which acquire lysosome-associated membrane protein 1 (LAMP1) and LAMP2. These structures fuse with lysosomes and acquire cathepsins and acid phosphatases to become autolysosomes.
AUTOPHAGY AND INNATE IMMUNITY
Intracellular bacteria and viruses must survive the vigorous defense responses of the host. Recent evidence suggests that autophagy is such a host cell response. Several studies show that both bacteria and viruses are vulnerable to autophagic destruction and that successful pathogens have evolved strategies to circumvent autophagy (2). New evidence indicates that autophagy plays a role in the degradation of both extracellular bacterial pathogens that invade the cell (e.g., group A Streptococcus) (4) and true intracellular bacterial pathogens (e.g., Mycobacterium tuberculosis and Shigella flexneri) (5–7). Furthermore, pathogens have developed methods to evade autophagy. S. flexneri encodes a virulence protein, IscB, that blocks bacterial colocalization with the autophagosome. Mutant bacteria lacking IscB demonstrate impaired growth in wild-type cells but not in Atg5−/− cells deficient in autophagy (7). These studies indicate that autophagy is involved in innate immunity and that autophagy can be antagonized by pathogen virulence factors. Such antagonism is likely to be a common survival strategy for diverse intracellular pathogens (8). M. tuberculosis resides long-term in the phagosome of macrophages by interfering with phagolysosome biogenesis (9). The inhibition of phagosome–lysosome fusion is mediated in part by mycobacterial lipids that mimic mammalian phosphatidylinositols and inhibit phosphatidylinositol 3-phosphate (PI3P)-dependent membrane trafficking mechanisms. This block can be overcome by activation of cellular autophagy, either through starvation or overexpression of p47 guanosine triphosphatase. Activation of autophagy reversed the usual acidification defect observed in mycobacterial-containing phagosomes, resulting in colocalization of mycobacterium-containing phagosomes with autophagosomes and lysosomes (5, 10). Additional recent studies have provided evidence that autophagy is involved in response to several other pathogens, including Rickettsia species, Listeria monocytogenes, and Salmonella typhimurium (2, 11, 12).
The above studies raise many important questions. What are the signaling pathways mediating the recognition and execution of autophagy associated with innate immunity? Importantly, how are the pathogens recognized by the autophagic machinery? Variable sizes of autophagosomes have been reported engulfing intracellular microorganisms (7, 13). It is not known if these are specialized autophagosomes or if they are possibly the result of fusion of multiple autophagosomes. If so, would the number of autophagosomes or the size of the autophagosome determine the cell anti-pathogen defense? Recent studies provide some insight about a newly discovered pathway of LPS-induced autophagy and how cells use the autophagic pathway to mediate cell killing against pathogens (6, 14). These studies identify toll-like receptors (TLRs) as mediators of autophagy associated with innate immunity.
SIGNALING BY TLRS
TLRs sense conserved structures found in a broad range of pathogens, causing innate immune responses that include the production of inflammatory cytokines, chemokines, and interferons. TLR signaling involves a family of five adaptor proteins, which couple to downstream protein kinases that ultimately lead to the activation of transcription factors such as nuclear factor-κB (NF-κB) and members of the IFN-regulatory factor (IRF) family. The key signaling domain, which is unique to the TLR system, is the Toll/IL-1 receptor (TIR) domain, which is located in each TLR and also in the adaptors. These adaptors are MyD88, MyD88-adaptor–like (MAL; also known as Toll/IL-1 receptor domain-containing adaptor protein [TIRAP]), TRIF [TIR-domain-containing adapter-inducing interferon-β], TRAM [TRIF-related adaptor molecule], and SARM [sterile-alpha and armadillo motif–containing protein] (15–17).
SIGNAL TRANSDUCTION BY LPS VIA TLR4
TLR4 signals in response to LPS. Studies using MyD88−/− mice revealed both MyD88-dependent and MyD88-independent pathways of TLR4 signaling. Activation of NF-κB and mitogen-activated protein kinase (MAPK) still occurred in these mice although in a delayed manner. In addition, the activation of IRF-3 and induction of IFN-β in response to LPS were all unaffected in MyD88-deficient mice. The studies point to a role for MyD88 in an early response to LPS (17). Mal is an essential adaptor that works together with MyD88 in TLR2 and TLR4. TRIF adaptor is responsible for the MyD88-independent effects of LPS. The activation of NF-κB in response to LPS in TRIF-deficient mice was almost normal, but when the cells were deficient in TRIF as well as MyD88, the NF-κB response to LPS was totally abolished. Furthermore, TRIF was shown to play a critical role in the activation of IRF-3 and induction of IFN-β by LPS. TRIF was also shown to be the sole adaptor used by TLR3 (16, 17).
