Abstract
Continuous renewal of intracellular components is required to preserve cellular functionality. In fact, failure to timely turnover proteins and organelles leads often to cell death and disease. Different pathways contribute to the degradation of intracellular components in lysosomes or autophagy. In this review, we focus on chaperone-mediated autophagy (CMA), a selective form of autophagy that modulates the turnover of a specific pool of soluble cytosolic proteins. Selectivity in CMA is conferred by the presence of a targeting motif in the cytosolic substrates that, upon recognition by a cytosolic chaperone, determines delivery to the lysosomal surface. Substrate proteins undergo unfolding and translocation across the lysosomal membrane before reaching the lumen, where they are rapidly degraded. Better molecular characterization of the different components of this pathway in recent years, along with the development of transgenic models with modified CMA activity and the identification of CMA dysfunction in different severe human pathologies and in aging, are all behind the recent regained interest in this catabolic pathway.
Keywords: aging, lysosomes, membrane proteins, proteases, protein translocation
Autophagy (from Greek, “self-eating”) is a highly regulated cellular process that mediates the degradation of intracellular components—single macromolecules and organelles—inside lysosomes (1). Under basal cellular conditions, the fine-tuned balance between protein synthesis and degradation contributes to the maintenance of cellular homeostasis and guarantees continuous renewal of proteome and organelles (2). In fact, studies in experimental animal models, or in certain pathologies, have revealed that failure of the autophagic system results in marked accumulation of abnormal proteins and defective organelles, which leads to functional failure, and often to cell death (1, 3). Added to this role in basal homeostasis, autophagy is also essential for cellular adaptation to environmental changes and in the cellular response to extracellular and intracellular stressors (4). This explains the involvement of autophagy in processes such as cellular growth, differentiation, development, and pathogen-to-host defense, among others (1).
Different types of autophagy have been described, depending on the mechanism that mediates the delivery of cytosolic cargo to lysosomes for degradation (Figure 1). The three main types of autophagy in mammals are macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (1, 5). In contrast to the first two pathways, which are conserved throughout the phylogenetic scale, increasing evidence supports that CMA appears later in evolution, probably to cover particular needs of the evolving species. In this review, we focus on the unique characteristics and cellular functions of CMA, the molecular components that participate in this type of autophagy, and how CMA malfunctioning underlies the pathogenesis of different human diseases and contributes to the phenotype of aging.
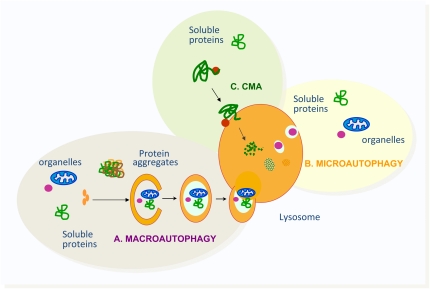
Figure 1.
Schematic model of the three main types of autophagy described in mammalian cells. (A) Macroautophagy degrades soluble proteins, organelles, and protein aggregates upon their sequestration in a double membrane vesicle that fuses with the lysosome to acquire the hydrolases required for proteolysis. (B) Microautophagy also degrades soluble proteins and organelles incorporated into lysosomes in small vesicles that form from invaginations of the lysosomal membrane. (C) Chaperone-mediated autophagy (CMA) only contributes to the degradation of a specific subset of soluble proteins, and does not require any type of membrane deformity for substrate delivery to lysosomes.
AUTOPHAGIC PATHWAYS: DIFFERENT ENTRIES INTO THE LYSOSOME
The three main autophagic pathways—macroautophagy, microautophagy and CMA—all contribute to lysosomal degradation, but differ in their regulation, the type of cargo preferentially degraded, and the mechanisms that contribute to targeting the cargo to the lysosomal compartment (1, 5–7).
Macroautophagy is the best-characterized autophagic process, and probably the one that contributes to the highest percentage of lysosomal degradation inside cells. In macroautophagy, cargo is secluded from the rest of the cytosol by a de novo–formed limiting membrane that elongates and seals on itself to generate a double membrane vesicle, known as autophagosome (Figure 1) (8, 9). The cytosolic components sequestered inside the autophagosome are degraded upon fusion of this vesicle with secondary lysosomes, which contribute a broad range of enzymes required for hydrolysis of the cargo. The diverse steps in macroautophagy are orchestrated by a group of proteins, generically known as Atg (autophagy-related) proteins, which were first characterized in yeast, and are conserved throughout the phylogenetic scale (10). Among these Atgs, two major cytosolic kinase complexes, the beclin-1/phosphatidylinositol kinase type III complex and mammalian target of rapamycin kinase and its auxiliary proteins, regulate initiation of macroautophagy, whereas proteins and lipids organized into two conjugation cascades are directly involved in the formation of autophagosomes (11). Genetic manipulations in ATG genes (knockout and overexpression) have helped the understanding of the physiological role of macroautophagy and the contribution of failure of this pathway to several pathologies (12–14). A detailed description of the molecular components that participate in macroautophagy, and in the regulation of this pathway, can be found in other contributions to this symposium, and in recent reviews (1, 8, 9). Cargo recognition in macroautophagy has been considered a nonselective process, and this is probably still true for soluble cytosolic proteins, because single protein molecules cannot be selectively identified and targeted for degradation through this pathway. However, macroautophagic selectivity is well supported for the recognition of particulate cargo, such as dysfunctional organelles or aggregated proteins, which can be sequestered by the limiting membrane with minimal inclusion in the autophagosome of nearby cytosolic components (15, 16). Although most of the initial studies described macroautophagy as an inducible pathway, only active under stress conditions such as nutritional starvation, recent studies support the existence of basal macroautophagy activity in most cells and tissues, and the importance of this continuous macroautophagy in maintenance of cellular homeostasis (12–14).
Less information is currently available on microautophagy, initially thought to be the only constitutive autophagic pathway in all cells, and responsible for continuous turnover of proteins and organelles (6). In microautophagy, cytosolic cargo is sequestered “in bulk” by direct engulfment in tubules or invaginations of the lysosomal membrane, which then pinch off into the lysosomal lumen for degradation (Figure 1) (6). This is a well-characterized process in yeast, where a subset of genes directly involved in microautophagy has been identified (17–19). However, our understanding of the molecular mechanism of microautophagy in mammals is rather limited, because homologs of the yeast genes have not been identified.
In contrast to the in-bulk sequestration of cytosolic components characteristic of macro- and microautophagy, soluble cytosolic proteins can be targeted selectively for degradation in lysosomes by CMA, as is described in more detail in the subsequent sections. In fact, characteristics that distinguish CMA from other forms of autophagy are the selective recognition of cargo by cytosolic chaperones, and the fact that substrates are not engulfed, but, instead, translocate across the lysosomal membrane in a receptor-mediated manner (Figure 1) (30). As in the case of macroautophagy, basal CMA activity can be detected in most types of mammalian cells, but maximal activation of this pathway is triggered in response to stressors, such as long-term starvation, oxidative stress, or exposure to toxic compounds that induce abnormal conformational changes in cytosolic proteins (20).
