Abstract
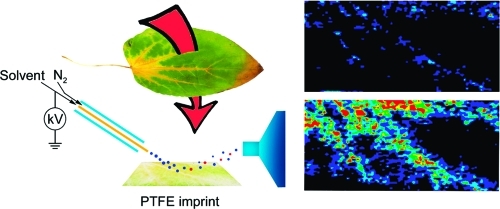
The ambient mass spectrometry technique, desorption electrospray ionization mass spectrometry (DESI-MS), is applied for the rapid identification and spatially resolved relative quantification of chlorophyll degradation products in complex senescent plant tissue matrixes. Polyfunctionalized nonfluorescent chlorophyll catabolites (NCCs), the “final” products of the chlorophyll degradation pathway, are detected directly from leaf tissues within seconds and structurally characterized by tandem mass spectrometry (MS/MS) and reactive-DESI experiments performed in situ. The sensitivity of DESI-MS analysis of these compounds from degreening leaves is enhanced by the introduction of an imprinting technique. Porous polytetrafluoroethylene (PTFE) is used as a substrate for imprinting the leaves, resulting in increased signal intensities compared with those obtained from direct leaf tissue analysis. This imprinting technique is used further to perform two-dimensional (2D) imaging mass spectrometry by DESI, producing well-resolved images of the spatial distribution of NCCs in senescent leaf tissues.
The seasonal metabolism of chlorophyll, evidenced by the autumnal degreening of deciduous plants, is a visible sign of life.(1) It is estimated that every year, more than 109 tons of chlorophyll are degraded.(2) It is remarkable, therefore, that the chemistry involved in chlorophyll breakdown began to be elucidated only within the last two decades.1,3−5 The first structural elucidation of a chlorophyll breakdown product in senescent barley leaves in 1991(3) revealed colorless tetrapyrroles, typified as nonfluorescent chlorophyll catabolites (NCCs), to be the “final” products of a degradation process during leaf senescence. To achieve deeper insights into common biochemical pathways of chlorophyll breakdown in higher plants, structural elucidation was subsequently focused on the characterization of NCCs and their fleetingly appearing precursors in different plant systems.4,6−8 The recent discovery of chlorophyll catabolites in fruit peels and flesh as well as their remarkable antioxidant properties(9) suggests that the chlorophyll degradation process may play a role in plant tissues that goes beyond the mere removal of cytotoxic chlorophyll during plant cell senescence.(10) To further evaluate this hypothesis, rapid and efficient analytical tools for direct identification and molecular profiling in plant tissues are needed.
Direct analysis of biological tissue by mass spectrometry and the even more demanding experiment of spatially resolved identification of chemical compounds in complex biological tissues are spawning powerful technologies.11−16 The ambient ionization technique known as desorption electrospray ionization mass spectrometry (DESI-MS) is of particular interest, since it offers a distinctive and simple approach based on analyte solubility both for direct analysis and for imaging mass spectrometry. Compared with traditional analytical methods that typically involve extensive sample preparation through extraction and chromatographic separation, ambient ionization methods involve the examination of samples under ordinary conditions with minimal to zero sample preparation.15−21 In DESI-MS, a stream of solvent droplets, charged by the application of a high voltage to the liquid, is driven by nitrogen gas and impacts the sample surface. The primary droplets produce a thin liquid film on the sample surface into which the compounds of interest are extracted. Further droplet collisions release analyte-containing secondary microdroplets, which follow the conventional desolvation processes of electrospray ionization as they move through an atmospheric pressure interface into the mass spectrometer.(22) With its simplicity, short analysis time (a few seconds) and high sensitivity, DESI-MS has been successfully used in direct targeted analysis of many different analytes, including fatty acids,(23) hormones,(24) and other small molecules.25−29 The direct characterization of plant tissues for alkaloids,(30) glycosides,(26) and Salvorin A(31) has also been successful.
In the case of spatially resolved identification of chemical compounds, DESI-MS imaging has become increasingly attractive, given its simplicity and the limited sample preparation steps compared with vacuum imaging techniques.32,33 Previous imaging applications of DESI include examination of biological tissue sections to image lipids and the imaging of natural products in algal tissue.34−38 Imprinting of biological samples onto hard surfaces prior to imaging experiments has been reported in the study of membrane lipids by SIMS;(33) NALDI;(39) and in the case of DESI, in the imaging of bacterial cultures.(20) These experiments enhance signal intensities relative to direct imaging. In the latter stages of preparing this manuscript, we became aware of the closely related work of Thunig et al.,(40) also reporting imprints of leaves on porous PTFE. Here, we report on the direct, rapid, and high-throughput analysis of chlorophyll degradation products from leaf tissues and the application of the imprinting technique in the analysis and imaging of these compounds from plant materials by DESI-MS.