In addition to NF-κB and IRF activation, TRIF mediates a third distinct signaling pathway in induction of apoptosis. TRIF is the only TLR adaptor that can mediate apoptosis (15). This pathway uses the C-terminal RHIM (RIP-homotypic interaction motif) of TRIF and seems to involve RIP (receptor interacting protein) 1, FADD (Fas-associated protein with death domain), and caspase-8 (18). Furthermore, the TRIF pathway was also shown to control TLR4-mediated bacterial-induced apoptosis of macrophages and dendritic cells (18, 19). In TLR4, another adaptor called TRAM is essential for the MyD88-independent response to LPS. SARM, the last of the five adaptors to be assigned a role in TLR signaling, is the most evolutionarily ancient, being the only member to have an ortholog in Caenorhabditis elegans. Inactivation of TIR-1 (C. elegans homolog of SARM) by small interfering RNA (siRNA) led to decreased worm survival in response to fungal infection (20). TIR-1 functions upstream of PMK-1 (p38 MAPK family 1), a worm p38 MAPK that is known to be important in innate immunity (21). More recently, SARM has been found to block TRIF-dependent transcription factors activation (22). Importantly, LPS enhances SARM expression in monocytes. Thus, SARM is the only TLR adaptor to provide negative feedback regulation in the signaling pathway.
Although much has been done in this area, significant gaps exist in our understanding of the specificity of the downstream signaling pathways and how they function in innate immunity. Recent studies (6, 14) link the TLR4 pathway to autophagy and determine several required adaptors and some underlying mechanisms. Future studies should provide a more comprehensive signaling pathway of autophagy associated with innate immunity.
Autophagy and TLRs
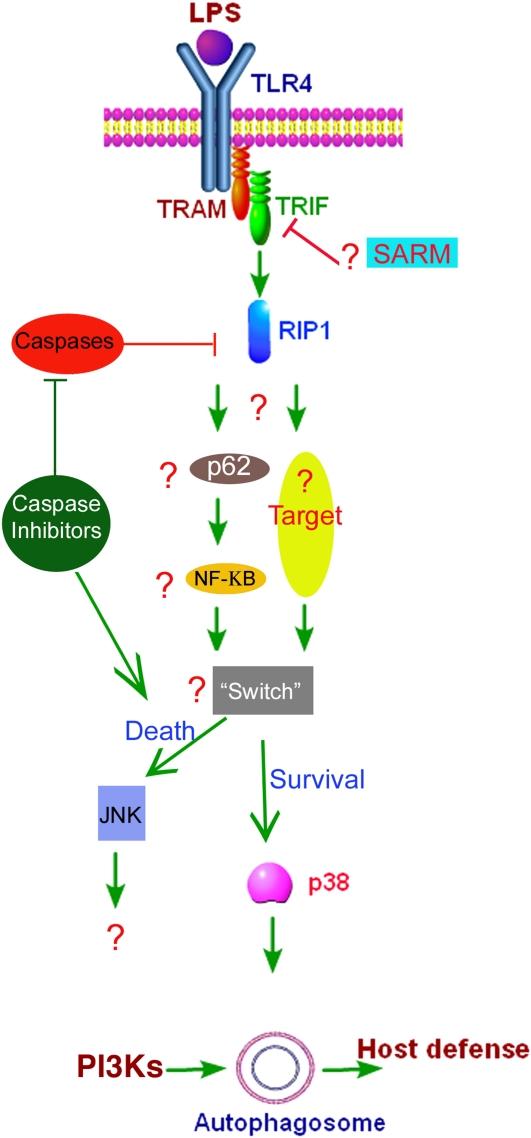
Recent studies show that TLR4 serves as a previously unrecognized environmental sensor for autophagy (6, 14, 23). The studies defined a new molecular pathway in which LPS induces autophagy in human and murine macrophages by a pathway regulated through a TRIF-dependent, myeloid differentiation factor 88 (MyD88)-independent TLR4 signaling. Receptor-interacting protein 1 and p38 mitogen-activated protein kinase are downstream components of this pathway (Figure 1). This signaling pathway does not affect cell viability, indicating that it is distinct from autophagic death-signaling pathway. Furthermore, LPS-induced autophagy could enhance mycobacterial colocalization with the autophagosomes. TRIF is responsible for the induction of IFN-inducible genes (16). These IFN-inducible genes are also induced by double-stranded RNA (dsRNA), signaling through TLR3. TLR3 is the only TLR other than TLR4 to use TRIF as an adaptor (16). Therefore, it is reasonable to envision a role for viral dsRNA and TLR3 in autophagy. Furthermore, most of the TRIF-interacting proteins, including RIP1, and the downstream signaling molecules and transcription factors have been found to be essentially the same for TRIF signaling whether mediated by TLR3 or TLR4. Few notable differences exist. TLR3 ligand is dsRNA and not LPS, TLR3 is intracellular, and TLR3 binds to TRIF directly, not via TRAM.
