Abstract
The epigenome consists of non–sequence-based modifications, such as DNA methylation, that are heritable during cell division and that may affect normal phenotypes and predisposition to disease. Here, we have performed an unbiased genome-scale analysis of ~4 million CpG sites in 74 individuals with comprehensive array-based relative methylation (CHARM) analysis. We found 227 regions that showed extreme interindividual variability [variably methylated regions (VMRs)] across the genome, which are enriched for developmental genes based on Gene Ontology analysis. Furthermore, half of these VMRs were stable within individuals over an average of 11 years, and these VMRs defined a personalized epigenomic signature. Four of these VMRs showed covariation with body mass index consistently at two study visits and were located in or near genes previously implicated in regulating body weight or diabetes. This work suggests an epigenetic strategy for identifying patients at risk of common disease.
INTRODUCTION
A role for epigenetics in common disease has long been suspected (1), and a strong relationship with cancer has been shown (2–4). We have argued that common disease involves both genetic and epigenetic factors and that epigenetic modification could mark environmental effects and mediate genetic effects (1). In addition to particular exposure-epigenetic relationships (5, 6), epigenetic changes with aging support the notion that there is an environmental component to epigenetic variation (7–10). Studies of identical twins show greater differences in global DNA methylation in older than in younger twins, consistent with an age-dependent progression of epigenetic change (8, 10). Global methylation changes over an 11-year span in participants of an Icelandic cohort, and age- and tissue-related alterations in some CpG islands from an array of 1413 arbitrarily chosen CpG sites near gene promoters, further corroborate the evidence for dynamic methylation patterns over time (9, 11). Other work, however, has suggested that epigenetic marks, or their maintenance, are themselves controlled by genes and are thus heritable in the traditional sense and associated with particular DNA variants (12). This would predict that methylation marks are stable rather than varying as controlled by changing environments.
Here we describe a genome-scale, gene-specific analysis of DNA methylation in the same individuals over time to identify a personalized epigenomic signature that may correlate with common genetic disease.
RESULTS
We report comprehensive high-throughput array-based relative methylation (CHARM) analyses on samples of the Age, Gene/Environment Susceptibility (AGES) study, assessing 4.5 million CpG sites genome-wide, which has been shown to identify differential DNA methylation without assumptions regarding where such changes would be, and uses arrays tiled through regions based on their relative CpG content, including all CpG islands, as well as CpG island “shores” that have been shown to be enriched in differential methylation (13).
In brief, the AGES study constitutes visit 7 (from 2002 to 2005) of the Reykjavik Study, which began with 18,000 residents of Reykjavik recruited in 1967. The AGES study recruited 5758 of the surviving members, between 69 and 96 years of age in 2002. Of these, 638 gave a DNA sample in 1991 as part of the Reykjavik Study visit 6 and there-fore had DNA from two time points, about 11 years apart, available for methylation analysis. We present data for 74 samples, a random set of those who had ample DNA remaining for both study visits. Descriptive statistics for these samples are given in Table 1.
Table 1.
Descriptive information [mean (SE)] for samples used in CHARM analyses at each time point.
| Visit 6 (1991) n = 48 |
Visit 7 (2002–2005) n = 64 |
|
|---|---|---|
| Age (years) | 74.08 (3.49) | 82.80 (3.45) |
| Sex (% male) | 33 | 31 |
| BMI (kg/m2) | 26.56 (3.81) | 26.01 (4.10) |
| Type 2 diabetes (%) | 8 | 11 |
| Glucose (mM) | 5.90 (0.59) | 5.79 (0.88) |
| Coronary events (%) | 10 | 14 |
| Waist circumference (cm) | – | 97.6 (10.9) |
| Fat (%) | – | 29.31 (7.89) |
| Hemoglobin A1C (%) | – | 5.69 (0.34) |
CHARM analysis of samples obtained from visit 7 identified 227 regions that met our criteria for polymorphic methylation patterns across individuals [variably methylated regions (VMRs)]. These represented regions of extreme variability across individuals defined by 10 or more consecutive probes with an average SD of >0.125 (table S1). These VMRs showed enrichment for development and morphogenesis categories (Table 2), including genes from all four HOX clusters. The appearance of developmental genes is predicted by our model that epigenetic variation would involve developmental genes, and this variability itself increases evolutionary fitness in an environmentally changing world (14).