Experimental Section
Chemicals and Reagents
Chloroform and dichloromethane were purchased from Sigma-Aldrich (Milkwaukee, WI), and methanol and acetonitrile (HPLC grade) were purchased from Mallinckrodt Baker Inc. (Phillipsburg, NJ). Deionized/distilled water was obtained using a Barnstead/Thermolyne deionizer unit (Barnstead Mega-Pure System, Dubuque, IA). The standard chlorophyll catabolite Cj-NCC-1 from Cercidiphyllum japonicum (Katsura tree) was isolated as described previously.(41) Porous PTFE sheets (thickness 1/16 in.) were purchased from SmallParts Inc. (Seattle, WA); Mitex PTFE filters (5 μm pores), from Millipore (Billerica, MA).
Plant Tissue Selection
Senescent leaves from three different deciduous trees were collected on the Purdue University campus prior to analysis. The collected species were Katsura tree or C. japonicum; American sweetgum or Liquidambar styraciflua; and hophornbeam or Ostrya virginiana.
Instrumentation
All MS experiments were performed using a Thermo Fisher Scientific LTQ mass spectrometer (San Jose, CA, USA). Typical instrumental parameters used included 3 microscans, 100 ms maximum ion injection time, ±15 V capillary voltage, 150 °C capillary temperature, and ±65 V tube lens voltage. Data were acquired and processed using Xcalibur 2.0 software (Thermo Fisher Scientific, San Jose, CA). The identification of analyte ions was confirmed by tandem mass spectrometry using collision-induced dissociation. An isolation window of 1.5 Th (mass/charge units) and a normalized collision energy of 20–30% (manufacturer’s unit) was selected.
Desorption Electrospray Ionization Mass Spectrometry (DESI-MS)
Direct DESI Analysis
The leaf surfaces were spotted with about 5–10 μL of a mixture of chloroform/dichloromethane (1:1, v/v) to partly remove the waxy layer. After evaporation of the solvent mixture, the leaf was mounted onto a microscope glass slide (Gold seal; Portsmouth, NH) using double-sided tape. DESI experiments were carried out using a prototype of the Prosolia Inc. (Indianapolis, IN) Omni Spray desorption electrospray ion source, configured as described previously.42,43 A sprayer-to-surface distance of 2 mm, a sprayer-to-inlet distance of 2.5 mm, an incident spray angle of 55°, and a collection angle of 10° were used. The spray voltage was set at 4–5 kV. Nitrogen gas (140 psi) was used for nebulization. The spray solvent, either 1% formic acid in methanol/water (70:30) or 0.1% ammonium acetate in methanol/water (70:30), was sprayed at a constant flow rate of 10–13 μL/min. Mass spectra were recorded for a few seconds during electrospray contact with the leaf surface. For tandem mass spectrometry (MS/MS) experiments, line scans across the tissue sample were performed, and data was acquired for ∼1 min.
Production of Leaf Imprints
The freshly collected senescent leaf was placed on top of the PTFE material with the lower epidermis facing the PTFE surface. The upper side of the leaf was then covered with a filter paper and a 10 mL round-bottom flask was used as a plunger to imprint the leaf sap onto the PTFE surface.
DESI Imaging Experiments
A 2D moving stage (Newport, Richmond, CA, USA) described elsewhere(44) was used. The leaf imprints were scanned in horizontal rows separated by 150 to 300 μm vertical steps until the entire tissue sample had been analyzed. The lines were scanned at a constant velocity in the range of 170–280 μm/s, and mass spectra were recorded over the m/z 400–900 range. Five microscans were averaged, and the scan time was fixed in the range of 0.7–1.4 s. Under these conditions, lateral spatial resolution in the range of 130–310 μm could be achieved in DESI-MS imaging. For individual settings and acquisition times, see the captions to Figure 5a and Supporting Information Figures S3 and S4. In-house software was used to convert the Xcalibur 2.0 mass spectra files (.raw) into a format compatible with BioMap (freeware, http://www.maldi-msi.org/), which was used to process the mass spectral data to generate two-dimensional spatially accurate ion images.
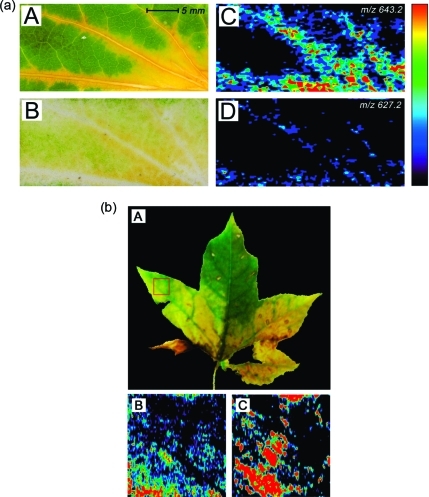
Figure 5.
