Abstract
Motivation: Several new de novo assembly tools have been developed recently to assemble short sequencing reads generated by next-generation sequencing platforms. However, the performance of these tools under various conditions has not been fully investigated, and sufficient information is not currently available for informed decisions to be made regarding the tool that would be most likely to produce the best performance under a specific set of conditions.
Results: We studied and compared the performance of commonly used de novo assembly tools specifically designed for next-generation sequencing data, including SSAKE, VCAKE, Euler-sr, Edena, Velvet, ABySS and SOAPdenovo. Tools were compared using several performance criteria, including N50 length, sequence coverage and assembly accuracy. Various properties of read data, including single-end/paired-end, sequence GC content, depth of coverage and base calling error rates, were investigated for their effects on the performance of different assembly tools. We also compared the computation time and memory usage of these seven tools. Based on the results of our comparison, the relative performance of individual tools are summarized and tentative guidelines for optimal selection of different assembly tools, under different conditions, are provided.
Contact: hdeng2@tulane.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Recently developed next-generation sequencing platforms, such as the Roche 454 GS-FLX System, Illumina Genome Analyzer and HiSeq 2000 system, and ABI SOLiD™ System, have revolutionized the field of biology and medical research (Schuster, 2008). Compared to traditional Sanger sequencing technology (Bentley, 2006; Sanger et al., 1977), these new sequencing platforms generate data much faster and produce much higher sequencing output, while decreasing costs by more than a thousand fold (Shendure and Ji, 2008). The ability to rapidly generate enormous numbers of sequence reads at markedly reduced prices has greatly extended the scope of economically feasible sequencing projects. The prospect of sequencing the entire human genome for a large number of samples has become a reality.
These new sequencing technologies also pose tremendous challenges to traditional de novo assembly tools designed for Sanger sequencing, as they are incapable of handling the millions to billions of short reads (35–400 bp each) generated by next-generation sequencing platforms (Dohm et al., 2007). Therefore, several novel de novo assembly tools have been developed, such as SSAKE (Warren et al., 2007), VCAKE (Jeck et al., 2007), SHARCGS (Dohm et al., 2007), Euler-sr (Chaisson and Pevzner, 2008), Edena (Hernandez et al., 2008), Velvet (Zerbino and Birney, 2008), Celera WGA Assembler (Miller et al., 2008), ABySS (Simpson et al., 2009) and SOAPdenovo (Li et al., 2009).
With the recent introduction of multiple de novo assembly tools, it has become necessary to systematically analyze their relative performance under various conditions so that researchers can select a tool that would produce optimal results according to the read properties and their specific requirements. Zhang et al. (2011) recently compared the performance of several of these tools for assembling sequences of different species. Although they evaluated multiple criteria such as runtime, RAM usage, N50 and assembly accuracy, their results were based on simulation reads using only a single depth of coverage (100×) and a single base call error rate (1.0%). Further investigation is necessary to determine whether, and how, these assembly tools are differentially affected by varying depths of coverage, sequencing errors, read lengths and extent of GC content of the sequence reads. Furthermore, the assembly performance of SOAPdenovo (v1.05) has dramatically improved for long read assembly. Consequently, sufficient information is not currently available for informed decisions to be made regarding the tool that would be most likely to produce the best results, based on variations in the practical conditions identified above.
Accordingly, in this study, we systematically studied and compared the performance of seven commonly used de novo assembly tools for next-generation sequencing technologies, using a number of metrics including N50 length (a standard measure of assembly connectivity, to be more specifically defined later), sequence coverage, assembly accuracy, computation time and computer memory requirement and usage. To imitate different practical conditions, we selected a number of experimentally derived benchmark sequences with different lengths and extent of GC content, and simulated single-end and paired-end reads with varying depths of coverage, base calling error rates and individual read lengths. Based on the results of our analyses, we have developed guidelines for optimal selection of different assembly tools under different practical conditions. Identifying and recognizing the various limitations of specific tools under different practical conditions may also provide useful guidance and direction for improving current tools and/or designing new high-performance tools.
2 METHODS AND MATERIALS
2.1 De novo sequencing tools
Seven tools, SSAKE (v3.7), VCAKE (vcakec_2.0), Euler-sr (v1.1.2), Edena (2.1.1), Velvet (v1.0.18), ABySS (v1.2.6) and SOAPdenovo (v1.05 for 64bit Linux), were selected for studies and comparative analyses. These tools are all publicly available, and most of these tools are currently often used to assemble short reads generated by next-generation sequencing platforms, such as Illumina Genome Analyzer (read length = 35–150 bp) and ABI SOLID (read length = 35–75 bp). Of these seven tools, all are capable of assembling single-end reads, but only SSAKE, Euler-sr, Velvet, ABySS and SOAPdenovo support paired-end reads assembly.
2.2 Benchmark sequences
Eight experimentally determined sequences (Table 1) were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/) and used as benchmark sequences to test the performance of the seven assembly tools. These sequences range from ~99 kb (base pair) to ~100 Mb, each with a different extent of GC content.
Table 1.