The three autophagic pathways do not function in the cell as completely independent entities, but, instead, their activation and function is coordinated as part of the global program for intracellular degradation and the cellular needs under different conditions (Figure 2). For example, starvation has been described to activate both macroautophagy and CMA (21). However, their activation does not occur simultaneously. Instead, macroautophagy is turned on during the first hours of nutrient deprivation; it reaches maximal activity in most cell types around 4–6 hours into the starvation state, and then gradually declines to basal levels. If starvation continues beyond that time, the decrease in macroautophagy is concomitant with a progressive increase in CMA activity (22). CMA reaches maximal activation in most cells around 12 hours, and remains active as long as starvation persists. This switch from macroautophagy to CMA may confer higher levels of selectivity when deciding the cellular components that can be hydrolyzed to obtain the amino acids required for cellular fueling and to maintain protein biosynthesis under these conditions. This synchronized mechanism of activation strongly supports the existence of molecular interactions among the different types of autophagy. For example, it is possible that activation of macroautophagy contributes to degradation of endogenous CMA inhibitors, hence contributing to the subsequent activation of this pathway as macroautophagy activity declines. Alternatively, some of the Atg proteins involved in macroautophagy could become CMA substrates under these conditions, and, consequently, the gradual increase in CMA activity may lead to their depletion and subsequent decrease in macroautophagy.
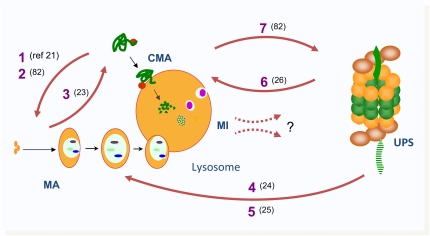
Figure 2.
Interactions among different proteolytic systems. Growing evidence supports multiple levels of interactions among autophagic pathways and between the autophagic system and the ubiquitin–proteasome system (UPS). Chronic blockage of CMA promotes up-regulation of macroautophagy (MA) [1] (21), whereas acute blockage leads to macroautophagic dysregulation [2] (79). Cells respond to blockage of MA by increasing CMA [3] (23). Acute blockage of the proteasome up-regulates MA [4] (24), whereas chronic blockage leads to macroautophagic dysregulation [5] (25). Some subunits of the proteasome are degraded by CMA [6] (26), which may explain why blockage of CMA is associated with proteasome dysregulation [7] (79). Interactions of microautophagy (MI) with other proteolytic systems remain undiscovered (?).
Further support for cross-talk among autophagic pathways has been provided in recent years when blockage of one autophagic pathway has shown to lead to activation of another one (Figure 2) (21, 23). Although autophagic pathways are not redundant, they can compensate for each other at least under basal cellular conditions. This compensation is only partial, as, for example, organelles normally degraded by macroautophagy cannot undergo degradation via CMA. Interestingly, although, in principle, macroautophagy should be able to degrade any CMA substrate, the loss of the characteristic selectivity of CMA seems still important, as cells with compromised CMA become vulnerable to a myriad of cellular stressors, despite the compensatory activation of macroautophagy observed in these cells (21). Little is known, as yet, on the possible relationship of macroautophagy and CMA with microautophagy, due, for the most part, to the limited information available on microautophagy in mammalian cells.
Temporal synchronization and cross-talk is not restricted to autophagic pathways, but extends to other proteolytic systems, such as the ubiquitin–proteasome system (Figure 2). Different cell types respond to acute blockage of the proteasome by up-regulating macroautophagy (24), whereas chronic blockage of this major cytosolic protease leads to dysregulation of inducible macroautophagy (25). Although still poorly studied, the fact that whole proteasome complexes are degraded by macroautophagy during starvation, and that subunits of the catalytic core of the proteasome undergo degradation via CMA, could underlie the basis of the cross-talk among these different proteolytic systems, essential for the maintenance of cellular homeostasis (26).
SELECTIVE AUTOPHAGY VIA CMA
As described in the previous section, the trademarks of CMA are the selective targeting to lysosomes of the subset of cytosolic proteins that undergo degradation by this pathway, and the direct translocation of these substrate proteins across the lysosomal membrane into the lysosomal lumen. In the following sections, we describe the molecular characteristics and components that contribute to the unique properties of this autophagic pathway.
The CMA Targeting Motif
Selectivity in CMA is conferred by the presence of a pentapeptide motif in the amino acid sequence of the substrate proteins that, when recognized by a cytosolic chaperone, results in targeting of substrates to lysosomes (27). This pentapeptide was first shown to be critical in the degradation of RNase A, the earliest CMA substrate identified (28), and it is shared by all substrate proteins identified to date. The pentapeptide motif is, as most intracellular targeting motifs, relaxed, and does not rely on the exact amino acid composition, but rather on the charge of the residues in the sequence (Figure 3). Studies with phage-display peptide libraries helped in establishing the basic requirements of the motif (27). The CMA targeting motif is always flanked by a glutamine (Q), and contains one acidic residue (D or E), a basic residue (K or R), a hydrophobic residue (F, I, L or V), and a fifth residue that can be basic or hydrophobic, but not negatively charged. In particular contexts, glutamine can be replaced by asparagine (N), preserving the similar affinity in the interaction with the chaperone. The motif does not depend on where it is located in the protein (N terminus, C terminus, or internally) as far as it is accessible to the chaperone. In fact, the presence of the motif in a protein does not necessary imply that it is continuously degraded by CMA. Often, when properly folded, the motif is hidden inside the protein core, and it only becomes exposed if the protein undergoes partial unfolding. In other proteins, the motif lies in the regions of intersection in between subunits, so that excess unassembled proteins can be eliminated via CMA. Although several cytosolic proteins have been shown to contain more than one CMA targeting motif, experimental studies with recombinant proteins have revealed that the presence of multiple motifs does not increase the affinity of the substrate protein for the chaperone, or its rate of degradation through CMA (20, 29). Multiplicity of sequences in the same protein may, instead, guarantee the removal of different protein fragments if the protein undergoes physiologic functional cleavage, as is the case with numerous proteins inside cells.
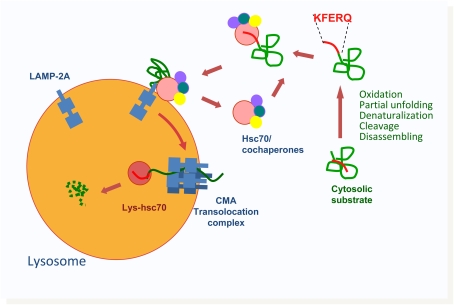
Figure 3.
Schematic model of chaperone-mediated autophagy (CMA). Cytosolic proteins substrate for CMA can undergo different modifications that lead to the exposure of the CMA-targeting motif and its subsequent recognition by cytosolic heat shock protein of 70 kD (Hsc70). The complex between the substrate protein, hsc70, and its cochaperones is delivered to the surface of the lysosomal membrane, where it interacts with lysosome-associated membrane protein (LAMP)-2A, the receptor for CMA. Once bound to LAMP-2A, substrate proteins undergo complete unfolding, and, assisted by a luminal form of hsc70 (Lys-hsc70), they cross the lysosomal membrane for rapid degradation in the lumen.