(a) Negative ion mode DESI imaging of a senescent Katsura tree leaf imprint on porous PTFE substrate. Spray solvent was 1% concentrated aqueous ammonia in methanol at a flow rate of 1.5 μL/min. Imaging parameters: 1.17 s/scan; 98 scans/horizontal row; 56 rows; pixel size was 310 × 250 μm; total acquisition time was 107 min. (A) 30.7 × 13.9 mm section of a photographic image taken from a senescent Katsura leaf. (B) 30.7 × 13.9 mm porous PTFE substrate with Katsura leaf imprint. (C, D) Ion images of the two chlorophyll catabolites7,45 in Katsura leaves at m/z 643.2 and 627.2. Both images are plotted on the same color scale, which is depicted on the right-hand side of the figure to visualize relative ion intensities from 0 (black) to 100 (red). (b) Negative ion mode DESI imaging of a senescent American sweetgum leaf imprint on porous PTFE substrate. Spray solvent was 1% concentrated aqueous ammonia in methanol at a flow rate of 1.5 μL/min. Imaging parameters: 0.72 s/scan; 119 scans/horizontal row; 50 rows; pixel size was 130 × 300 μm; total acquisition time was 71 min. (A) Photographic image taken from a senescent American sweetgum leaf. The rastered 15 × 15 mm section is highlighted in red. (B, C) Ion images of the two chlorophyll catabolites found in American sweetgum(46) at m/z 643.2 and 627.2. Both images are plotted on the same color scale, which is depicted on the right-hand side of panel a to visualize relative ion intensities from 0 (black) to 100 (red).
Results and Discussion
Direct Identification of Chlorophyll Catabolites in Senescent Leaves by DESI-MS
Direct DESI Analysis of the Leaf Surface
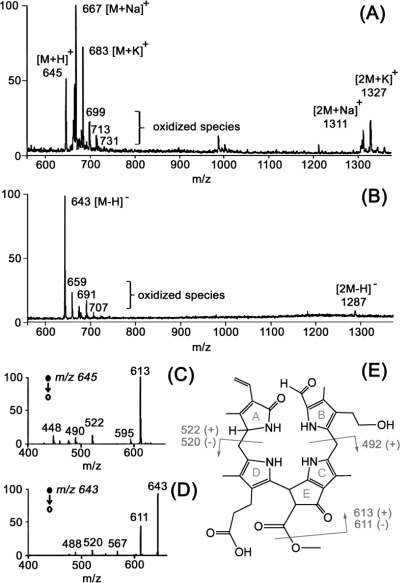
Three higher plants, which were easily accessed on the Purdue University campus, were selected for these experiments. The Katsura tree (C. japonicum) and American Sweetgum (L. styraciflua) are widespread deciduous trees whose chlorophyll degradation products are identified, structurally characterized and described in the literature.7,45,46 The third, the hophornbeam tree (O. virginiana), has not been studied before. Spray solvent composition and DESI source geometry were optimized for both positive and negative ion modes using as a reference compound the pure, isolated chlorophyll catabolite Cj-NCC-1 (molecular mass, 644 Da) from C. japonicum, dissolved in methanol.7,45 Chlorophyll catabolites are known to be linear tetrapyrroles with an even number of nitrogen atoms;(47) hence, the protonated molecules are expected to have odd masses between 600 and 1100 Da. A sample spot on PTFE surface of ∼3 mm in diameter was analyzed using methanol/water (20:80, v/v) as spray solvent. The polyfunctionalized linear tetrapyrrole, Cj-NCC-1, was found to readily undergo ionization in the positive as well as the negative ion mode and could be detected using MS/MS product ion scans at concentrations as low as 10 ppb, corresponding to 20 pg absolute amount. Collision-induced dissociation (CID) experiments showed typical fragmentation patterns, which are characteristic of tetrapyrrolic chlorophyll degradation products(9) (Figure 1). At concentrations above 500 ppm, dimeric ions were observed.
Figure 1.
(A, B) DESI-MS spectra of a 3 mm spot of Cj-NCC-1 standard on a PTFE surface. Spray solvent was methanol/water (20:80) at a flow rate of 3 μL/min. (A) Positive ion mode DESI-MS spectrum. Protonated, sodiated, and potassiated molecules were detected at m/z 645, 667, and 683, respectively; the dimer, at m/z 1311. (B) Negative ion mode DESI-MS. Ions corresponding to deprotonated molecules were observed at m/z 643, and its dimer, at m/z 1287. (C) Positive ion mode DESI-MS/MS spectrum of the protonated molecule, m/z 645. (D) Negative ion mode DESI-MS/MS spectrum of the deprotonated molecule, m/z 643. (E) Constitutional formula of Cj-NCC-1,(45) characteristic CID fragmentations observed in positive ion mode (+) and negative ion mode (−) are highlighted in gray.
The optimized DESI conditions were then used to directly analyze freshly collected senescent leaves of C. japonicum.(45) Pretreatment of the leaf surfaces with about 5–10 μL of a mixture of chloroform/dichloromethane (1:1, v/v) prior to analysis turned out to be necessary for the removal of the waxy top layer. After solvent evaporation, the DESI spray was rastered across the leaf surface until the entire leaf section had been sampled. Signals at m/z 645 as well as at m/z 683, corresponding to [M + H]+ and [M + K]+ of the catabolite Cj-NCC-1, were observed in the full scan mass spectrum (Supporting Information, Figure S2). The detection of Cj-NCC-1 in the senescent leaf was confirmed by CID fragmentation of the ions m/z 645 and 683, which showed fragment ions with relative intensities consistent with those of the isolated Cj-NCC-1 catabolite (Figure 1). A neutral loss of methanol (loss of 32 Da) corresponded to the presence of a methyl ester group, and a loss of 123 Da indicated a vinyl group to be substituted on ring A of the tetrapyrrole.