Information for the eight benchmark sequences used in this study
| Species | GenBank | Chr. | Seq len (bp) | GC (%) |
|---|---|---|---|---|
| D.mel | AC018485 | 2L | 99 441 | 36.90 |
| H.inf | NC_007146 | — | 1 914 490 | 38.16 |
| T.bru | AE017150 | 2 | 1 193 948 | 44.38 |
| H.sap | NT_037622 | 4 | 1 413 146 | 49.81 |
| E.coli | NC_009800 | — | 4 643 538 | 50.82 |
| C.ele | NC_003283 | V | 20 919 568 | 35.43 |
| H.sap | NT_007819 | 7 | 50 360 631 | 41.03 |
| H.sap | NT_005612 | 3 | 100 537 107 | 38.96 |
D.mel: Drosophila melanogaster, H.inf: Haemophilus influenza, T.bru: Trypanosoma brucei, H.sap: Homo sapiens, E.coli: Escherichia coli, C.ele: Caenorhabditis elegans; GenBank: GenBank accession number; GC: percentage of GC contents reported by Tandem repeats finder (v4.40, http://tandem.bu.edu/trf/trf.html). H.inf and E.coli are the complete genomes. For clarity, H.sap-1 was used to refer to NT_037622, H.sap-2 was NT_007819 and H.sap-3 was NT_005612.
2.3 Sequencing read simulations
Simulated single-end and paired-end reads were generated from benchmark sequences with several variable parameters, including depth of coverage, base calling error rate (BCER) and individual read length. Depth of coverage is the average number of reads by which any position of an assembly is independently determined (Taudien et al., 2006). BCER is the estimated probability of error for each base call (Ewing and Green, 1998).
Single-end reads simulation method was the same as that used previously (Dohm et al., 2007), that is, each read was generated as a DNA fragment of the preset read length from any position in the benchmark sequence with equal probability. Each base of the read was then randomly and independently changed into another base with probability of BCER. In paired-end read simulation, a fragment with length of fragment size was randomly obtained from the benchmark sequence, then two reads of the preset read length were generated simultaneously from the two ends of this fragment, which were considered as one pair. We applied the fragment size distribution based on the empirical distribution of the experimental read dataset of the E.coli library (GenBank accession no. SRX000429) (Supplementary Fig. S1). The simulation of base calling errors was the same as that of single-end read errors.
The total number of reads was determined by the following formula:
To study and compare the seven selected de novo assembly tools, sequencing reads were simulated as follows.
To determine how assembly performance was affected by different depths of coverage and GC contents, single-end reads (BCER=0.6%, read length=35, 50 and 75 bp) and paired-end reads (BCER=0.6%, read length=35 bp*2, 75 bp*2, 125 bp*2) were generated from four benchmark sequences (sequences 1–4 in Table 1), in which GC content was ~36–50%.
To determine how the assembly performance was affected by different BCER, sequencing reads were generated with BCER set to 0.0, 0.2, 0.4, 0.6, 0.8 and 1.0%. Three benchmark sequences (sequences 1–3 in Table 1) were selected for the simulation. In single-end reads assembly, read length was 35 bp, and depth of coverage was set to 30× and 70×. In paired−end reads assembly, read length was 35bp*2 and depth of coverage was 30× and 70×.
To compare required computational demand (runtime and computer memory usage) of the seven tools, four benchmark sequences with gradually increasing lengths ranging from ~5 million bp to ~100 million bp (sequences 5–8 in Table 1) were selected for simulation. BCER was set to 0.6%, individual read lengths were set to 35 bp for single-end and 35bp*2 for paired-end reads, and depth of coverage was set to 70×.
2.4 Runtime settings
Runtime parameters for the seven assembly tools were generally set to the default or recommended values of each method with a few exceptions: for VCAKE, the runtime parameter c was set as 0.7 in order to make it consistent with SSAKE. [Each base call in VCAKE was dependent on a voting result; when the votes were totaled and the base proportion exceeded a threshold, c, that base was added to the output contig (Jeck et al., 2007).] Parameter k for Velvet, ABySS, SOAPdenovo and parameter m for Edena should vary with read length in order to get good N50 lengths. Since no clear default settings for these parameters were presented in the manuals for the corresponding tool, we established values for k and m that produced relatively optimal N50 lengths, based on our own preliminary empirical testing of conditions for each tool. Specific values of the parameters k and m are provided in Supplementary Table S1.
Most of the assembly was carried out on a cluster with eight computer nodes, with each node consisting of dual Quad-Core (2.40 GHz) processors and 12 GB RAM. Comparison tests of required computational demand were performed on a server with dual Quad-Core (2.40 GHz) Processors and 32 GB RAM.
2.5 Performance evaluation
The seven selected de novo assembly tools were applied to assemble the simulated sequencing reads into contigs. In paired-end assembly, tools that support paired-end reads performed an additional step of scaffold construction to get the final output contigs. Contigs with lengths >100 bp were used to evaluate the performance of each tool. Each simulation and assembly was conducted five times, and the assembly results were set as the average values.