Figure 1.
Model of LPS-induced signaling pathway for autophagy as predicted from our recent data (6) and from recent studies. LPSs induce autophagy by stimulation of the TLR4-TIR-domain-containing adapter-inducing interferon-β (TRIF)-receptor interacting protein (RIP) 11 pathway. The signaling components directly downstream of RIP still need to be identified. Nevertheless, p38 mitogen-activated protein kinase is required for LPS-induced autophagy and for survival. This pathway seems to be in contrast to the c-Jun-N-terminal kinase (JNK)-mediated autophagic pathway previously described (25). Reproduced by permission from Reference 23.
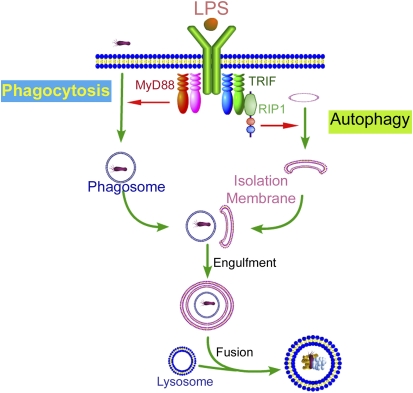
Although induction of autophagy by starvation is complete at 2 hours (2, 5), LPS induction of autophagy requires 8 to 16 hours. We have proposed a model to suggest that the delayed response in autophagosome formation might be coupled to the macrophage need to first internalize the pathogen in the phagosome (Figure 2). It is intriguing to note that among all TLRs, only TLR4 uses both the myeloid differentiation factor 88 (MyD88)-dependent and MyD88-independent pathways. The discovery of TRIF-dependent induction of autophagy may offer a mechanistic rationale for the need for two TLR4-mediated pathways. A plausible explanation could be that a close cooperation between the two arms for TLR4 is needed for innate immunity. For the cell to control a pathogen, two processes will need to be performed in sequence: phagocytosis for pathogen internalization and then autophagy, which would involve fusion of the phagosome with autophagosome, which matures and fuses with lysosomes. A previous study suggested that TLR4 MyD88-dependent pathway is needed for phagocytosis (24). A cooperation model would suggest that TLR4-MyD88, a fast response pathway, would be in charge of phagocytosis and that TLR4-TRIF, a slower response pathway, would be in charge of autophagy.
Figure 2.
Proposed model time kinetics of host defense revealing. Rapid phagocytosis of invading pathogens, mediated by toll-like receptor (TLR)-4 myeloid differentiation factor 88 (MyD88)-dependent pathway (B24), is followed by a slower TLR4 TIR-domain-containing adapter-inducing interferon-β (TRIF)-dependent activation of autophagy. Autophagosomes encapsulate infected phagosomes before fusing with lysosomes. This leads to the effective removal of the sequestered pathogen after fusion with the lytic vesicles.