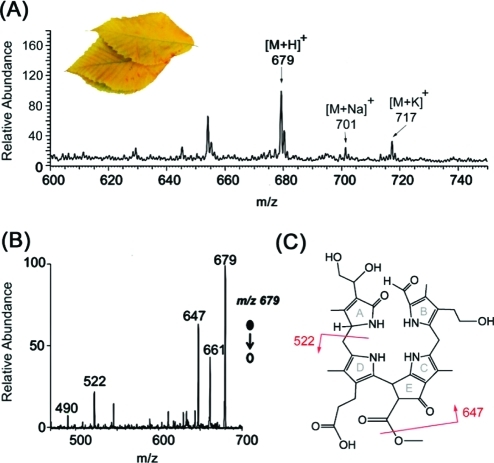
Similar DESI conditions were then used to analyze senescent leaves of the hophornbeam tree. As can be seen in Figure 2, the DESI-MS spectrum showed signals at m/z 679, 701, and 717 as the DESI sprayer was rastered across the pretreated leaf surface. These signals indicated the presence of a compound similar to Hv-NCC-1, the first identified and characterized chlorophyll catabolite from barley leaves (Hordeum vulgare),(3) or So-NCC-2, identified in spinach (Spinacia oleracea).(48) These catabolites differ from Cj-NCC-1 by the presence of a dihydroxyethyl substituent on ring A (for a constitutional structure, see Figure 2, panel C). The ions at m/z 679, 701, and 717 correspond to the protonated, sodiated, and potassiated molecules of this compound, respectively, as further verified by fragmentation of the protonated molecule, m/z 679 (Figure 2, panel B). The characteristic loss of methanol (fragment at m/z 647) was typical of the presence of a methyl ester, whereas a loss of 157 Da (fragment at m/z 522) indicated the cleavage of ring A substituted with a diol functionality. Further confirmation of the proposed structure of the observed catabolite “Ov-NCC-1” from O. virginiana leaves was achieved using reactive DESI experiments. (The terminology Ov-NCC-1 corresponds to Ostrya virginiananon-fluorescent chlorophyll catabolite 1; as can be seen in Supporting Information Figure S1, chlorophyll degradation products are typically named with respect to their retention behavior in RP-HPLC).
Figure 2.
(A) Positive ion mode DESI-MS spectrum of the direct analysis of a hophornbeam (O. virginiana) leaf. Spray solvent was methanol/water (80:20) at a flow rate of 3 μL/min. Protonated, sodiated, and potassiated molecules were detected at m/z 679, 701, and 717, respectively. (B) DESI-MS/MS spectrum of the isolated protonated molecule, m/z 679. (C) Proposed structure of the chlorophyll catabolite corresponding to the ion at m/z 679, the Ov-NCC-1. The structure shown is proposed considering previous data.3,48 Characteristic fragmentations due to the losses of methanol (fragment at m/z 647) and ring A (fragment at m/z 522) are marked.
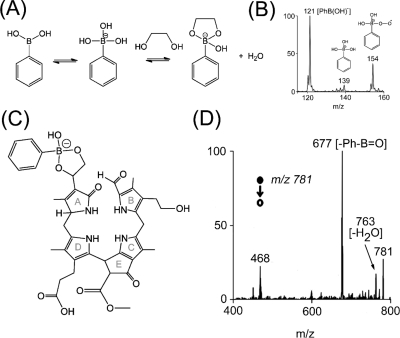
Reactive DESI, a variant of the basic DESI experiment, improves the sensitivity and selectivity of target molecules detection by implementing rapid chemical reactions at the sample spot concurrently with acquisition of mass spectra.49,50 The experiment is carried out by dissolving a suitable chemical reagent into the spray solvent. Reactions selected are chosen to occur rapidly and produce products that are ionized efficiently or have a permanent charge.51,52 In this study, the presence of a diol functionality (as a substituent on ring A) in the proposed structure of Ov-NCC-1 was further confirmed by reactive DESI-MS experiments on the senescent O. virginiana leaf extract. The chemical reagent of choice was an aqueous solution of phenylboronic acid (PhB(OH)2) adjusted to slightly basic pH with aqueous ammonia, as explained elsewhere.(53) In the presence of a cis-diol functionality, a cyclization reaction (see Figure 3; panel A) with phenylboronate anions (PhB(OH)3–; m/z 139) occurs, and the product of the reaction can be detected directly by mass spectrometry in the negative ion mode.(53) As depicted in Figure 3 (panel D), a signal at m/z 781 was observed, and the fragmentation patterns recorded under MS/MS conditions confirm the reaction of phenylboronic acid with the diol substituent of the chlorophyll catabolite; hence, confirming the structure predicted from the MS/MS data of the underivatized catabolite (Figure 2).
Figure 3.