The performance of each tool was measured by a number of metrics, including N50 length, sequence coverage, assembly error rate, computation time and computer memory usage. N50 length is the longest length such that at least 50% of all base pairs are contained in contigs of this length or larger (Lander et al., 2001). N50 length provides a standard measure of assembly connectivity, reflecting the nature of the bulk of the assembly rather than the cutoff which defines the smallest reportable assembly unit (Jaffe et al., 2003). Higher N50 length indicate better performance of the assembly tool. Sequence coverage refers to the percentage of the benchmark sequence covered by output contigs. In the calculation of assembly error rates, we aligned the output contigs to the benchmark sequence, and calculated the number of mismatched bases from alignment results. The assembly error rate was the percentage of these mismatched bases in the total bases of aligned contigs in the reference sequence. Sequence coverage and assembly error rates were analyzed by blastz (Schwartz et al., 2003).
3 RESULTS
3.1 Assembly performance affected by depth of coverage and GC content
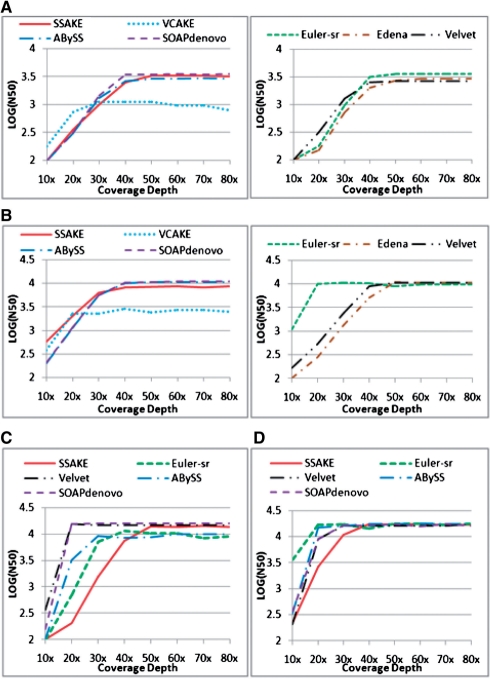
To determine whether, and how, the assembly performance of the seven tools was differentially affected by the depth of coverage and extent of GC content in the source sequences, these tools were used to assemble simulated sequence reads (BCER=0.6%) generated from different benchmark sequences (GC content=~36–50%) at different depths of coverage. Assembly performance of the seven tools is illustrated in Figure 1 and Tables 2–5. Figure 1 and Tables 4 and 5 present test results for part of a benchmark sequence as an example, but similar results were obtained for the other benchmark sequences tested (Supplementary Tables S2–9).
Fig. 1.
Comparison of the effect of various coverage depths on N50 length in T.bru assembly when BCER was 0.6%. (A) Single-end reads assembly, read length (RL)=35 bp; (B) single-end assembly, RL=75 bp; (C) paired-end reads assembly, RL=35 bp; (D) paired-end assembly, RL=75 bp.
Table 2.
Comparison of N50 lengths in assembly of single-end reads when depth of coverage was 70× and BCER was 0.6%
| Seq | RL (bp) | SS | VC | Eu | Ed | Ve | AB | SO |
|---|---|---|---|---|---|---|---|---|
| D.mel | 35 | 6717 | 2215 | 9064 | 4917 | 4085 | 4087 | 4145 |
| H.inf | 25 558 | 2669 | 26 491 | 19 231 | 17 988 | 18 547 | 22 036 | |
| T.bru | 3264 | 963 | 3528 | 2934 | 2667 | 3014 | 3504 | |
| H.sap-1 | 1177 | 653 | 1393 | 1053 | 910 | 961 | 1202 | |
| D.mel | 75 | 28 646 | 3683 | 23 676 | 22 695 | 22 679 | 22 673 | 25 115 |
| H.inf | 46 069 | 3235 | 38 667 | 38 724 | 38 715 | 38 361 | 42 778 | |
| T.bru | 8205 | 2682 | 9733 | 10 847 | 10 682 | 10 814 | 11 108 | |
| H.sap-1 | 2706 | 691 | 2169 | 4315 | 3810 | 3358 | 5227 |
RL, read length; Seq, benchmark sequence; SS, SSAKE; VC, VCAKE; Eu, Euler-sr; Ed, Edena; Ve, Velvet; AB, ABySS; SO, SOAPdenovo.
Table 3.
Comparison of N50 length in assembly of paired-end reads when depth of coverage was 70× and BCER was 0.6%
| Seq. (GC %) | RL (bp) | SS | Eu | Ve | AB | SO |
|---|---|---|---|---|---|---|
| D.mel (36.90) | 35 | 29 771 | 27 326 | 28 604 | 29 892 | 30 308 |
| H.inf (38.16) | 91 821 | 90 275 | 92 349 | 93 956 | 1 19 805 | |
| T.bru (44.38) | 14 470 | 9498 | 14 948 | 9998 | 15 598 | |
| H.sap-1 (49.81) | 3188 | 3116 | 4730 | 4281 | 14 972 | |
| D.mel (36.90) | 75 | 29 963 | 29 029 | 29 676 | 30 923 | 30 863 |
| H.inf (38.16) | 1 22 151 | 1 07 232 | 1 20 699 | 1 20 175 | 1 20 886 | |
| T.bru (44.38) | 16 768 | 17 051 | 16 094 | 17 566 | 16 326 | |
| H.sap-1 (49.81) | 7436 | 4041 | 34 592 | 33 429 | 34 265 |
GC, GC content.
