Abstract
Motivation: A plugin for the Java-based PathVisio pathway editor has been developed to help users draw diagrams of bioregulatory networks according to the Molecular Interaction Map (MIM) notation. Together with the core PathVisio application, this plugin presents a simple to use and cross-platform application for the construction of complex MIM diagrams with the ability to annotate diagram elements with comments, literature references and links to external databases. This tool extends the capabilities of the PathVisio pathway editor by providing both MIM-specific glyphs and support for a MIM-specific markup language file format for exchange with other MIM-compatible tools and diagram validation.
Availability: The PathVisio-MIM plugin is freely available and works with versions of PathVisio 2.0.11 and later on Windows, Mac OS X and Linux. Information about MIM notation and the MIMML format is available at http://discover.nci.nih.gov/mim. The plugin, along with diagram examples, instructions and Java source code, may be downloaded at http://discover.nci.nih.gov/mim/mim_pathvisio.html.
Contact: margot@discover.nci.nih.gov; augustin@mail.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Understanding biological pathways requires knowledge of both the biological components of a pathway and the interactions between its components. Improvements in molecular biology techniques have fueled a surge in this type of information. This has made the organization of biological knowledge along with consistent mechanisms for how this information is exchanged among researchers of utmost importance.
One of the most common means of relating information about biological pathways is through the use of pathway diagrams which are oftentimes more ‘natural’ to readers. The Molecular Interaction Map (MIM) notation is a graphical notation for depicting diagrams of biochemical and cellular processes designed to alleviate ambiguity that exists in many diagrams currently found in publications (Kohn et al., 2006). MIM diagrams are similar to road maps or electronic circuit diagrams, and are used to define entities and their interactions in biological systems.
The MIM notation has been described extensively in the past, but until now it has only limited software support. Software is important to make the creation of large-scale diagrams, typical to MIM diagrams, feasible. Previously, creation of MIM diagrams was conducted using general graphics products, such as Adobe Illustrator. Some missing features in these tools include easy access to MIM glyphs, connectors that remain attached to elements as they are rearranged, connectors with routing capability and functionality to associate annotations to specific elements. A comparison of several interaction network and pathway diagram editors is provided in Table 1 in the Supplementary Material.
PathVisio is a full-featured diagram editor for biological pathways with the ability to change many attributes of diagram elements: data nodes (color, size and labels) and connecting lines (color and arrowheads) (van Iersel et al., 2008). PathVisio also allows elements to be linked to external databases and references, and it is capable of producing graphics in a variety of formats including SVG, PNG and PDF. PathVisio is a Java-based application available on all major computing platforms (e.g. Windows, Mac OS X and Linux). Additionally, PathVisio has provided some of the graphical elements necessary for the MIM notation since PathVisio 1.1.
Since these earlier versions, continued development of the MIM support has been conducted as an independent plugin. By moving MIM functionality into a plugin, the user interface can be highly customized to MIM support, while the core program remains open to unformatted pathway diagrams. The major objective of this project is to provide complete support for the MIM notation in a simple to use graphical editor and for it to serve as one starting point for future MIM software development.
2 IMPLEMENTATION
PathVisio provides a plugin interface that allows the majority of the editor to be extended and customized; changes were also introduced into the core software as necessary to facilitate MIM-specific features. Here, we present the support for the MIM notation within PathVisio.
2.1 PathVisio-MIM features
The MIM notation has evolved, since its inception in 1999, as it has become necessary to support additional biological constructs. An updated specification (available at http://discover.nci.nih.gov/mim/) was recently created that is meant to facilitate software implementation; readers should refer to Luna et al. (2011) for information regarding the MIM glyphs as implemented in this plugin. The PathVisio-MIM plugin provides full implementation of the updated specification and the following features.