Sequence analysis of the cytosolic proteome has revealed that about 30% of cytosolic proteins might be potential substrates for CMA (27). However, it is possible that this amount is an underestimation, because particular post-translational modifications, such as deamidation, phosphorylation, acetylation, etc., could provide the charge missing in a four–amino acid sequence (29). This possibility of modulating chaperone recognition of the substrates by post-translational modifications adds an additional level of regulation to CMA.
MOLECULAR PLAYERS IN CMA
Chaperones and Cochaperones in the Cytoplasm
CMA substrates are recognized first in the cytoplasm by the heat shock cognate protein of 70 kD (hsc70), the constitutively expressed member of the 70-kD family of chaperones (Figure 3) (30). This is actually the same chaperone responsible for disassembly of clathrin from coated vesicles and for folding of unfolded cytosolic proteins upon recognition of exposed hydrophobic regions. It is unknown what determines the multiplicity of functions of the chaperones, but the particular array of cochaperones that bind to hsc70 in each condition is probably behind the final fate of the substrate protein. A subset of cochaperones, hsp90, hsp40, Bcl-2 associate athanogene 2 (Bag-1), hsc70-hsp90 organizing protein (Hop), and hsc70-interacting protein (Hip), has been shown to interact with the CMA substrate–chaperone complex at the lysosomal membrane (31). Some of the cochaperones may not be directly involved in substrate targeting, but rather participate in the unfolding step required before the substrate can translocate across the lysosomal membrane (29).
Hsc70 is the only chaperone that interacts directly with the substrate in a manner regulated by ATP/ADP binding cycles. Further studies are required to understand the way in which targeting and release are synchronized, and whether other cytosolic proteins could also participate in targeting of CMA substrates. In that respect, a recent study has proposed the participation of Carboxyl terminus of hsc70-interacting prtoein (CHIP), another multifunctional cochaperone, in lysosomal degradation of cytosolic proteins, although the autophagic process followed by the CHIP-interacting substrates has not been elucidated (32).
A Receptor for CMA Substrates at the Lysosomal Membrane
Once at the lysosomal surface, the substrate–chaperone complex binds to the membrane, and, after unfolding the substrate, is translocated into the lumen (Figure 3). Binding to the lysosomal membrane was shown to be a saturable process that could be competed for by adding other CMA substrates, and reduced by protease treatment of the lysosomal membrane, supporting the existence of a protein receptor at the membrane (33–35). Combination of different biochemical procedures in lysosomes incubated with well characterized CMA substrates led to the identification of the lysosome-associated membrane protein type 2A (LAMP-2A) as a CMA receptor (36). LAMP-2A is a single-span membrane protein with a very heavily glycosylated luminal region and a short (12–amino acid) C-terminus tail exposed on the surface of the lysosomes, where substrate proteins bind. LAMP-2A is one of the three splice variants of the lamp2 gene, all of which contain identical luminal regions, but different transmembrane and cytosolic tails (37). Incubation of lysosomes with synthetic peptides of the amino acid composition of the C terminus of each of the LAMP-2 variants, or with antibodies specific for each of these variants, revealed that CMA substrates only bind to the cytosolic tail of LAMP-2A through four positively charged residues not present in the other LAMP-2 variants (38). Specific knockdown of each of the LAMP-2 variants in fibroblasts in culture have confirmed that only LAMP-2A, and not LAMP-2B or -2C, is required for CMA (21). A mouse model with complete knockout of the lamp2 gene revealed a dramatic phenotype that includes, among other things, massive accumulation of autophagic vacuoles, impaired lysosomal biogenesis, and altered cholesterol trafficking (39, 40), supporting the concept that the divergent transmembrane and cytosolic tails of these proteins determine functional differences and their involvement in different cellular processes. In fact, splicing of lamp2 and the relative abundance of each variant changes depending on the cell type and stage of development. The proposed involvement of at least one of the three LAMP-2 proteins in macroautophagy, in light of the massive accumulation of autophagic vacuoles observed in different tissues of lamp-2 knockout mice, makes this gene and its splicing a possible attractive switch in the regulation of the cross-talk between macroautophagy and CMA.
Binding of CMA substrate proteins to the cytosolic tail of LAMP-2A is limiting for this pathway (41). In fact, overexpression of LAMP-2A in cultured cells increases CMA activity, whereas selective knockdown of this LAMP-2 variant blocks this autophagic pathway (21, 36). An almost linear correlation exists between changes in CMA activity under different physiologic and pathologic conditions and levels of LAMP-2A at the lysosomal membrane (41). Surprisingly, transcriptional up-regulation of LAMP-2A is unusual—in fact, it has only been described during the activation of CMA in response to mild oxidative stress (42). In most instances, LAMP-2A levels are controlled directly at the lysosomal compartment, and do not require de novo synthesis of the protein (Figure 4) (41). Changes in the half-life of LAMP-2A once in the lysosomal compartment (through regulated cleavage by two membrane proteases), or in the distribution of LAMP-2A between the lysosomal membrane and matrix, tightly control the availability of the cytosolic tail of this receptor at the surface of the lysosome (41). Cathepsin A has been identified as one of the two proteases that mediate the cleavage of LAMP-2A in the region between its transmembrane and cytosolic tail, leading to the release of a truncated form of LAMP-2A into the lysosomal lumen, where it is rapidly degraded by the resident proteases (Figure 4) (43). In fact, the up-regulation of CMA observed in mice knocked out for cathepsin A or in cells of patients with galactosialidosis (who lack cathepsin A) results from decreased rates of degradation of the lysosomal receptor (43).
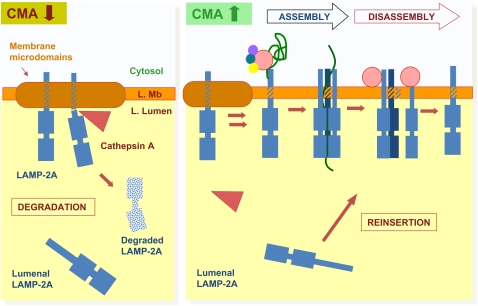
Figure 4.
Local regulation of chaperone-mediated autophagy (CMA) activity in lysosomes. Under conditions of low CMA activity (left), lysosome-associated membrane protein (LAMP)-2A is recruited to lysosomal membrane (L. Mb) microdomains, where it undergoes partial cleavage by cathepsin A, followed by rapid degradation of this truncated product in the lysosomal lumen (L. Lumen). There is a fraction of lysosomal LAMP-2A that resides in the lumen and is not accessible for substrate binding or translocation. When CMA is activated (right), association of LAMP-2A with the membrane microdomains decreases, favoring, instead, binding of substrate proteins to the cytosolic tail of this receptor. Substrate binding promotes multimerization of LAMP-2A to form a translocation complex. Once the substrate is released into the lysosomal lumen, hsc70 present at the lysosomal membrane promotes disassembly of LAMP-2A from the translocation complex. When maximal activation of CMA is required, the pool of LAMP-2A resident in the lysosomal lumen can be mobilized toward the membrane to contribute to substrate binding/uptake.