Table 4.
Comparison of sequence coverage and assembly error rates in assembly of single-end reads with various GC contents and depths of coverage (BCER=0.6%)
| RL | Seq | DC | SS | VC | Eu | Ed | Ve | AB | SO | |
|---|---|---|---|---|---|---|---|---|---|---|
| (bp) | (GC%) | |||||||||
| SC (%) | 35 | D.mel (36.90) | 30× | 79.48 | 78.76 | 75.44 | 77.17 | 77.43 | 77.55 | 78.70 |
| 50× | 79.74 | 77.60 | 76.33 | 78.55 | 77.97 | 78.29 | 78.59 | |||
| 70× | 79.54 | 77.70 | 76.40 | 78.33 | 77.86 | 78.06 | 78.62 | |||
| T.bru (44.38) | 30× | 72.64 | 71.01 | 68.07 | 67.02 | 67.78 | 67.19 | 68.74 | ||
| 50× | 73.16 | 70.39 | 68.40 | 67.45 | 67.94 | 67.21 | 68.65 | |||
| 70× | 73.56 | 70.40 | 68.59 | 67.27 | 67.58 | 67.15 | 68.76 | |||
| 75 | D.mel (36.90) | 30× | 80.93 | 79.44 | 78.94 | 78.41 | 79.93 | 79.92 | 80.09 | |
| 50× | 80.13 | 79.45 | 78.69 | 80.58 | 79.83 | 79.82 | 80.49 | |||
| 70× | 80.99 | 79.83 | 79.40 | 80.02 | 79.99 | 79.83 | 80.82 | |||
| T.bru (44.38) | 30× | 77.92 | 77.12 | 71.02 | 75.67 | 74.84 | 74.83 | 76.99 | ||
| 50× | 77.57 | 78.43 | 70.60 | 76.20 | 74.91 | 74.66 | 76.59 | |||
| 70× | 78.68 | 78.38 | 71.48 | 76.99 | 74.86 | 75.02 | 76.70 | |||
| AER (%) | 35 | D.mel (36.90) | 30× | 0.31 | 0.27 | 0.34 | 0.23 | 0.28 | 0.26 | 0.32 |
| 50× | 0.39 | 0.29 | 0.36 | 0.33 | 0.29 | 0.23 | 0.32 | |||
| 70× | 0.32 | 0.24 | 0.38 | 0.26 | 0.23 | 0.26 | 0.39 | |||
| T.bru (44.38) | 30× | 0.27 | 0.17 | 0.25 | 0.08 | 0.07 | 0.04 | 0.16 | ||
| 50× | 0.32 | 0.14 | 0.26 | 0.07 | 0.05 | 0.04 | 0.14 | |||
| 70× | 0.33 | 0.16 | 0.26 | 0.09 | 0.06 | 0.04 | 0.10 | |||
| 75 | D.mel (36.90) | 30× | 0.42 | 0.75 | 0.42 | 0.53 | 0.28 | 0.23 | 0.39 | |
| 50× | 0.42 | 0.76 | 0.45 | 0.49 | 0.37 | 0.29 | 0.41 | |||
| 70× | 0.45 | 0.79 | 0.49 | 0.53 | 0.35 | 0.31 | 0.43 | |||
| T.bru (44.38) | 30× | 0.63 | 0.67 | 0.47 | 0.59 | 0.46 | 0.42 | 0.66 | ||
| 50× | 0.56 | 0.84 | 0.52 | 0.46 | 0.49 | 0.48 | 0.63 | |||
| 70× | 0.65 | 0.88 | 0.48 | 0.53 | 0.46 | 0.49 | 0.67 |
SC, sequence coverage; AER, assembly error rate.
Table 5.