(A) Rapid and reversible covalent complexation of phenylboronic acid with 1,2-diols to form cyclic boronates in basic aqueous media.(53) (B) Reactive species generated in solution and detected during reactive DESI-MS. (C) Proposed structure of the cyclization product of phenylboronic acid and the chlorophyll catabolite. (D) Negative ion mode reactive DESI-MS/MS spectrum of a hophornbeam (O. virginiana) leaf extract spotted on a PTFE surface. Spray solvent at a flow rate of 3 μL/min was 3 mM phenylboronic acid reduced to pH 9 using aqueous ammonia. The deprotonated cyclization product of phenylboronic acid and the chlorophyll catabolite at m/z 781 showed characteristic loss of Ph-B=O.
DESI Analysis of Leaf Imprints on PTFE Surfaces
Imprinting of biological samples onto uniform hard surfaces has been used previously to improve the sensitivity of mass spectrometric imaging techniques.29,33 Prior to DESI-MS leaf imaging, leaf imprints of senescent plants were prepared and analyzed to examine any changes in signal intensities, compared with results from direct leaf analysis experiments. Preliminary direct leaf analysis experiments showed that despite surface pretreatment, chlorophyll catabolite related ions were abundant only at certain hot spots on the leaf surface in the positive as well as negative ion mode.
A possible explanation is provided by the anatomy of angiosperm leaves. Upper and lower regions of a leaf contain disimilar amounts of chlorophylls, and epidermal cells are covered by waxy leaf cuticles.(54) Although a surface pretreatment with dichloromethane/chloroform prior to analysis improved the signal intensity, epidermal cells and residual waxes constituted a mechanical barrier to extraction by charged microdroplets. The imprinting technique similar to that described for thin film DESI imaging(29) was therefore employed to break through this barrier. The choice of porous PTFE as a substrate for imprints was based on previous reports, which showed that the use of this material for DESI analysis makes the desorption of the analytes more efficient, resulting in more intense and stable signals compared with other surfaces.(55) The freshly collected senescent leaf was placed on porous PTFE with the lower epidermis facing the PTFE surface. A plunger was then used to imprint the leaf sap onto the PTFE material, and DESI analysis was carried out on the imprinted PTFE surface.
As a result of this transfer of the relevant compounds onto the PTFE surface, ion intensities from the DESI analysis of the PTFE leaf imprints increased by at least 1 order of magnitude compared with direct analysis. Ion intensitites depended on the area of the leaf being sampled, whether yellow (high NCC concentration) or green (low NCC concentration). Negative ion mode spectra obtained from the analysis of imprints made from senescent Katsura as well as hophornbeam leaves are shown (Figure 4, panels A and D). Among the most abundant signals in the mass region of interest (m/z 600–900) are those of chlorophyll catabolites. With a few exceptions,10,54,56 chlorophyll catabolites feature a free propionic acid side chain functionality, which is formed by saponification of the phytyl ester during chlorophyll degradation. It is easily deprotonated and, therefore, responsible for favorable ionization properties in the negative ion mode.
Figure 4.
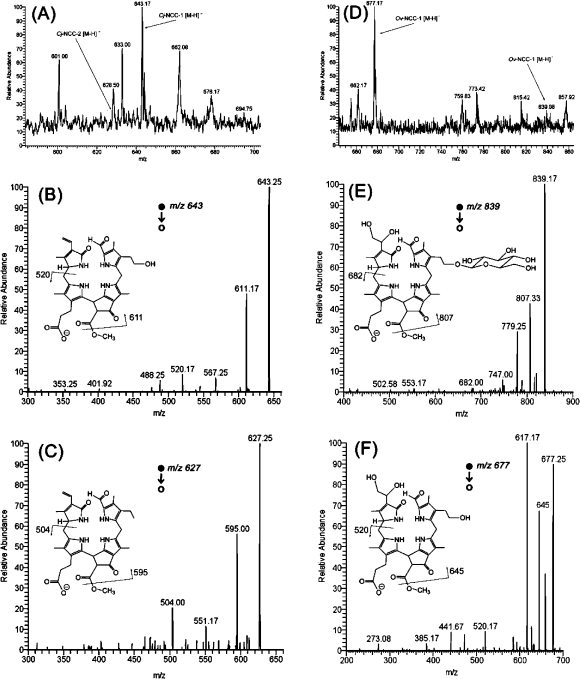
Negative ion mode DESI-MS spectra of PTFE leaf imprints. Spray solvent was 1% concentrated aqueous ammonia in methanol at a flow rate of 3 μL/min. (A) DESI-MS spectrum of a Katsura tree (C. japonicum) leaf imprint. (B, C) DESI-MS/MS spectra of the deprotonated molecules, m/z 643 and 627, corresponding to the two known chlorophyll catabolites in C. japonicum.7,45 Characteristic fragmentations due to the losses of methanol, and ring A are highlighted in the depicted structures. (D) DESI-MS spectrum of a hophornbeam (O. virginiana) leaf imprint. (E, F) DESI-MS/MS spectra of the deprotonated molecules, m/z 677 and 839, corresponding to the two chlorophyll catabolites found in O. virginiana. Characteristic fragmentations due to the losses of methanol, and ring A are highlighted in the inserted structures (proposed considering data for constitutionally identical compounds described in the literature3,48,57,58).