In clear contrast to the detailed characterization of the role of LAMP-2A in substrate binding, the mechanisms behind the translocation of substrate proteins across the lysosomal membrane are, as yet, poorly understood. Much evidence supports direct translocation across the lysosomal membrane, rather than engulfment by invaginations of the membrane. Thus, CMA uptake is a saturable process that requires binding to a receptor protein; invaginations have never been observed when this transport is reproduced in vitro, conjugation or cross-linking of substrate proteins to bigger structures, such as gold particles, prevents their uptake, and substrate proteins need to be completely unfolded before reaching the lysosomal lumen (22, 34–36, 44). By analogy with other protein translocation systems, involvement of a multispan membrane protein to create a discontinuity in the lysosomal membrane is expected. However, to date, proteomic analysis of proteins associated with LAMP-2A at the lysosomal membrane has not rendered any such multispan membrane proteins. Recent studies support the existence of a unique mechanism for translocation of substrate proteins across the lysosomal membrane via CMA that involves multimerization of LAMP-2A (Figure 4) (45). Thus, we have found that binding of substrate proteins to the cytosolic tail of monomeric forms of LAMP-2A drives its multimerization to form a 700-kD complex at the lysosomal membrane (45). Formation of this complex is essential for substrate translocation, because mutations in the transmembrane region of LAMP-2A that prevent multimerization do not affect substrate binding, but block substrate translocation. Despite our initial predictions that this complex sat as a stable translocon at the lysosomal membrane, capped by cytosolic and luminal chaperones, we have found that the CMA translocation complex forms only transiently, and that, once the substrate crosses the membrane, LAMP-2A rapidly disassembles in a process mediated by the hsc70 present on the cytosolic side of the lysosomal membrane (Figure 4) (45). Cytosolic and lysosomal chaperones only associate with lower-order complexes of substrate and LAMP-2A, but are no longer present in the 700-kD complex required for translocation. The transitory nature of this translocation complex may arise from the need to prevent leakage of lytic lysosomal components into the cytosol, and may help to preserve the lysosomal acid pH (45). This translocation mechanism, unique to lysosomes, underscores the importance of lateral mobility of LAMP-2A molecules at the lysosomal membrane. In fact, we have recently identified that this lateral mobility, and the subsequent multimerization, is, in part, regulated through the dynamic association of LAMP-2A to membrane microdomains of defined lipid and protein composition (46). LAMP-2A associated with these cholesterol-enriched regions is prone to degradation, as cathepsin A binds preferentially to the luminal part of these regions. In contrast, activation of CMA triggers exit of LAMP-2A out of the discrete membrane microdomains and the subsequent multimerization of the receptor protein required for substrate translocation (Figure 4) (46).
Lysosomal Chaperones: Beyond the Lysosomal Membrane
Although, in contrast to other translocation systems, luminal chaperones do not form part of a stable translocon unit in CMA, they are still essential for substrate uptake. In fact, a form of hsc70 resident in the lysosomal lumen (lys-hsc70) is needed for complete translocation of substrate proteins into lysosomes (Figure 3) (47). Incubation of cultured fibroblasts with blocking antibodies against hsc70 that reach the lysosomal lumen through endocytosis exerts a strong inhibitor effect on CMA (47). Furthermore, levels of lys-hsc70 have helped identify subgroups of lysosomes that manifest different ability to perform CMA (33). Only those lysosomes containing hsc70 in their lumen are competent for uptake of CMA substrates. Interestingly, the percentage of hsc70-containing lysosomes, which is no more than 40% under resting conditions, escalates to 80% in liver under conditions in which CMA is up-regulated, such as during prolonged starvation or mild oxidative stress (33, 42). This increase in the amount of lysosomes competent for CMA is a consequence, at least in part, of changes in the luminal acidification in these organelles. Thus, lys-hsc70 is stable in the lysosomal lumen at a pH of around 5.2, but a slight increase in the lysosomal pH is enough to destabilize this protein and make it amenable to degradation by the abundant lysosomal proteases (33, 42). Many factors could contribute to transient changes in lysosomal pH and the subsequent destabilization of hsc70. Among them, we have recently identified that fusion of lysosomes with autophagosomes when macroautophagy is maximally activated contributes to a dissipation of pH enough to render hsc70 unstable, and results in decreased CMA activity (23). The pH dependence of lys-hsc70 may thus constitute a novel regulatory node in the cross-talk between macroautophagy and CMA.
The mechanism by which lys-hsc70 mediates substrate translocation remains unclear. This chaperone can act either actively, by facilitating substrate internalization in an energy-dependent manner, or passively, by binding the portion of substrate already translocated and preventing its retrograde movement to the cytoplasm. Also unknown is the pathway followed by lys-hsc70 to reach the lysosomal lumen. Blockage of macroautophagy or CMA does not reduce the amount of lys-hsc70 (21, 23), discarding the involvement of these autophagic pathways in the delivery of hsc70 to lysosomes. It is possible that hsc70 reaches the lysosome through fusion with late endocytic compartments, where hsc70 has also been detected. Whether other luminal chaperones are required for substrate translocation is currently unknown.
WHAT IS THE PHYSIOLOGICAL ROLE OF CMA?
CMA was originally described as a starvation-induced process; however, recent studies also support an important role of this autophagic pathway in cellular homeostasis and in the cellular response to very different stressors.
CMA and Nutritional Stress
Although some level of basal CMA activity can be detected in almost all cell types, maximal activation of this pathway is attained during prolonged starvation (in animals) (22) or serum removal (in cultured cells) (28). As described in previous sections, the initial cellular response to starvation is the activation of macroautophagy, which will randomly degrade cellular components to obtain essential macromolecules. However, macroautophagic activity is not usually maintained beyond 6–8 hours of starvation. In conditions of persistent starvation, cells switch to the most selective degradation provided by CMA through which only nonessential proteins undergo proteolysis. For example, a considerable number of enzymes involved in glycolysis—a process no longer required during starvation—contain in their sequence CMA-targeting motifs and undergo degradation under these conditions (48). It is also possible that activation of CMA during prolonged nutritional stress may play a regulatory role by contributing to modulation of the activity of other cellular pathways. For example, the described degradation of some of the catalytic subunits of the 20S proteasome by CMA may be behind the decrease in proteasome-dependent degradation observed under these conditions (26). Likewise, degradation by CMA of regulators of transcription factors such as IkBα has been shown to contribute to some of the starvation-induced changes in transcriptional activity (49).
The time course of activation of CMA in response to starvation is different from tissue to tissue (50). Thus, for example, in heart, maximal activation of CMA is attained in less than 12 hours, but the reasons for these differences remain unknown. Further studies are also required to unveil the signaling mechanisms that induce activation of CMA during nutritional stress.