AUTOPHAGY AS A PROSURVIVAL OR A PRODEATH PATHWAY
There has been an intense controversy regarding the role of autophagy as a prosurvival or prodeath mechanism. For example, whereas inhibition of apoptosis enhances autophagic cell death (25), autophagy is essential for restricting cell death to infected sites in plants (26). The term “autophagic cell death” has been used to describe a form of programmed cell death morphologically distinct from apoptosis and presumed to result from excessive levels of cellular autophagy. Evidence for autophagic cell death includes visualization of autophagosomes in dying cells, prevention of cell death by autophagic inhibition, and that under some conditions inhibition of apoptosis can lead to caspase-independent autophagic cell death. Several studies have shown that the pharmacologic inhibitor of autophagy 3-MA delays or partially inhibits starvation- or stress-induced death in several cell and animal models. However, 3-MA can inhibit other PI3Ks other than class III specific for autophagy. Furthermore, in these studies, it is difficult to determine if autophagy led to cell death or if it was merely associated with cell death, for instance to degrade and clean up damaged organelles (8).
Two important studies provide genetic evidence that the autophagic pathway is involved in cell death. RNA knockdown by siRNA directed against two autophagy genes, Atg7 and beclin 1, blocked cell death in cells treated with the caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp(OMe)-fluoromethylketone (zVAD) (25). Furthermore, siRNA against autophagy genes Atg5 and beclin 1 blocked death of bax−/−, bak−/− murine fibroblasts treated with staurosporine or etoposide (1). Although these findings implicate autophagy in cell death, they still raise the question: can autophagy be a death mechanism in cells whose apoptotic machinery is intact? The interpretation of the above studies has been further complicated by the more recent finding that calpain-mediated cleavage of the autophagy gene Atg5 switches autophagy to apoptosis (27). Truncated Atg5 translocated from the cytosol to mitochondria, associated with the antiapoptotic molecule Bcl-xL and triggered cytochrome c release and caspase activation, thus providing a molecular link between autophagy and apoptosis.
A case for the role of autophagy in promoting cell survival is equally strong. This notion is supported by studies demonstrating increased death in cells or organisms lacking autophagy genes. The prosurvival function of autophagy is best characterized in nutrient deficiency. Degradation of membrane lipids and proteins by the autolysosomes generates free fatty acids and amino acids that can be reused to fuel mitochondrial ATP energy production and maintain protein synthesis. This recycling function of autophagy is linked mechanistically to its ability to sustain life during starvation (8). Mice deficient in autophagy (Atg5−/− or Atg7−/−) die during the neonatal period, when the placental blood supply is interrupted resulting in a form of starvation (28). Similar examples of starvation-induced death in autophagy-deficient cells have been shown for unicellular organisms and plants (26, 29). Furthermore, a critical role for autophagy in maintaining cell survival in face of growth factors withdrawal has been recently shown (30).
The above contradicting studies probably suggest that autophagy can be either prosurvival or prodeath, depending on the context of autophagy induction. For example, in the context of innate immunity, autophagy can serve as a prosurvival mechanism to save infected cells, but under specific circumstances autophagy could serve as a prodeath mechanism to limit the spread of infection (26, 31). The major gap in our knowledge is related to effector mechanisms that dictate the switching in autophagy role between cell survival and cell death. For example, in LPS-stimulated macrophages autophagy is a prosurvival mechanism (6). However, caspase inhibition in LPS-stimulated autophagy results in cell death (31). The LPS prosurvival pathway is mediated by p38 MAPK (6), whereas the cell death pathway seems to be mediated by c-Jun-N-terminal kinase (JNK) pathway (25, 31).
RIP1 AT THE CROSSROADS OF CELL SURVIVAL AND DEATH
Our recent study suggests that RIP1 is required for LPS-induced autophagy (6). Although p38 MAPK activation is required for LPS-induced autophagy, the fact that p38 MAPK is also activated even to a larger extent by MyD88 pathway that is not required for autophagy suggests that p38 MAPK is not a direct downstream target for RIP1 and there are still unidentified signaling components downstream of RIP1 (Figure 1). RIP1 is a serine-threonine kinase, yet the substrate and function of its kinase activity are not known. Of the three distinct domains of RIP1 (an aminoterminal kinase domain, an intermediary domain, and a carboxyterminal death domain) only intermediary domain and the death domain are required for NF-κB activation (32, 33). Interestingly, RIP1 kinase activity has been shown to be essential for caspase-independent cell death by Fas (32). Thus, considering the recently recognized role of RIP1 in TLR4-mediated autophagy, it is important to determine which RIP1 domain is required for its role in autophagy. Identification of RIP1 downstream substrates for its kinase activity or identification of RIP1-interacting proteins is likely to yield important information about the downstream signaling components in LPS-induced autophagy.