The identification of the compounds responsible for the peaks observed in the mass spectra was based on MS/MS experiments (Figure 4, panels B, C and E, F). Linear tetrapyrrolic compounds generally show characteristic fragmentation patterns under CID conditions. A neutral loss of methanol (32 Da) found in all four MS/MS spectra depicted in Figure 4 (Panels B, C, E, and F) indicated the presence of a methyl ester functionality at the 132 position of the catabolites (as in chlorophyll a). The loss of ring A is often observed and reflects either the presence of a vinyl substituent (123 Da) or a dihydroxyethyl group (157 Da). The loss of a glycose unit in the case of a glycosylated ring B is important, but it can be observed only when the protonated molecule is analyzed in the positive ion mode.
The gain in sensitivity obtained from the use of leaf imprints also allowed for the detection of NCCs that are typically present in relatively lower concentrations and therefore barely detectable directly from leaf tissues. According to previous reports, the natural abundances of the two known chlorophyll degradation products of the Katsura tree (Cj-NCC-1 and Cj-NCC-2) are different.7,45Cj-NCC-1 represents more than 95% of the two described chlorophyll catabolites, which is reflected by the different ion intensities obtained by DESI-MS analysis of the leaf imprint. Although the signal of the deprotonated major catabolite Cj-NCC-1 is intense at m/z 643 (see Figure 4, panel A), the signal of the minor abundance Cj-NCC-2 at m/z 627 was barely visible in the full scan mode. Nevertheless, the ions at m/z 627 were isolated and fragmented to give a high-quality spectrum (Figure 4, panel C). A different pair of catabolites was detected in imprints of senescent leaves from the hophornbeam tree. The signals of the two deprotonated molecules at m/z 677 and 839 were detected at a ratio of 5:1 (Figure 4, panel B). Subsequent HPLC analysis of extracts of senescent hophornbeam leaves shows the same compounds, although in different ratios within the plant tissue (see Supporting Information Figure S1), the peak m/z 677 being more intense than m/z 839.
Imaging Chlorophyll Catabolites in Senescent Plant Tissues by Desorption Electrospray Ionization Mass spectrometry
For imaging experiments, the fresh imprints of senescent leaves were prepared as described. DESI-MS imaging was performed on the imprints in the negative ion mode, detecting ions in the m/z 400–900 range. A typical image section of a senescent Katsura tree leaf is shown in Figure 5 (panel A). The degreening of the leaf can be seen in the regions near the veins of the leaf. Panel B shows the PTFE imprint of a selected leaf section, and ion images of two deprotonated chlorophyll catabolites in senescent leaves of the Katsura tree, m/z 643.2 and 627.2, are depicted in panels C and D, respectively. Each selected ion image is plotted on the same color scale (depicted on the right side of Figure 5). As expected, the most intense signals are those of the [M – H]− ions of the chlorophyll catabolites and are found in the degreened areas of the leaf. Small features depicting the beginning of senescence are reflected by ion intensities. Light microscopic images of leaf imprints (see Supporting Information Figure S5) indicated that the loss of spatial resolution due to the imprinting procedure could be kept in a range below 200 μm, which corresponds to the actual lateral resolution using the described DESI imaging method. A comparison of the images of the two ions also confirmed that catabolite Cj-NCC-1 (m/z 643.2) is more abundant in senescing Katsura tree leaves than catabolite Cj-NCC-2 (m/z 627.2). Therefore, DESI-MS imaging may also represent a useful tool for the determination of relative amounts of biologically relevant compounds in plant tissues. This qualitative agreement of relative concentration differences with spatial intensity distributions was also reported in a recent DESI imaging study of animal tissue.(36)
DESI imaging on the senescent leaves of American sweetgum and hophornbeam also yielded informative ion images (Figure 5 and Supporting Information Figures S3 and S4). In three different types of leaves, six linear tetrapyrrolic compounds were detected, identified, and imaged by DESI-MS. These correspond to four structurally different catabolites found in higher plants.3,7,45,48,57,58 The results show the detectability of chlorophyll catabolites with different functional groups, suggesting that this methodology can be utilized to image a broad variety of biologically relevant polyfunctionalized molecules.
Conclusions
DESI-MS allows the rapid identification, structural elucidation, and spatially resolved determination of biologically relevant polyfunctionalized compounds in plant tissues. Specifically, the detection and the spatial distribution of NCCs, the final products of the chlorophyll degradation process, have been determined in several species. The sensitivity of DESI-MS for leaf tissue analysis was enhanced by the introduction of an imprinting process using porous PTFE materials as substrate; this also allowed for the identification of additional secondary chlorophyll catabolites that present in relatively lower concentrations in these leaf matrices. Further DESI imaging experiments provided relevant information on the distribution of these compounds within senescent leaves, and confirmed their relative concentrations. The results suggest the potential use of DESI-MS and DESI-MS imaging as tools for the direct analysis of polyfunctionalized compounds in plant materials.
Acknowledgments
Financial support was provided by the Austrian Science Foundation (Project L472 to B.K.) and the National Science Foundation (CHE NSF 0848650, and NSF 0852740-DBI to R.G.C.). The authors thank Celine Bland for contributions to the preliminary DESI experiments and Dr. Ayanna Jackson for valuable discussions. The first two authors contributed equally to this work.