CMA and Oxidative Stress
The fact that degradation rates of particular CMA substrates decreased in the presence of antioxidants (49) provided the first clue for an involvement of CMA in the degradation of oxidized proteins inside cells. Further studies confirmed that, in fact, oxidized cytosolic proteins can be detected in the lysosomal lumen during chronic oxidative stress, and that oxidation of well characterized CMA substrate proteins increased their degradation through this pathway (42). Studies both in cultured cells (42, 51) and in vivo in different rodent organs (52) support the idea that up-regulation of CMA is part of the cellular response to oxidative stress, and that impaired CMA activity—either experimentally induced (21) or that associated with age (52)—results in considerable accumulation of oxidized proteins inside cells.
Increased CMA of substrate proteins during oxidative stress is probably a consequence of oxidation-induced changes both in the substrate proteins themselves and in the CMA machinery. Thus, the partial level of unfolding often associated with protein oxidation may favor the recognition of CMA substrate proteins by the chaperone and/or their translocation across the lysosomal membrane—by reducing the time required for complete unfolding—as higher rates of substrate lysosomal uptake have been observed under those conditions (42). In addition, independent of this effect on the substrate proteins, levels of LAMP-2A in lysosomes increase markedly in cells and organs exposed to oxidative stress (42). Interestingly, in contrast to nutritional stress, where up-regulation of CMA is attained through changes in the turnover and dynamics of LAMP-2A in the lysosomal compartment (41), the increased levels of LAMP-2A in lysosomes during oxidative stress results from transcriptional up-regulation of the lamp2 gene (42). However, both mechanisms can overlap in certain tissues, depending on the cellular conditions. Thus, recent studies have shown that the increase in circulating ketone bodies associated with prolonged nutritional stress induces CMA activation through the pro-oxidant effect that this metabolic by-product exerts on cells (51).
Oxidized proteins are often degraded through the ubiquitin/proteasome system (53, 54). How the degradative fate of an oxidized protein—through the proteasome or by CMA—is determined remains unknown. It is unlikely that different subsets of cellular proteins undergo degradation by one pathway or another, because, for example, antioxidants can decrease the degradation of IkBα by both the proteasome and CMA. In addition, although, in principle, CMA degradation would be reserved to only those oxidized proteins that contain a KFERQ-like motif in their sequence, it has been proposed that oxidation of particular residues may help complete a CMA targeting motif. Thus, for example, oxidation of a histidine may complete a motif only missing the negatively charged residue (55). It is thus more plausible that the same protein, when oxidized, may undergo degradation via the proteasome or CMA, depending on the cellular conditions.
Specialized Functions of CMA
The study of CMA in particular cell types and the characterization of the degradation of specific cellular proteins by CMA are behind the cell type–specific functions recently proposed for this pathway. Thus, for example, CMA activity has been linked to the regulation of cellular proliferation in tubular kidney cells through the degradation of Pax-2 (56). Levels of this transcription factor, an essential regulator of kidney cell proliferation and differentiation, are controlled through its degradation by CMA. Consequently, changes in CMA activity may modulate kidney organogenesis and growth. Similarly, a role for CMA in antigen presentation has been proposed in dendritic cells. Macroautophagy has been shown to contribute to both the presentation of endogenous peptides on major histocompatibility complex (MHC) class II molecules (57), as well as the presentation mediated by MHC class I molecules (58). Although relatively limited information is available on the contribution of CMA to immunity, recent studies have shown that reduction of LAMP-2A or hsc70 levels decreases presentation via MHC class II (59). Interestingly, pharmacological inhibition of hsp90, which will reduce CMA activity, also resulted in decreased antigen presentation in a second independent study (60). Antigen processing and loading usually occurs in endosomes rather than in secondary lysosomes, as the lower proteolytic capacity of the former compartment allows preservation of the presenting peptides and their loading on MHC class II molecules. However, to date, CMA has only been described to take place in secondary lysosomes. Future studies are required to determine whether this hsc70-mediated presentation of antigens takes place in late endosomes or in lysosomes.
CMA AND AGING
Altered cellular homeostasis and reduced ability to respond to stress are characteristics of almost all tissues in old organisms. The important contribution of autophagy to these two cellular processes explains the recent interest in the study of changes in autophagic function with age. In fact, both macroautophagy and CMA activity have been described to decrease with age (61–63). In light of the previously described physiological functions of CMA, the age-dependent decline in the activity of this pathway with age may contribute, at least in part, to the accumulation of damaged proteins in aging organisms, and could be behind the inefficient response to stressors orchestrated by aging cells (Figure 5).
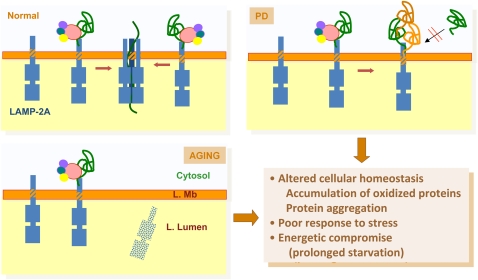
Figure 5.
Pathology of chaperone-mediated autophagy (CMA). This model depicts normal CMA and two different conditions in which the activity of this process is compromised. In Parkinson's disease (PD), abnormal association of pathogenic proteins to lysosome-associated membrane protein (LAMP)-2A at the lysosomal membrane (L. Mb) promotes formation of irreversible protein oligomers that compromise uptake and degradation of other proteins through CMA. In aging, compromised stability of LAMP-2A at the lysosomal membrane results in reduced levels of this receptor protein, and the consequent decrease in substrate binding and uptake. Some of the consequences of CMA failure in pathologic conditions and in aging are listed. L. Lumen = lysosomal lumen.
In contrast to the major morphological changes that the lysosomal system (in particular, the one related to macroautophagy) undergoes with age (expansion of the cellular acid compartment, accumulation of autophagic vacuoles, presence of undegraded products in the lysosomal lumen in the form of lipofuscin), no major changes in morphology have been identified with age in the subset of lysosomes able to perform CMA (63). Functional analysis has revealed, however, that the ability of substrate proteins to bind and translocate across the membranes of these compartments is severely impaired in lysosomes from organs from old organisms, or even from senescent cells in culture (63, 64). A systematic analysis of each of the steps and components involved in CMA has identified an age-dependent decrease in the lysosomal levels of LAMP-2A as mainly responsible for the functional impairment of this pathway in old organisms (Figure 5) (63). Lysosomal levels of LAMP-2A start to decline in a gradual manner by middle age. Interestingly, the functional decline in CMA presents a delay with respect to the reduction of LAMP-2A levels, because cells initially compensate for the lower number of LAMP-2A molecules per lysosome by recruiting a higher number of lysosomes to perform CMA (i.e., increase in the levels of hsc70-containing lysosomes). However, this compensation is only transient, and, eventually, the functional compromise on CMA becomes evident (63). Reduced levels of LAMP-2A do not result from decreased transcriptional activity, problems in protein synthesis, or altered trafficking of LAMP-2A to lysosomes in aging cells (65). Instead, the stability of LAMP-2A is severely impaired in old cells once it reaches the lysosomal compartment, leading to a decrease in the levels of this receptor in lysosomes (Figure 5) (65). Although the basis for LAMP-2A instability requires further investigation, changes in the lipid composition of the lysosomal membrane and their subsequent effect on the dynamics of LAMP-2A in this compartment likely contribute to the reduced levels of this receptor in old lysosomes (65).