In the signaling pathways mediating autophagy in response to TLR stimulation, there are still many downstream targets to be determined and their roles to be elucidated (Figure 1). The role of NF-κB in autophagy in general or autophagic cell death in particular still remains to be revealed. Recent studies by two independent groups have shown that ubiquitination of RIP1 on lysine 377 in the intermediate domain is required for RIP1 activation of NF-κB by TNF-α (34, 35). Furthermore, it was found that the polyubiquitin chain of RIP1 serves as a scaffold for binding of NF-κB essential modulator (NEMO), a regulatory subunit in the IκB kinase (IKK) complex required for NF-κB activation (34). P62 (also known as sequestosome 1) is a multimodule adaptor protein that has been shown to interact with atypical protein kinase C and with RIP1, participating in NF-κB activation in both pathways (36). Interestingly, p62, which also has been to shown to interact with RIP1 in the TNF pathway (37), has a ubiquitin-binding domain that binds polyubiquitin chains (36). p62 has been shown to associate with autophagosomes and is believed to play a role in targeting ubiquitinated proteins to the autophagosome (38). The exact role of p62 in autophagy, however, remains to be elucidated.
RIP3 AS A MOLECULAR SWITCH FOR CELL DEATH BY NECROPTOSIS
The term necroptosis has been recently used to specifically describe death receptor–induced cell death that occurs in the absence of sufficient caspase activity (39, 40). When apoptotic cell death is blocked by pan-caspase inhibitors, such as zVAD, the cell uses necroptosis as an alternative cell death pathway (31, 39). Necroptosis has also been described as autophagic cell death, but, as discussed above, the role of autophagy in this process remains uncertain. As indicated above, RIP1 has been implicated in the regulation of both apoptosis and necroptosis. Three recent independent studies have identified RIP3 kinase to be required for cells to undergo necroptosis (41–43). The expression of RIP3 renders cells permissive to necroptosis, whereas cells deficient in RIP3 are resistant to this form of cell death. Preliminary evidence suggests that RIP3 exerts its effect by affecting mitochondrial metabolic pathways leading to increased reactive oxygen species, which are required for necroptosis by RIP3 (40, 43). However, as is the case with RIP1, the downstream targets for RIP3, including the substrate for RIP3 kinase activity, remain to be elucidated.
AUTOPHAGY AND ADAPTIVE IMMUNITY
The role of autophagy in adaptive immune responses is being unraveled in recent years and it appears to be much more extensive than originally anticipated. Because autophagy enhances delivery of pathogen-related peptides to the lysosomes, its enhancing effect on antigen presentation by major histocompatibility complex class II (MHCII) has been shown both in vitro (44) and in vivo (45). The autophagy effect on antigen presentation has been effectively used to enhance vaccine efficacy of bacillus Calmette-Guérin (BCG). The variable efficacy of BCG vaccination against tuberculosis prompted efforts to increase its efficacy. A recent study used autophagy to enhance vaccine efficacy against tuberculosis using the mouse model (45). It examined the effect of autophagy on the processing of immunodominant mycobacterial antigen Ag85B by antigen-presenting cells (APCs), macrophages, and dendritic cells. Rapamycin-induced autophagy enhanced Ag85B presentation by APCs infected with wild-type H37Rv, H37Rv-derived ΔfbpA mutant, or BCG. Furthermore, rapamycin enhanced colocalization of mycobacteria with autophagosomes and lysosomes. Rapamycin-enhanced antigen presentation was attenuated when autophagy was suppressed by 3-methyladenine or by siRNA against beclin-1. Importantly, mice immunized with rapamycin-treated dendritic cells (DCs) infected with either ΔfbpA or BCG showed enhanced Th1-mediated protection when challenged with virulent M. tuberculosis. Furthermore, overexpression of Ag85B in BCG induced autophagy in APCs and enhanced immunogenicity in mice suggesting that vaccine efficacy can be enhanced by augmenting autophagy-mediated antigen presentation (45).