Comparison of sequence coverage and assembly error rates in assembly of paired-end reads with various GC contents and depths of coverage (BCER=0.6%)
| RL (bp) | Seq (GC%) | DC | SS | Eu | Ve | AB | SO | |
|---|---|---|---|---|---|---|---|---|
| SC (%) | 35 | D.mel (36.90) | 30× | 77.05 | 71.12 | 78.75 | 79.53 | 78.85 |
| 50× | 79.16 | 71.98 | 79.03 | 79.65 | 78.52 | |||
| 70× | 79.11 | 70.61 | 78.95 | 79.71 | 78.92 | |||
| T.bru (44.38) | 30× | 72.69 | 71.07 | 71.37 | 73.46 | 70.07 | ||
| 50× | 72.50 | 71.67 | 71.50 | 73.11 | 70.34 | |||
| 70× | 73.59 | 69.08 | 71.32 | 73.27 | 70.78 | |||
| 75 | D.mel (36.90) | 30× | 79.77 | 71.31 | 79.18 | 81.00 | 80.20 | |
| 50× | 79.88 | 70.17 | 78.79 | 80.82 | 80.36 | |||
| 70× | 79.73 | 69.54 | 78.43 | 81.59 | 79.69 | |||
| T.bru (44.38) | 30× | 77.19 | 71.56 | 76.52 | 81.97 | 76.25 | ||
| 50× | 78.29 | 70.12 | 76.66 | 81.97 | 76.54 | |||
| 70× | 78.49 | 70.86 | 78.17 | 81.10 | 76.55 | |||
| AER (%) | 35 | D.mel (36.90) | 30× | 0.33 | 0.54 | 0.17 | 0.14 | 0.28 |
| 50× | 0.32 | 0.52 | 0.19 | 0.16 | 0.29 | |||
| 70× | 0.34 | 0.44 | 0.21 | 0.19 | 0.33 | |||
| T.bru (44.38) | 30× | 0.30 | 1.30 | 0.25 | 0.19 | 0.21 | ||
| 50× | 0.34 | 0.87 | 0.22 | 0.17 | 0.23 | |||
| 70× | 0.40 | 0.73 | 0.21 | 0.21 | 0.20 | |||
| 75 | D.mel (36.90) | 30× | 0.47 | 0.37 | 0.27 | 0.13 | 0.35 | |
| 50× | 0.41 | 0.48 | 0.21 | 0.14 | 0.42 | |||
| 70× | 0.62 | 0.38 | 0.23 | 0.16 | 0.35 | |||
| T.bru (44.38) | 30× | 0.52 | 0.79 | 0.55 | 0.45 | 0.39 | ||
| 50× | 0.52 | 0.61 | 0.57 | 0.39 | 0.43 | |||
| 70× | 0.61 | 0.72 | 0.61 | 0.42 | 0.42 |
With increasing depths of coverage, the performance of these seven tools showed some interesting patterns (Fig. 1) in assembly connectivity measured by N50 length. Although there was an initial increase in N50 lengths with increasing depth of coverage, N50 lengths reached a plateau when the depth of coverage reached a certain threshold. For simplicity, DCAP will be used here to refer to the depth of coverage at which the N50 length plateau was reached.
In single-end assembly, DCAP for SSAKE and Edena (~50×) was greater than that for VCAKE, Velvet, ABySS and SOAPdenovo (30–40×); DCAPs for Euler-sr varied with read length (~50× when read length was 35 bp and ~20× when read length was 75 bp). In paired-end assembly, DCAPs for most tools were lower than those observed in single-end assembly. DCAPs for SSAKE (~40×) was still greater than that for Velvet, ABySS and SOAPdenovo (20–30×); DCAPs for Euler-sr varied with read length (~40× when read length was 35bp*2 and ~20× when read length was 75bp*2).
To compare N50 values among the various tools, we chose N50 values at a depth of coverage of 70×, because this exceeded the DCAP for all tools (Tables 2 and 3). General observations for N50 values of these tools under these various conditions are described below. Comparison results varied with different read lengths and GC content. Sequences with a GC content of 36.90 and 38.16% are referred to as ‘Low GC content’, whereas, those with a GC content of 44.38 and 49.81% are referred to as ‘High GC content’. Similarly, ‘short read’ and ‘long read’ refer to 35 and 75 bp read lengths, respectively.
In single-end reads assembly, with:
low GC content and short read: N50EULER-sr≥N50SSAKE>N50SOAPdenovo≈N50Edena>N50Velvet≈N50ABySS>N50VCAKE;
low GC content and long read: N50SSAKE>N50SOAPdenovo>N50Edena≈N50Velvet≈N50ABySS≈N50EULER-sr>N50VCAKE;
high GC content and short read: N50EULER-sr≥N50SOAPdenovo≈N50SSAKE>N50Edena≈N50Velvet≈N50ABySS>N50VCAKE; and
high GC content and long read: N50SOAPdenovo>N50Edena≥N50Velvet≈N50ABySS>N50SSAKE>N50EULER-sr>N50VCAKE.
In paired-end reads assembly:
SOAPdenovo generated the greatest N50 lengths in almost all tests;
SSAKE generated relatively high N50 lengths when GC content was low;
N50 lengths for Velvet and ABySS were comparable to one another for all tests;
N50 lengths for Velvet and ABySS were comparable to SOAPdenovo when assembling long reads; and
N50 lengths for Euler-sr were the lowest for almost all tests.
3.2 Assembly performance with regard to sequence coverage and assembly error rate
Using benchmark sequences D.mel and T.bru as examples, we compared assembly performance of the seven tools with regard to sequence coverage and assembly error rate (Tables 4 and 5). Generally, long reads resulted in high sequence coverage and assembly error rates.
In single-end reads assembly:
SSAKE and VCAKE were comparable to one another, and provided higher sequence coverage than the other tools. Sequence coverage for SOAPdenovo was a little lower, but very close to SSAKE when assembling long reads (75 bp);
Edena, Velvet and ABySS were clustered together, with slightly lower sequence coverage than SOAPdenovo;
Euler generated the lowest sequence coverage for almost all tests;
ABySS showed the lowest assembly error rates for almost all tests; and
SSAKE, VCAKE, SOAPdenovo and Euler-sr generated higher assembly error rates than Edena, Velvet and ABySS.