A novel transgenic mouse model with regulatable expression of LAMP-2A in liver has been pivotal to understanding the contribution of the decline of CMA with age to the phenotype of aging. Activation of the expression of the exogenous LAMP-2A transgene in these animals in middle life (right at the time when the levels of endogenous LAMP-2A start to decline) has been shown to be efficient in preserving normal levels of LAMP-2A in the same animals until late in life, and consequently maintain rates of CMA activity comparable to those observed in young animals (52). In support of a critical role for CMA in maintenance of cellular homeostasis, livers from old animals with preserved CMA activity show lower levels of oxidized and aggregate proteins, and less accumulation of lipofuscin. Lower rates of cell death upon exposure of old animals to different stressors confirm the importance of CMA as part of the cellular response to stress (52). Finally, we have found that liver function in old animals with preserved CMA is comparable to that in young animals, supporting the importance of proper cellular homeostasis and of CMA in organ function, and the contribution of the altered CMA activity with age to the functional decline in old organisms. Current efforts in our laboratory aim at determining whether a similar restoration of CMA function can be achieved in all organs, and if preventing the decline in the activity of this pathway in all tissues and organs would lead to increases health span in old organisms.
CMA AND DISEASE
The better molecular characterization of CMA, along with the identification of new substrate proteins for this type of autophagy, have helped in establishing connections between CMA dysfunction and different pathological conditions.
Kidney Diseases and Nephropathies
The first connection between kidney dysfunction and CMA possibly dates to more than 20 years ago, when an increment in the amount of lys-hsc70 was noticed in acute tubular necrosis induced by inorganic mercury (66). However, the first direct link between CMA and kidney disease is more recent, and was established with the toxic-induced hyaline droplet nephropathy (67). Chronic exposure to environmental toxins promotes accumulation of α2-microglobulin in the form of hyaline droplets in rat tubular epithelial cells, leading eventually to loss of cell function and cell death (68). The cytosolic form of α2-microglobulin—a main target of the toxic insult—is normally degraded by CMA. Consequently, this pathway is up-regulated as part of the cellular response to the increasing levels of damaged α2-microglobulin upon exposure to the environmental toxin (67). However, if the exposure to toxins persists, CMA up-regulation becomes insufficient for the removal of the altered protein, which starts accumulating in the tubular cells, thus triggering functional kidney failure. Although still unexplored, it is likely that progression of kidney pathology in other protein conformational disorders could also depend on the ability of CMA to accommodate the increasing load of pathogenic protein.
Independently of this role of CMA in quality control in kidney, the described importance of CMA in regulating cellular growth in this organ has also been recently associated with the pathogenesis of the hypertrophic kidney in diabetes (69). Levels of intracellular proteins containing the CMA-targeting motif have been shown to be abnormally increased in kidneys of mouse models of acute diabetes mellitus, supporting a decrease in CMA activity in this organ (69). Reductions in levels of LAMP-2A and hsc70 have also been described in these conditions (69), although the cause of these changes remains unknown. Further studies are required to determine whether CMA failure is also behind the kidney hypertrophy observed in other pathologic conditions, such as unilateral nephrectomy or chronic kidney damage.
Lysosomal Storage Disorders
Lysosomal storage disorders (LSDs) are a group of genetic diseases resulting from loss of a specific lysosomal enzyme activity, and the consequent accumulation of its substrates in the lysosomal compartment (70). The enzymatic impairment can result from the malfunctioning of a specific enzyme in lysosomes, or from failure in its delivery to this compartment. CMA, like any other type of autophagy, is likely to be indirectly affected in these pathologies, because lysosomes are the final compartment for all autophagic pathways. However, a direct connection to CMA has been recently established with two different LSDs: galactosialidosis and mucolipidosis type IV. Patients with galactosialidosis lack cathepsin A, a protein that acts as chaperone for different lysosomal enzymes, but that has also been recently shown to participate in LAMP-2A turnover (43). The inability to properly degrade LAMP-2A in the cells from these patients results in abnormally high rates of CMA. In the case of patients with mucolipidosis type IV, who bear a mutation in the transient receptor potential mucolipin-1, CMA activity decreases (71). The fact that hsc70 interacts with this receptor has led to the proposition that altered docking of hsc70 at the lysosomal membrane could be behind the observed decrease in CMA. However, another possible explanation that requires further testing is that the small molecule channeling activity of this membrane protein is required for CMA.
Although the classification as LSD has been traditionally restricted to defects in enzymatic activity, recently, alterations in nonenzymatic lysosomal proteins, such as membrane proteins, have also been shown to result in lysosome malfunctioning and substrate accumulation. Among these new forms of LSD, of particular relevance for CMA is Danon disease, a vacuolar myopathy that originates from a primary defect in the lamp2 gene (72). Although, as indicated in previous sections, elimination of the three protein variants of this gene will result in a complex phenotype (including accumulation of autophagic vacuoles and problems in lysosomal biogenesis and in cholesterol trafficking), it is anticipated that patients with Danon disease will also have reduced CMA activity.
Neurodegenerative Disorders and Neuronal Homeostasis
The most common neurodegenerative disorders belong to a subgroup of pathologies known as protein conformational disorders, in which mutations or post-translational modifications of a particular protein favor its organization into oligomeric structures toxic for the neuronal cells. During the early stages of the pathology, the pathogenic proteins are kept “in check” through continuous degradation by the proteolytic systems, because high cytosolic levels of these proteins are a precipitating factor in their aggregation (2). However, as the ability of the proteolytic systems to get rid of these proteins decreases, due to the pathology itself or to aging, these proteins start to accumulate inside cells exerting their toxic effect. This also explains why many of these diseases are considered age-related disorders.
Degradation of these pathogenic proteins thus occurs through different pathways, depending on their aggregation status. Thus, when still present as soluble single molecules, both the ubiquitin–proteasome system and CMA can contribute to their removal, whereas, once organized into irreversible oligomeric structures, macroautophagy is the only suitable method of degradation.
An additional, interesting aspect of these pathogenic proteins is that often, when organized into irreversible oligomers, not only do they not undergo proper degradation through the ubiquitin–proteasome system or CMA, but they also have a negative effect on the function of these proteolytic pathways. In that respect, we have recently described that α-synuclein, the protein that accumulates in the affected neurons in Parkinson's disease, is a bona fide CMA substrate, but, when mutated or post-translationally modified, exerts an inhibitory effect on CMA (73, 74). Pathogenesis in the disease thus results not only from the combined effect of the defective degradation of α-synuclein and its accumulation as oligomers or toxic aggregates in the cells, but also from the blockage on CMA that will decrease turnover and elimination of oxidized and damaged proteins, and will render cells susceptible to multiple stressors (Figure 5). This “double hit” predicts that, if at least the CMA blockage were prevented, cellular conditions should improve. This concept has been recently tested using forms of mutant α-synuclein in which the CMA-targeting motif has been altered so that they can no longer be targeted for degradation through this pathway. As predicted, toxicity of these variants was markedly less, because CMA blockage did not occur (75). Although the mechanism by which pathogenic α-synuclein proteins block CMA is not completely elucidated, it is known that these forms of the protein bind to the lysosomal membrane through LAMP-2A with higher affinity than any other previously known substrate. In addition, the high concentration of these proteins in these regions of the membrane seems to favor their organization into irreversible oligomers that affect lysosomal function (Figure 5) (74).