In addition to autophagy effect on antigen presentation, autophagy was recently shown to affect central tolerance via thymic selection of the T-cell repertoire (46) and to modulate homeostasis of T cells (47), B cells (48), and Paneth cells in intestinal epithelium (49). It is clear that this area of investigation will greatly expand in the near future to elucidate the multifaceted effects of autophagy on the adaptive immune system. Furthermore, the process of autophagy itself can be modulated by factors that skew the immune response (e.g., Th1- or Th2-inducing cytokines). This latter phenomenon is important in the context of immunity in diseases with Th1 or Th2 bias. This topic is further discussed below regarding autophagy and viral association with asthma.
AUTOPHAGY AND ASTHMA
Asthma is currently considered as a chronic airway inflammatory disease with a multifactorial pathogenesis. The pathogenesis of asthma involves bronchoconstriction, neuronal control of the airways, allergic responses, cytokine responses, inflammation including viral-induced and Th2 style–associated, innate immune factors, and genetic predisposition (50). Of the above factors, viral-mediated inflammation and Th2 cytokine immune responses have gained much interest. Thus, airway inflammation is the most widely accepted paradigm for asthma pathogenesis. It has also become evident that there are distinct patterns of airway inflammation. Based on the behavior of the adaptive immune responses in mice, these inflammatory patterns are categorized as Th1 or Th2. Evidence of a Th2 inflammatory profile was shown in mouse models of asthma and in samples from subjects with asthma (50, 51). The Th2 inflammatory pattern is driven by IL-4 and is characterized by CD4+ T cell secretion of IL-4, IL-5, and IL-13 (52).
Viruses and Asthma
The association of viral infections with human asthma posed a challenge to the Th2 hypothesis. Respiratory viral infections that classically activate Th1 responses can serve as acute triggers of asthmatic attacks and can produce chronic asthma phenotype (53, 54). An attempt to reconcile these observations with those of Th2 cytokine profile suggests that an initial Th1 immune response is required for the development of Th2 response (50, 55). It has been also suggested that viral infections can produce permanent alterations in the host immune system toward the asthma phenotype (54).
Respiratory viral infections are detected in 75 to 85% of asthma exacerbations in both children and adults (56–58). However, the mechanisms of viral-induced asthma exacerbations or etiologic role of viruses in chronic asthma are still under investigation (50, 58). Recent studies have shown that airway epithelial cells of subjects with asthma, cultured in vitro, had lowered innate immunity against rhinovirus infection (59). It was also observed that cells from subjects with asthma had a profoundly abnormal response to viral infection, resulting in efficient viral replication. In contrast, cells from healthy control subjects were resistant to infection (60). These observations are in agreement with the increased susceptibility to viral infection observed in clinical studies (61). Importantly, cells from subjects with asthma were also found to be resistant to viral-induced apoptosis (50). The latter finding is important in the context of airway inflammation because viral induction of apoptosis serves to abort infection by allowing the infected cell to be phagocytosed without release of inflammatory mediators. In contrast, an infected cell failing to undergo apoptosis will replicate virus and undergo cell necrosis, releasing large amounts of proinflammatory mediators and releasing a large number of progeny viruses to continue infection of neighboring cells (58, 60). Although several recent hypotheses have been proposed to explain the increased viral susceptibly by asthmatic cells, the underlying mechanisms are still poorly understood.
Th1 and Th2 Cytokines and Autophagy
In addition to the role of autophagy in innate immunity, recent reports suggest that autophagy is modulated by cytokines of the adaptive immune response (62). IFN-γ (Th1 cytokine) up-regulates autophagy (5). In contrast, IL-4 and IL-13 have been shown to inhibit starvation-induced autophagy in macrophages (62). The underlying mechanism is possibly mediated by the stimulatory effect of IL-4 and IL-13 on type I PI3K (63) that activates TOR, the major cellular inhibitor of autophagy (2). In this context, IL-13 was used as an autophagy inhibitor in the original studies examining type I PI3K role in autophagy (64). However, TOR signal is quite complex and a recent study suggested that IL-4 enhanced autophagy-mediated removal of protein aggregates in HeLa cells (13). The above studies suggest that Th1 and Th2 cytokines have modulatory roles in autophagy. Given the complexity of immune profile of cells in subjects with asthma, particularly in the context of viral infections, experimental data are needed to determine if autophagy is modulated in these cells and the role of each cytokine in this regulation.