In paired-end reads assembly:
sequence coverage comparisons had the following relationships: SCABySS>SCSOAPdenovo≈SCSSAKE≈SCVelvet>SCEuler-sr;
ABySS showed the lowest assembly error rates for almost all tests;
SOAPdenovo generated more assembly errors than Velvet in assembly of sequences with low GC content (e.g. D.mel) but fewer assembly errors than Velvet in assembly of high GC content sequence (e.g. T.bru). The assembly error rate for SOAPdenovo and Velvet were both lower than SSAKE; and
Euler-sr generated the highest assembly error rates for almost all tests.
3.3 Assembly performance affected by different BCER
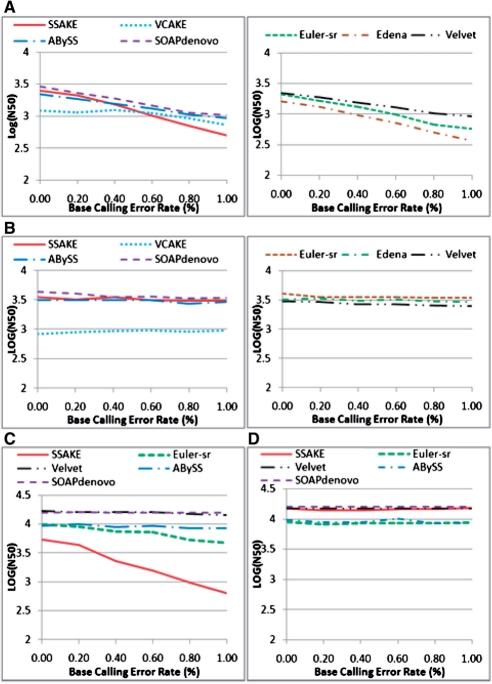
To determine whether, and how, assembly performance of the seven tools was differentially affected by changes in BCER, these tools were applied to assemble sequencing reads simulated from three benchmark sequences (D.mel, H.inf and T.bru) with variable BCER (0.0–1.0%, with a 0.2% incremental change at every step).
Since similar results were obtained with the three benchmark sequences (Supplementary Tables S10–15), we present the results for sequence T.bru as an example (Fig. 2).
Fig. 2.
Comparison of the effects of various BCER on N50 length in T.bru assembly when read length was 35 bp. (A) Single-end reads assembly, depth of coverage (DC)=30×; (B) single-end assembly, DC=70×; (C) paired−end reads assembly, DC=30×; (D) paired-end assembly, DC=70×.
N50 lengths for all seven tools showed decreasing trends, with increases in BCER, but generated different patterns.
When depth of coverage was below the DCAP of a tested tool, N50 lengths for the specific tool decreased exponentially with increases in BCER. When depth of coverage was below the DCAP (e.g. 30×), increases in BCER produced more significant decreases in N50 lengths for SSAKE, Edena and Euler-sr than for Velvet, ABySS and SOAPdenovo.
When depth of coverage exceeded the corresponding DCAP, however, N50 lengths were essentially unaffected by changes in BCER.
For instance, in Figure 2A, N50 lengths decreased with increasing BCER when depth of coverage was at 30× for all tools, but were essentially unaffected by changes in BCER when depth of coverage exceeded their DCAP (e.g. 70×, Fig. 2B and D).
Similarly, for paired-end assembly at a depth of coverage of 30×, N50 lengths for SSAKE and Euler-sr decreased exponentially with increases in BCER, but N50 lengths for Velvet, ABySS and SOAPdenovo remained stable as BCER increased (Fig. 2C). Thus, the pattern described above is sustained, because 30× is below DCAP of SSAKE and Euler−sr (~50×), but exceeded DCAP of Velvet, ABySS and SOAPdenovo (20–30×).
3.4 Computational demand
When selecting a tool for de novo sequence assembly, computational demand by the tool should also be considered. This is particularly important when analyzing large genome sequence data (e.g. human genomes) for large samples. The utility of a tool can be seriously limited if it takes up excessive memory space, consumes too much CPU time and exceeds reasonable execution time. Consequently, we compared the runtime (RT) and resident memory usage (RM) required for the seven tools to assemble large datasets. The test results are presented in Table 6.
It was not feasible to use some of these tools to assemble large sequences because memory required for the assembly process was beyond our computer power. For instance, SSAKE could not assemble sequences >20 Mb (C.ele, H.sap-2 and H.sap-3). VCAKE and Euler-sr could not assemble sequences >50 mega bps (H.sap-2, H.sap-3). Edena could not assemble sequence >100 mega bps (H.sap-3). Velvet could not assemble paired-end reads of the H.sap-3 sequence.