α-Synuclein is not the only protein that, when altered, interferes with CMA activity. Mutant forms of ubiquitin carboxyl-terminal esterase L1, another protein involved in Parkinson's disease pathogenesis, have also been shown to interact abnormally with LAMP-2A, and consequently alter CMA activity (76). Furthermore, a mutant form of Tau, the protein that organizes in toxic filaments in Alzheimer's disease, is also targeted to lysosomes via CMA. Once at the membrane, it undergoes only partial translocation, resulting in CMA blockage (77). In this case, the toxic effect goes beyond that resulting from CMA blockage, because cleavage of partially translocated Tau proteins by the lysosomal enzymes produces shorter Tau fragments, which are highly amyloidogenic. These fragments organize into toxic oligomers on the surface of lysosomes, compromising the stability of their membranes. Leakage of lysosomal enzymes to the cytosol under these conditions is likely one of the events that precede cell death under these conditions (77).
An additional connection of CMA with neurodegeneration has been established in a recent work that identified degradation of myocyte enhancer factor (MEF) 2D, a transcriptional factor that promotes neuronal survival by CMA. Degradation of MEF2D by CMA may thus contribute to modulating neuronal homeostasis. Levels of MEF2D are abnormally elevated in brains from patients with Parkinson's disease and α-synuclein transgenic mice (78), supporting the concept that alterations in neuronal homeostasis can also be an additional consequence of CMA blockage in Parkinson's disease.
CONCLUSIONS
The better molecular characterization of the components that participate in CMA has allowed the identification of pathologic conditions in which CMA activity is abnormal, thus linking this pathway with different diseases. In addition, genetic manipulation of the limiting component of this pathway—namely, LAMP-2A—has permitted up-regulation and down-regulation of CMA in intact cells, and thus the ability to start learning about the physiological relevance of this selective form of autophagy in cellular homeostasis and the response to stress. However, despite these recent advances, there are still numerous questions that remain unanswered about this pathway. For example, how does interaction of substrate proteins with hsc70 lead to targeting to the lysosomal membrane? What is the role of the different cochaperones known to interact with the hsc70–substrate protein complex? Is there a binding motif in the sequence of the substrate proteins required for interaction with LAMP-2A? What are the additional components of the translocation complex? What is the relationship between CMA-competent and -incompetent lysosomes? Do they correspond to different biogenic stages of the same organelle, or are they generated independently? Furthermore, despite the exquisite dissection of the components at the lysosomal membrane that participate in CMA, and their tight regulation, little is known about the cellular signaling mechanisms that control this pathway. What is the major signaling complex that activates CMA during stress? Are there endogenous inhibitors of this pathway? How does the regulation of CMA intersect with the better-characterized regulation of other autophagic processes? Future efforts should also be directed to a better understanding of the mechanisms that modulate the cross-talk between autophagic pathways and the development of pharmacological agents that can modulate CMA activity, as these could have therapeutic applications in conditions with reduced CMA activity, such as some of the described neurodegenerative disorders or aging.
Acknowledgments
The authors thank Dr. Susmita Kaushik and Dr. Fernando Macian for critically reading the manuscript.
Supported by National Institutes of Health grants from National Institute on Aging (AG021904, AG031782), National Institute of Diabetes and Digestive and Kidney Diseases (DK041918), National Institute on Neurological Disorders and Stroke (NS038370), a Glenn Foundation Award, and a Hirsch/Weill-Caulier Career Scientist Award.
Conflict of Interest Statement: E.B. has received funding from noncommercial entities for research with NIH ($50,001–$100,000). A.M.C. has received funding from noncommercial entities for research with the National Institutes of Health (NIH) ($100,001 or more), the Glenn Foundation ($50,001–$100,000), and Hirsch/Weill-Caulier ($50,001–$100,000).
References
- 1.Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature 2008;451:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci 2009;64:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin Z-H, Ravikumar B, Stefanis L, Tolkovsky AM. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 2005;1:11–22. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 2007;7:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol 2004;14:70–77. [DOI] [PubMed] [Google Scholar]
- 6.Dice J. Lysosomal pathways of protein degradation. Austin, TX: Landes Bioscience; 2000.
- 7.Shintani T, Klionsky D. Autophagy in health and disease: a double-edged sword. Science 2004;306:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol 2009;10:458–467. [DOI] [PubMed] [Google Scholar]
- 9.Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Narayanan U, Renna M, et al. Mammalian macroautophagy at a glance. J Cell Sci 2009;122:1707–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003;5:539–545. [DOI] [PubMed] [Google Scholar]
- 11.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 2004;15:231–236. [DOI] [PubMed] [Google Scholar]
- 12.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006;441:885–889. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005;169:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006;441:880–884. [DOI] [PubMed] [Google Scholar]
- 15.Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in alpha-1-antitrypsin deficiency. Cell Death Differ 2009;16:39–45. [DOI] [PubMed] [Google Scholar]
- 16.Gorman AM. Neuronal cell death in neurodegenerative diseases: Recurring themes around protein handling. J Cell Mol Med 2008;12:2263–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sattler T, Mayer A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J Cell Biol 2000;151:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb D. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 2003;14:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai Y, Koller A, Rangell L, Keller G, Subramani S. Peroxisome degradation by microautophagy in pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol 1998;141:625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massey A, Zhang C, Cuervo A. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol 2006;73:205–235. [DOI] [PubMed] [Google Scholar]
- 21.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA 2006;103:5905–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuervo A, Knecht E, Terlecky S, Dice J. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol 1995;269:C1200–C1208. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik S, Massey A, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 2008;19:2179–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA 2005;102:13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Q, Dimayuga E, Martin S, Bruce-Keller A, Nukala V, Cuervo A, Keller J. Characterization of chronic low-level proteasome inhibition on neural homeostasis. J Neurochem 2003;86:489–497. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem 1995;227:792–800. [DOI] [PubMed] [Google Scholar]
- 27.Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 1990;15:305–309. [DOI] [PubMed] [Google Scholar]
- 28.Dice JF, Chiang H-L, Spencer EP, Backer JM. Regulation of catabolism of microinjected ribonuclease A: identification of residues 7–11 as the essential pentapeptide. J Biol Chem 1986;262:6853–6859. [PubMed] [Google Scholar]