The Noncanonical Role of Atg5 Members as Suppressor of Innate Antiviral Immune Signaling
It has been recently found that a key regulator of the autophagic process, the Atg12-Atg5 conjugate, associates with the signaling molecules retinoic acid-inducible gene I (RIG-I) and interferon-β promoter stimulator 1 (IPS-1), which are essential for recognition of RNA virus infection and which transmit signals to up-regulate type I IFNs. Interestingly, the Atg12-Atg5 conjugate seemed to negatively regulate the type I IFN modulating pathway through direct interaction with caspase recruitment domains presented by RIG-1 and IPS-1 (65, 66). Thus, in contrast to the antipathogenic properties of autophagic processes, Atg 5 appeared to promote RNA virus replication by inhibiting innate antivirus immune responses. These findings identify a rather paradoxical role for Atg5 in immunity against viruses. They also emphasize that autophagy genes can have important functions not related to autophagy.
In sum, autophagy has been shown to be critical for many processes in innate and adaptive immunity. It is not surprising that autophagy regulation and aberration in that process will prove to contribute to many human diseases. These revelations will provide scientists with opportunities to develop therapeutic strategies for these diseases affected by autophagy.
Supported by National Heart, Lung and Blood Institute, National Institute of Allergy and Infectious Diseases, grants HL069033, HL075421, HL080205, HL084322, and AI070973 (N.T.E.) and American Heart Association.
Conflict of Interest Statement: Neither author has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004;306:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol 2004;2:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001;2:211–216. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science 2004;306:1037–1040. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo ML, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004;119:753–766. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Jagannath C, Liu X-D, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007;27:134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science 2005;307:727–731. [DOI] [PubMed] [Google Scholar]
- 8.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005;120:159–162. [DOI] [PubMed] [Google Scholar]
- 9.Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol 2004;20:367–394. [DOI] [PubMed] [Google Scholar]
- 10.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006;313:1438–1441. [DOI] [PubMed] [Google Scholar]
- 11.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 2006;281:11374–11383. [DOI] [PubMed] [Google Scholar]
- 12.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 2003;5:455–468. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 2006;172:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008;27:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. Toll-like receptor downstream signaling. Arthritis Res Ther 2005;7:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signaling. Nat Rev Immunol 2007;7:353–364. [DOI] [PubMed] [Google Scholar]
- 17.Palsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 2004;113:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruckdeschel K, Pfaffinger G, Haase R, Sing A, Weighardt H, Häcker G, Holzmann B, Heesemann J. Signaling of apoptosis through TLRs critically involves toll/IL-1 receptor domain-containing adapter inducing IFN-beta, but not MyD88, in bacteria-infected murine macrophages. J Immunol 2004;173:3320–3328. [DOI] [PubMed] [Google Scholar]
- 19.De Trez C, Pajak B, Brait M, Glaichenhaus N, Urbain J, Moser M, Lauvau G, Muraille E. TLR4 and Toll-IL-1 receptor domain-containing adapter-inducing IFN-β, but not MyD88, regulate Escherichia coli-induced dendritic cell maturation and apoptosis in vivo. J Immunol 2005;175:839–846. [DOI] [PubMed] [Google Scholar]
- 20.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol 2004;5:488–494. [DOI] [PubMed] [Google Scholar]
- 21.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA 2004;101:6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol 2006;7:1074–1081. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Liu XD, Gong X, Eissa NT. Signaling pathway of autophagy associated with innate immunity. Autophagy 2008;4:110–112. [DOI] [PubMed] [Google Scholar]
- 24.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science 2004;304:1014–1018. [DOI] [PubMed] [Google Scholar]
- 25.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin1 program of autophagic cell death by caspase-8. Science 2004;304:1500–1502. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005;121:567–577. [DOI] [PubMed] [Google Scholar]
- 27.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006;8:1124–1132. [DOI] [PubMed] [Google Scholar]
- 28.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032–1036. [DOI] [PubMed] [Google Scholar]
- 29.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 2003;278:17636–17645. [DOI] [PubMed] [Google Scholar]