Runtime and RM usage varied dramatically in this test. For all tools, there was an approximately linear increase in memory consumption with increasing benchmark sequence lengths, with RMSSAKE> RMVCAKE> RMEdena> RMEuler-sr> RMVelvet > RMABySS≥ RMSOAPdenovo in single-end reads assembly and RMSSAKE> RMEuler-sr> RMSOAPdenovo> RMABySS in paired-end reads assembly.
The runtime of these tools also increased approximately linearly with increasing benchmark sequence lengths, with RTSSAKE> RTVCAKE> RTEuler-sr> RTEdena> RTABySS> RTVelvet> RTSOAPdenovo.
Runtime and RM usage for Velvet sometimes became abnormal in paired-end reads assembly of large genomes. For example, in paired-end reads assembly of H.sap-2, Velvet consumed much more memory and runtime than ABySS and SOAPdenovo; in paired-end reads assembly of H.sap-3, Velvet could not even finish the assembly.
In general, SOAPdenovo and ABySS were more efficient than other tools in terms of runtime and memory usage. SSAKE consumed the greatest amount of computational resources.
In this test, we also analyzed N50 lengths, sequence coverage and assembly error rate. The results were consistent with several conclusions in previous sections (Supplementary Table S16).
Table 6.
Comparison of runtime and RAM in the computational demand test
| Bench.Seq (length: bp) | E.coli (4.6M) | C.ele (20.9M) | H.sap-2 (50.3M) | H.sap-3 (100.5M) | |
|---|---|---|---|---|---|
| Runtime (s) | |||||
| SE | SSAKE | 2776 | – | – | – |
| VCAKE | 1672 | 16 742 | – | – | |
| Euler-sr | 1689 | 11 961 | 29 622 | – | |
| Edena | 895 | 8450 | 17 043 | – | |
| Velvet | 205 | 1003 | 2786 | 6098 | |
| ABySS | 265 | 1300 | 3307 | 6608 | |
| SOAPdenovo | 62 | 253 | 560 | 1029 | |
| PE | SSAKE | 9163 | – | – | – |
| Euler-sr | 1455 | 15 068 | – | – | |
| Velvet | 229 | 1351 | 55 581 | – | |
| ABySS | 458 | 3081 | 9199 | 21 683 | |
| SOAPdenovo | 78 | 374 | 889 | 2257 | |
| RAM (MB) | |||||
| SE | SSAKE | 9933 | – | – | – |
| VCAKE | 4099 | 17 408 | – | – | |
| Euler-sr | 1536 | 7065 | 13 312 | – | |
| Edena | 1741 | 7557 | 30 720 | – | |
| Velvet | 1229 | 4045 | 9830 | 22 528 | |
| ABySS | 1126 | 3993 | 8909 | 18 432 | |
| SOAPdenovo | 935 | 2867 | 8089 | 18 227 | |
| PE | SSAKE | 16 384 | – | – | – |
| Euler-sr | 1638 | 7578 | – | – | |
| Velvet | 1331 | 5324 | 30 720 | – | |
| ABySS | 950 | 4505 | 9830 | 18 432 | |
| SOAPdenovo | 1638 | 5939 | 10 342 | 19 456 | |
Bench.Seq, benchmark sequence; SE, single-end reads assembly; PE, paired-end reads assembly. ‘–’ denotes the RAM of computer is not enough or runtime is too long (>10 days) to get assembly results.
4 CONCLUSIONS AND DISCUSSIONS
This study compared seven publically available and commonly used de novo assembly tools: SSAKE, VCAKE, Euler-sr, Edena, Velvet, ABySS and SOAPdenovo. These tools are specifically designed to assemble large numbers of short reads generated by next-generation sequencing platforms.
In analyzing these tools, stronger performance is indicated by higher N50 values, higher sequence coverage, lower assembly error rates and lower computational resource consumption (to enable assembly of larger genomes). The performance of different assembly tools was dependent, to some extent, on the test conditions. Based on the results of our investigation, we propose the following guidelines for tool selection. Generally, SSAKE, Edena and Euler-sr need higher depths of coverage (~50×) than Velvet, ABySS and SOAPdenovo (~30×) to generate higher N50 lengths; SOAPdenovo was the fastest of all tools, and ABySS almost always consumed the least memory space. We have developed a tentative reference/guidelines for selecting optimal de novo tools under varying conditions (Table 7). Specific comments regarding the performance of individual tools under different conditions are summarized below.
Table 7.