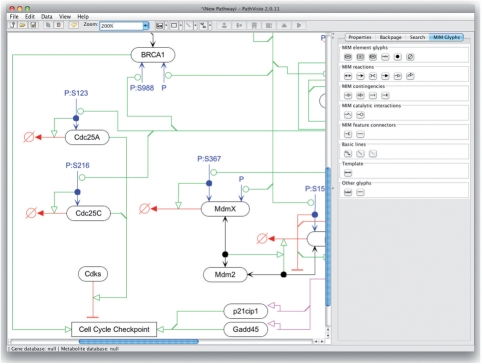
MIM Glyphs: the plugin introduces a new side panel that contains the MIM glyphs separated into the categories laid out by the specification; these are shown in Figure 1. Interaction glyphs drawn through this interface use orthogonal line connectors that are recommended for MIM diagrams. Support for the additional MIM glyphs, such as complexes, branching connectors and the glyph representing intermolecular interactions are provided through user interactions outside the MIM panel; for instance, the product of a binding interaction added through the right-click menu. Instructional videos are provided for usage of these glyphs (http://discover.nci.nih.gov/mim/mim_pathvisio.html).
Fig. 1.
Screenshot of PathVisio diagram editor with PathVisio-MIM plugin loaded. MIM-specific glyphs are shown in the right-hand side panel.
Annotations: interactions on diagrams may be annotated with comments and literature reference information. These annotations allow users to document the sources of interactions and help readers of MIM diagrams find the original resources to gain a greater understanding of a particular biological system. Additionally, entities may be cross-referenced with identifiers from a wide range of external databases, such as Entrez Gene, HUGO gene names, etc.
Support for MIMML: diagrams may be stored as either the native PathVisio GPML format or in the MIM Markup Language (MIMML) format that was recently developed (Luna et al., 2011). The MIMML format is intended as a common exchange mechanism between MIM software, and is supported by another MIM editor, MIMTool (http://code.google.com/p/mimtool/; Edes, M. et al., manuscript in preparation). Additionally, this format is used for the validation of MIM diagrams according to syntax rules outlined by the MIM specification. Import and export functionality in the plugin is provided through a MIM application programming interface (API); export functionality only exports MIM-specific elements to help ensure diagrams are in accordance with the MIM notation and exchangeable with other MIM software. A potential use for the import capability is in loading MIMML documents from other tools for conversion into formats that PathVisio supports.
3 CONCLUSION
The PathVisio-MIM plugin helps biologists draw diagrams in accordance with the MIM notation. This combined with the many features of PathVisio will aid in making the MIM notation accessible to a wider audience. To encourage usage of this software and the notation by first-time users, we provide example files, written documentation and instructional videos on the project web site.
The plugin takes advantage of the extensible nature of PathVisio to provide support of all the major features of the MIM notation and may serve as an example for MIM support in other software. Code additions that were made in the core PathVisio editor should facilitate the support of other notations in PathVisio, such as the Systems Biology Graphical Notation (SBGN) or the modified Edinburgh Pathway notation (Freeman et al., 2010; Le Novère et al., 2009); support for the SBGN notation is currently in development via a separate plugin. Support for these notations or other notations within the same software tool presents a basis for translating diagrams between notations. Future development of the plugin is expected as additions are made to the MIM specification. Also, we expect the addition of validation support in PathVisio for rulesets, such as the one developed for MIMML.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Allan Kutchinsky and Michael Blinov for helpful discussions and the PathVisio community.
Funding: This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the Netherlands Consortium for Systems Biology (NCSB), which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
Conflict of Interest: none declared.
REFERENCES
- Freeman T.C., et al. The mEPN scheme: an intuitive and flexible graphical system for rendering biological pathways. BMC Syst. Biol. 2010;4:65. doi: 10.1186/1752-0509-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn K.W., et al. Molecular interaction maps of bioregulatory networks: a general rubric for systems biology. Mol. Biol. Cell. 2006;17:1–13. doi: 10.1091/mbc.E05-09-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novère N., et al. The systems biology graphical notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Luna A., et al. A formal MIM specification and tools for the common exchange of MIM diagrams: an XML-based format, an API, and a validation method. BMC Bioinformatics. 2011;12:167. doi: 10.1186/1471-2105-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel M.P., et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.