Supporting Information Available
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Kräutler B.; Matile P. Acc. Chem. Res. 1999, 32, 35–43. [Google Scholar]
- Brown S. B.; Houghton J. D.; Hendry G. A. F. In Chlorophylls; Scheer H., Ed.; CRP-Press: Boca Raton, FL, 1991; pp 465–489. [Google Scholar]
- Kräutler B.; Jaun B.; Bortlik K.; Schellenberg M.; Matile P. Angew. Chem. Int. Ed. 1991, 30, 1315–1318. [Google Scholar]
- Kräutler B.; Hörtensteiner S. In Chlorophylls and Bacteriochlorophylls. Biochemistry, Biophysics, Functions and Applications, Advances in Photosynthesis and Respiration; Grimm B., Porra R., Rüdiger W., Scheer H., Eds.; Springer: Dordrecht, The Netherlands, 2006; Vol. 25; pp 237–260. [Google Scholar]
- Hörtensteiner S.; Kräutler B.. Biochim. Biophys. Acta 2010, DOI:10.1016/j.bbabio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Mühlecker W.; Ongania K. H.; Kräutler B.; Matile P.; Hörtensteiner S. Angew. Chem. Int. Ed. 1997, 36, 401–404. [Google Scholar]
- Oberhuber M.; Berghold J.; Breuker K.; Hörtensteiner S.; Kräutler B. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 6910–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinska A.; Anders I.; Aubry S.; Schenk N.; Tapernoux-Lüthi E.; Müller T.; Kräutler B.; Hörtensteiner S. Plant Cell 2007, 19, 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T.; Ulrich M.; Ongania K.-H.; Kräutler B. Angew. Chem. Int. Ed. 2007, 46, 8699–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S.; Müller T.; Ebert M.-O.; Jockusch S.; Turro N. J.; Kräutler B. Angew. Chem. Int. Ed. 2008, 47, 8954–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer S. G.; Kraft M. L.; Weber P. K. Annu. Rev. Biophys. 2009, 38, 53–74. [DOI] [PubMed] [Google Scholar]
- McDonnell L. A.; Heeren R. M. Mass Spectrom. Rev. 2007, 26, 606–643. [DOI] [PubMed] [Google Scholar]
- Pacholski M. L.; Winograd N. Chem. Rev. 1999, 99, 2977–3006. [DOI] [PubMed] [Google Scholar]
- Seeley E. H.; Caprioli R. M. Proteomics Clin. Appl. 2008, 2, 1435–1443. [DOI] [PubMed] [Google Scholar]
- Li Y.; Shrestha B.; Vertes A. Anal. Chem. 2008, 80, 407–420. [DOI] [PubMed] [Google Scholar]
- Pol J.; Vidova V.; Kruppa G.; Kobliha V.; Novak P.; Lemr K.; Kotiaho T.; Kostiainen R.; Havlicek V.; Volny M. Anal. Chem. 2009, 81, 8479–8487. [DOI] [PubMed] [Google Scholar]
- Takats Z.; Wiseman J. M.; Gologan B.; Cooks R. G. Science 2004, 306, 471–473. [DOI] [PubMed] [Google Scholar]
- Cooks R. G.; Ouyang Z.; Takats Z.; Wiseman J. M. Science 2006, 311, 1566–1570. [DOI] [PubMed] [Google Scholar]
- Harris G. A.; Nyadong L.; Fernandez F. M. Analyst 2008, 133, 1297–301. [DOI] [PubMed] [Google Scholar]
- Van Berkel G. J.; Pasilis S. P.; Ovchinnikova O. J. Mass Spectrom. 2008, 43, 1161–1180. [DOI] [PubMed] [Google Scholar]
- Venter A.; Nefliu M.; Cooks R. G. TRAC: Trends Anal. Chem. 2008, 27, 284–290. [Google Scholar]
- Costa A. B.; Cooks R. G. Chem. Phys. Lett. 2008, 464, 1–8. [Google Scholar]
- Song Y.; Talaty N.; Tao W. A.; Pan Z.; Cooks R. G. Chem. Commun. (Cambridge) 2007, 61–63. [DOI] [PubMed] [Google Scholar]
- Huang G.; Chen H.; Zhang X.; Cooks R. G.; Ouyang Z. Anal. Chem. 2007, 79, 8327–8332. [DOI] [PubMed] [Google Scholar]
- Cooks R. G.; Manicke N. E.; Dill A. L.; Ifa D. R.; Eberlin L. S.; Costa A. B.; Wang H.; Huang G.; Ouyang Z. Faraday Discuss. 2011, 149, 247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. U.; Tata A.; Wu C.; Perry R. H.; Haas G.; West L.; Cooks R. G. Analyst 2009, 134, 867–874. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Gumaelius L. M.; Eberlin L. S.; Manicke N. E.; Cooks R. G. Analyst 2007, 132, 461–467. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Manicke N. E.; Dill A. L.; Cooks R. G. Science 2008, 321, 805. [DOI] [PubMed] [Google Scholar]