Recommendations for de novo tool selection under varying conditions
| Read property |
Small genome |
Large genome |
||||||
|---|---|---|---|---|---|---|---|---|
| GC | Read | High N50 | High SC | Low AER | High N50 | High SC | Low AER | |
| SE | Low | Short | Eu, SS | SS | Ed, AB, Ve | Eu, SO, Ed | SO, Ed, AB, Ve | Ed, AB, Ve |
| Long | SS, SO | SS | AB, Ve | SO | SO, Ed, AB, Ve | AB, Ve | ||
| High | Short | Eu, SO | SS, SO | AB, Ve, Ed | SO, Eu | SO | AB, Ve, Ed | |
| Long | SO, Ed, AB, Ve | SS, SO | AB, Ve | SO, Ed | SO | AB, Ve | ||
| PE | Low | Short | SO, SS, AB, Ve | AB, SS, Ve, SO | AB, Ve, SO | SO, AB, Ve | AB, SO, Ve | AB, Ve, SO |
| Long | SO, SS | AB, SS, SO, Ve | AB, Ve, SO | SO, AB, Ve | AB, SO, Ve | AB, Ve, SO | ||
| High | Short | SO | AB | AB, Ve, SO | SO | AB | AB, Ve, SO | |
| Long | SO, AB, Ve | AB | AB, Ve, SO | SO, AB, Ve | AB | AB, Ve, SO | ||
Requirements of assembly performance includes high N50, high sequence coverage (SC), low assembly error rate (AER). For different requirements, we recommend some de novo tools with order of priority according to properties of sequence reads, including single-end/paired-end, GC content, read length and sequence length. SE, single end reads; PE, paired end reads; Eu, Euler-sr; SS, SSAKE; Ed, Edena; AB, ABySS; Ve, Velvet; SO, SOAPdenovo.
SSAKE provided good sequence coverage, and also generated good N50 lengths when assembling sequences with low GC content. On the other hand, SSAKE tended to generate more assembly errors and needed more depth of coverage to reach DCAP than most of the other tools tested. The time and memory usage of SSAKE was also the highest of the tools tested. Our results indicated that assembly of large sequences (e.g. Homo sapiens) was not feasible with SSAKE.
VCAKE produced the shortest N50 lengths in most situations, and the sequence coverage by VCAKE was comparable to SSAKE. VCAKE also generated many assembly errors, even higher than that of SSAKE under certain test conditions. The computational resources required to run VCAKE were a little less than those required for SSAKE.
In assembling single-end short reads, Euler-sr produced the longest N50 values, but it also generated high assembly error rates, comparable to that of SSAKE. In addition, sequence coverage of Euler-sr was the lowest under most test situations. Euler-sr consumed intermediate computational resources.
Under most conditions tested, Velvet and ABySS show similar assembly performance; they generated similar N50 lengths, their DCAPs were relatively low and they required acceptable computational resources. Consequently, it is feasible to use these tools for assembling large sequences, such as those obtained for Homo sapiens. ABySS produced fewer assembly errors, and consumed a little less memory and more runtime than Velvet. When assembling paired-end reads, ABySS produced the highest assembly coverage of all tools tested. When assembling larger genomes, Velvet sometimes used exceptionally high runtimes and memory.
Edena needs a high depth of coverage, comparable to SSAKE, to reach the DCAP. It produced similar, or greater, N50 values to Velvet in most single-end assemblies, and generated assembly error rates that were comparable to Velvet. The computation demands of Edena were intermediate, between SSAKE and ABySS.
SOAPdenovo was the fastest assembler. Its DCAP was as low as that of ABySS and it produced among the highest N50 values in paired-end read assembly, and relatively high N50 values in single-end assembly. SOAPdenovo generated higher assembly error rates and lower sequence coverage than ABySS. It also consumed more memory than ABySS when assembling paired-end reads. The appropriate setting for SOAPdenovo (SOAPdenovo31mer, SOAPdenovo63mer and SOAPdenovo127mer that support kmer ≤31,≤63 and ≤127, respectively) must be selected based on read length. SOAPdenovo63mer/SOAPdenovo127mer consumed two/four times as much RAM as SOAPdenovo31mer.
In light of our results, investigators may choose the most appropriate assembly tool(s) to use based on their specific experimental setting and available computational resources. Our results may also serve as a reference, when designing sequencing projects, for selecting targeted depths of coverage, control levels of sequencing error rates, etc. Given the rapid increase in use of next-generation sequencing technologies, our results should be of value to both empiricists, during experimental design, and to bio-informaticians who seek guidance for selecting appropriate assembly tool(s) for data analyses and who attempt improvement of the assembly tools.
Funding: Shanghai Leading Academic Discipline Project (S30501 in part); startup fund from Shanghai University of Science and Technology. The investigators of this work were partially supported by grants from NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049 and R03TW008221); Franklin D. Dickson/Missouri Endowment from University of Missouri–Kansas City and the Edward G. Schlieder Endowment from Tulane University.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Chaisson M.J., Pevzner P.A. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J.C., et al. SHARCGS, a fast and highly accurate short-read assembly algorithm for de novo genomic sequencing. Genome Res. 2007;17:1697–1706. doi: 10.1101/gr.6435207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Hernandez D., et al. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe D.B., et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., et al. Extending assembly of short DNA sequences to handle error. Bioinformatics. 2007;23:2942–2944. doi: 10.1093/bioinformatics/btm451. [DOI] [PubMed] [Google Scholar]
- Lander E.S., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li R., et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.R., et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., et al. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Schwartz S., et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Simpson J.T., et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudien S., et al. Should the draft chimpanzee sequence be finished? Trends Genet. 2006;22:122–125. doi: 10.1016/j.tig.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Warren R.L., et al. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23:500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., et al. A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies. PLoS One. 2011;6:e17915. doi: 10.1371/journal.pone.0017915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.