- 30.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005;120:237–248. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem 2006;281:8788–8795. [DOI] [PubMed] [Google Scholar]
- 32.Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 2000;1:489–495. [DOI] [PubMed] [Google Scholar]
- 33.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 34.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFa requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 2006;22:245–257. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J Biol Chem 2006;281:13636–13643. [DOI] [PubMed] [Google Scholar]
- 36.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci 2007;32:95–100. [DOI] [PubMed] [Google Scholar]
- 37.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J 1999;18:3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009;137:1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008;135:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Declercq W, Berghe TV, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell 2009;138:229–232. [DOI] [PubMed] [Google Scholar]
- 41.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Cell 2009;137:1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 2009;137:1100–1111. [DOI] [PubMed] [Google Scholar]
- 43.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009;325:332–336. [DOI] [PubMed] [Google Scholar]
- 44.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2007;26:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med 2009;15:267–276. [DOI] [PubMed] [Google Scholar]
- 46.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 2008;455:396–400. [DOI] [PubMed] [Google Scholar]
- 47.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol 2009;182:4046–4055. [DOI] [PubMed] [Google Scholar]
- 48.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy 2008;4:309–314. [DOI] [PubMed] [Google Scholar]
- 49.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter MJ, Holtzman MJ. A centennial history of research on asthma pathogenesis. Am J Respir Cell Mol Biol 2005;32:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson DS, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley AM, Corrigan C, Durham SR, Kay AB. Predominant Th2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med 1992;326:298–304. [DOI] [PubMed] [Google Scholar]
- 52.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone:I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 1986;136:2348–2357. [PubMed] [Google Scholar]
- 53.Lemanske RF Jr. Viruses and asthma: inception, exacerbation, and possible prevention. J Pediatr 2003;142(2, Suppl)S3–S8. [DOI] [PubMed] [Google Scholar]
- 54.Walter MJ, Morton JD, Kajiwara N, Agapov E, Holtzman MJ. Viral induction of a chronic asthma phenotype and genetic segregation from the acute response. J Clin Invest 2002;110:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Randolph DA, Carruthers CJ, Szabo SJ, Murphy KM, Chaplin DD. Modulation of airway inflammation by passive transfer of allergen-specific Th1 and Th2 cells in a mouse model of asthma. J Immunol 1999;162:2375–2383. [PubMed] [Google Scholar]
- 56.Chauhan AJ, Inskip HM, Linaker CH, Smith S, Schreiber J, Johnston SL, Holgate ST. Personal exposure to nitrogen dioxide (NO2) and the severity of virus-induced asthma in children. Lancet 2003;361:1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grissell TV, Powell H, Shafren DR, Boyle MJ, Hensley MJ, Jones PD, Whitehead BF, Gibson PG. Interleukin-10 gene expression in acute virus-induced asthma. Am J Respir Crit Care Med 2005;172:433–439. [DOI] [PubMed] [Google Scholar]
- 58.Johnston SL. Innate immunity in the pathogenesis of virus-induced asthma exacerbations. Proc Am Thorac Soc 2007;4:267–270. [DOI] [PubMed] [Google Scholar]
- 59.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PA, Bartlett NW, Kebadze T, Mallia P, Stanciu LA, Parker HL, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med 2006;12:1023–1026. [DOI] [PubMed] [Google Scholar]
- 60.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 2005;201:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corne JM, Marshall C, Smith S, Schreiber J, Sanderson G, Holgate ST, Johnston SL. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet 2002;359:831–834. [DOI] [PubMed] [Google Scholar]
- 62.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 2007;27:505–517. [DOI] [PubMed] [Google Scholar]
- 63.Wright K, Ward SG, Kolios G, Westwick J. Activation of phosphatidylinositol 3-kinase by interleukin-13. An inhibitory signal for inducible nitric-oxide synthase expression in epithelial cell line HT-29. J Biol Chem 1997;272:12626–12633. [DOI] [PubMed] [Google Scholar]
- 64.Petiot A, Ogier-Denis E, Blommaart EFC, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000;275:992–998. [DOI] [PubMed] [Google Scholar]
- 65.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA 2007;104:14050–14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy 2008;4:67–69. [DOI] [PubMed] [Google Scholar]