- Watrous J.; Hendricks N.; Meehan M.; Dorrestein P. C. Anal. Chem. 2010, 82, 1598–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaty N.; Takats Z.; Cooks R. G. Analyst 2005, 130, 1624–1633. [DOI] [PubMed] [Google Scholar]
- Kennedy J. H.; Wiseman J. M. Rapid Commun. Mass Spectrom. 2010, 24, 1305–1311. [DOI] [PubMed] [Google Scholar]
- Li Y.; Shrestha B.; Vertes A. Anal. Chem. 2007, 79, 523–532. [DOI] [PubMed] [Google Scholar]
- Sjovall P.; Lausmaa J.; Nygren H.; Carlsson L.; Malmberg P. Anal. Chem. 2003, 75, 3429–3434. [DOI] [PubMed] [Google Scholar]
- Eberlin L. S.; Dill A. L.; Golby A. J.; Ligon K. L.; Wiseman J. M.; Cooks R. G.; Agar N. Y. Angew. Chem., Int. Ed. 2010, 49, 5953–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin L. S.; Ifa D. R.; Wu C.; Cooks R. G. Angew. Chem., Int. Ed. 2010, 49, 873–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin L. S.; Dill A. L.; Costa A. B.; Ifa D. R.; Cheng L.; Masterson T.; Koch M.; Ratliff T. L.; Cooks R. G. Anal. Chem. 2010, 82, 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod M.; Shi Y. Z.; Cheng J. X.; Cooks R. G. J. Am. Soc. Mass Spectrom. 2010, 21, 1177–1189. [DOI] [PubMed] [Google Scholar]
- Lane A. L.; Nyadong L.; Galhena A. S.; Shearer T. L.; Stout E. P.; Parry R. M.; Kwasnik M.; Wang M. D.; Hay M. E.; Fernandez F. M.; Kubanek J. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 7314–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidova V.; Novak P.; Strohalm M.; Pol J.; Havlicek V.; Volny M. Anal. Chem. 2010, 82, 4994–4997. [DOI] [PubMed] [Google Scholar]
- Thunig J.; Hansen S. H.; Janfelt C. Anal. Chem. 2011, 83, 3256–3259. [DOI] [PubMed] [Google Scholar]
- Moser S.; Ulrich M.; Müller T.; Kräutler B. Photochem. Photobiol. Sci. 2008, 7, 1577–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks R. G.; Ouyang Z.; Takats Z.; Wiseman J. M. Science 2006, 311, 1566–1570. [DOI] [PubMed] [Google Scholar]
- Takats Z.; Wiseman J. M.; Cooks R. G. J. Mass Spectrom. 2005, 40, 1261–1275. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Wiseman J. M.; Song Q. Y.; Cooks R. G. Int. J. Mass Spectrom. 2007, 259, 8–15. [Google Scholar]
- Curty C.; Engel N. Phytochemistry 1996, 42, 1531–1536. [Google Scholar]
- Iturraspe J.; Moyano N.; Frydman B. J. Org. Chem. 1995, 60, 6664–6665. [Google Scholar]
- Moser S.; Müller T.; Oberhuber M.; Kräutler B. Eur. J. Org. Chem. 2009, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghold J.; Breuker K.; Oberhuber M.; Hörtensteiner S.; Kräutler B. Photosynth. Res. 2002, 74, 109–119. [DOI] [PubMed] [Google Scholar]
- Nyadong L.; Green M. D.; De Jesus V. R.; Newton P. N.; Fernandez F. M. Anal. Chem. 2007, 79, 2150–2157. [DOI] [PubMed] [Google Scholar]
- Wu C. P.; Ifa D. R.; Manicke N. E.; Cooks R. G. Anal. Chem. 2009, 81, 7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K. L.; Unger S. E.; Vincze A.; Cooks R. G.; Keough T. J. Am. Chem. Soc. 1982, 104, 1507–1511. [Google Scholar]
- Sonsmann G.; Romer A.; Schomburg D. J. Am. Soc. Mass Spectrom. 2002, 13, 47–58. [DOI] [PubMed] [Google Scholar]
- Chen H.; Cotte-Rodriguez I.; Cooks R. G. Chem. Commun. (Cambridge) 2006, 597–599. [DOI] [PubMed] [Google Scholar]
- Kräutler B.; Banala S.; Moser S.; Vergeiner C.; Müller T.; Lütz C.; Holzinger A. FEBS Lett. 2010, 584, 4215–4221. [DOI] [PubMed] [Google Scholar]
- Ifa D. R.; Manicke N. E.; Rusine A. L.; Cooks R. G. Rapid Commun. Mass Spectrom. 2008, 22, 503–510. [DOI] [PubMed] [Google Scholar]
- Moser S.; Müller T.; Holzinger A.; Lütz C.; Jockusch S.; Turro N. J.; Kräutler B. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 15538–15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghold J.; Eichmüller C.; Hörtensteiner S.; Kräutler B. Chem. Biodiversity 2004, 1, 657–668. [DOI] [PubMed] [Google Scholar]
- Berghold J.; Müller T.; Ulrich M.; Hörtensteiner S.; Kräutler B. Chem. Monthly 2006, 137, 751–763. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.