- 29.Dice J. Chaperone-mediated autophagy. Autophagy 2007;3:295–299. [DOI] [PubMed] [Google Scholar]
- 30.Chiang H, Terlecky S, Plant C, Dice J. A role for a 70 kDa heat shock protein in lysosomal degradation of intracellular protein. Science 1989;246:382–385. [DOI] [PubMed] [Google Scholar]
- 31.Agarraberes F, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci 2001;114:2491–2499. [DOI] [PubMed] [Google Scholar]
- 32.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem 2005;280:23727–23734. [DOI] [PubMed] [Google Scholar]
- 33.Cuervo A, Dice J, Knecht E. A lysosomal population responsible for the hsc73-mediated degradation of cytosolic proteins in lysosomes. J Biol Chem 1997;272:5606–5615. [DOI] [PubMed] [Google Scholar]
- 34.Cuervo A, Terlecky S, Dice J, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem 1994;269:26374–26380. [PubMed] [Google Scholar]
- 35.Terlecky S, Dice J. Polypeptide import and degradation by isolated lysosomes. J Biol Chem 1993;268:23490–23495. [PubMed] [Google Scholar]
- 36.Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996;273:501–503. [DOI] [PubMed] [Google Scholar]
- 37.Gough NR, Hatem CL, Fambrough DM. The family of LAMP-2 proteins arises by alternative splicing from a single gene: characterization of the avian LAMP-2 gene and identification of mammalian homologs of LAMP-2B and LAMP-2C. DNA Cell Biol 1995;14:863–867. [DOI] [PubMed] [Google Scholar]
- 38.Cuervo A, Dice J. Unique properties of LAMP2A compared to other LAMP2 isoforms. J Cell Sci 2000;113:4441–4450. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka Y, Guhde G, Suter A, Eskelinen E-L, Hartmann D, Lullmann-Rauch R, Janssen P, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2–deficient mice. Nature 2000;406:902–906. [DOI] [PubMed] [Google Scholar]
- 40.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lüllmann-Rauch R, Hartmann D, Heeren J, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 2004;15:3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuervo A, Dice J. Regulation of LAMP2A levels in the lysosomal membrane. Traffic 2000;1:570–583. [DOI] [PubMed] [Google Scholar]
- 42.Kiffin R, Christian C, Knecht E, Cuervo A. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 2004;15:4829–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuervo AM, Mann L, Bonten E, d'Azzo A, Dice J. Cathepsin a regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J 2003;22:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem 2000;275:27447–27456. [DOI] [PubMed] [Google Scholar]
- 45.Bandhyopadhyay U, Kaushik S, Vartikovsky L, Cuervo AM. Dynamic organization of the receptor for chaperone-mediated autophagy at the lysosomal membrane. Mol Cell Biol 2008;28:5747–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J 2006;25:3921–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 1997;137:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aniento F, Roche E, Cuervo AM, Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem 1993;268:10463–10470. [PubMed] [Google Scholar]
- 49.Cuervo AM, Hu W, Lim B, Dice JF. Iκb is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell 1998;9:1995–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wing S, Chiang HL, Goldberg AL, Dice JF. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J 1991;275:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem 2005;280:25864–25870. [DOI] [PubMed] [Google Scholar]
- 52.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 2008;14:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 1999;68:1015–1068. [DOI] [PubMed] [Google Scholar]
- 54.Friguet B, Bulteau A, Chondrogianni N, Conconi M, Petropoulos I. Protein degradation by the proteasome and its implications in aging. Ann N Y Acad Sci 2000;908:143–154. [DOI] [PubMed] [Google Scholar]
- 55.Gracy R, Talent J, Zvaigzne A. Molecular wear and tear leads to terminal marking and the unstable isoforms of aging. J Exp Zool 1998;282:18–27. [PubMed] [Google Scholar]
- 56.Franch H, Sooparb S, Du J, Brown N. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem 2001;276:19126–19131. [DOI] [PubMed] [Google Scholar]
- 57.Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Müller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA 2005;102:7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol 2009;10:480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. LAMP-2A facilitates MHC class II presentation of cytoplasmic antigens. Immunity 2005;22:571–581. [DOI] [PubMed] [Google Scholar]
- 60.Rajagopal D, Bal V, Mayor S, George A, Rath S. A role for the hsp90 molecular chaperone family in antigen presentation to lymphocytes via major histocompatibility complex class II molecules. Eur J Immunol 2006;36:828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol 2001;56:B375–B383. [DOI] [PubMed] [Google Scholar]
- 62.Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol 2003;38:519–527. [DOI] [PubMed] [Google Scholar]
- 63.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem 2000;275:31505–31513. [DOI] [PubMed] [Google Scholar]
- 64.Neff N, Bourret L, Miao P, Dice JF. Degradation of proteins microinjected into IMR-90 human diploid fibroblasts. J Cell Biol 1981;91:184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey A, Martinez-Vicente M, Cuervo A. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 2007;120:782–791. [DOI] [PubMed] [Google Scholar]
- 66.Hernadez-Pando R, Pedraza-Chaverri J, Orozco-Estevez H, Silva-Serna P, Moreno I, Rondan-Zarate A, Elinos M, Correa-Rotter R, Larriva-Sahd J. Histological and subcellular distribution of 65 and 70 kD heat shock proteins in experimental nephrotoxic injury. Exp Toxicol Pathol 1995;47:501–508. [DOI] [PubMed] [Google Scholar]
- 67.Cuervo A, Hildebrand H, Bomhard E, Dice J. Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int 1999;55:529–545. [DOI] [PubMed] [Google Scholar]
- 68.Lehman-Mckeeman L, Rodriguez P, Caudill D, Fey M, Eddy C, Asquith T. Hyaline droplet nephropathy resulting from exposure to 3,5,5-trimethylhexanoyloxybenzene sulfonate. Toxicol Appl Pharmacol 1991;107:429–438. [DOI] [PubMed] [Google Scholar]
- 69.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int 2004;65:2135–2144. [DOI] [PubMed] [Google Scholar]
- 70.Neufeld E. Lysosomal storage diseases. Annu Rev Biochem 1991;60:257–280. [DOI] [PubMed] [Google Scholar]
- 71.Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, Slaugenhaupt SA. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol 2009;219:344–353. [DOI] [PubMed] [Google Scholar]
- 72.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000;406:906–910. [DOI] [PubMed] [Google Scholar]
- 73.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004;305:1292–1295. [DOI] [PubMed] [Google Scholar]
- 74.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol 2007;6:352–361. [DOI] [PubMed] [Google Scholar]
- 75.Xilouri M, Vogiatzi T, Vekrellis K, Park D, Stefanis L. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One [serial on the Internet]. 2009. [accessed May 13, 2009];4:e5515. Available from: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0005515. [DOI] [PMC free article] [PubMed]
- 76.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between parkinson disease-associated mutant uch-l1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem 2008;283:23731–23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Martinez-Vicente M, Krüger U, Kaushik S, Wong E, Mandelkow E, Cuervo A, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet 2009;18:4153–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka J, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 2009;323:124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy 2008;4:442–456. [DOI] [PubMed] [Google Scholar]