Table 2.
Gene Ontology analysis of the 227 VMRs revealed enrichment for the development and morphogenesis categories (P < 0.01).
| P | FDR | Odds ratio |
Observed count |
Expected count |
GO term | Genes |
|---|---|---|---|---|---|---|
| 0.0011 | 0.22 | 7.04 | 5 | 0.79 | Anterior/posterior pattern formation | HOXA5, HOXB6, HOXD8, HOXC10, HOXA1 |
| 0.0019 | 0.22 | 43.31 | 2 | 0.07 | Blastoderm segmentation | HOXB6, HOXD8 |
| 0.0019 | 0.22 | 43.31 | 2 | 0.07 | Determined anterior/posterior axis, embryo | HOXB6, HOXD8 |
| 0.0082 | 0.25 | 17.31 | 2 | 0.14 | Neuron recognition | FOXG1, NTM |
| 0.0086 | 0.25 | 3.63 | 6 | 1.77 | Pattern specification process | HOXA5, FOXG1, LEF1, HOXC10, MYF6, HOXA1 |
| 0.0096 | 0.25 | 7.47 | 3 | 0.44 | Placenta development | ESX1, LEF1, CDX4 |
| 0.0096 | 0.25 | 15.74 | 2 | 0.15 | Intra-Golgi vesicle-mediated transport | COPZ1, GABARAPL2 |
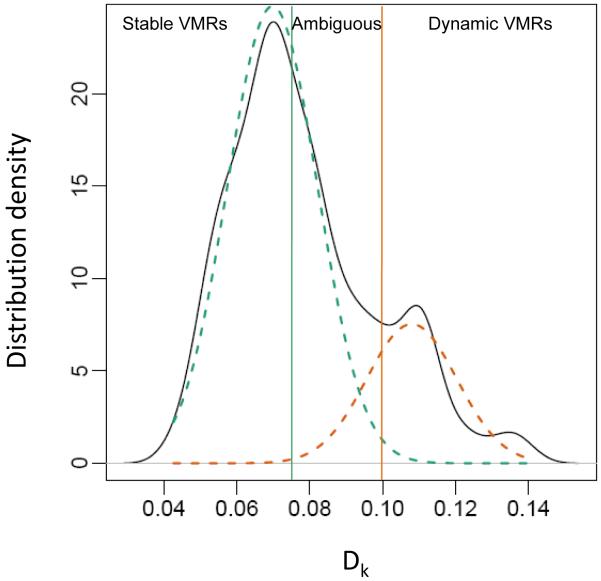
Next, to determine whether methylation at these regions changed within individuals over time, we analyzed the distribution of the absolute value of average within-person change in methylation over time per VMR and found two underlying distributions (Fig. 1). These data fit a two-component mixture model, with 41 VMRs easily classified into the higher intraindividual difference group (probability of membership in orange distribution, >0.99; Fig. 1), defined as dynamic VMRs; 119 VMRs easily classified into the lower distribution (probability of green distribution, >0.99), defined as stable VMRs; and 67 residing in the overlapping region labeled ambiguous, with respect to intraindividual change over time. Thus, about half of the regions that are variably methylated across individuals appear to be stable over time within individuals.
Fig. 1.
Distribution of intraindividual change in methylation over time at VMRs. Mixture distribution analysis shows that Dk, the average absolute value of intraindividual differences in methylation over time for VMR k, fits two underlying curves. Green, stable showing little change; orange, dynamic showing larger changes. Ambiguous is intermediate in Dk.
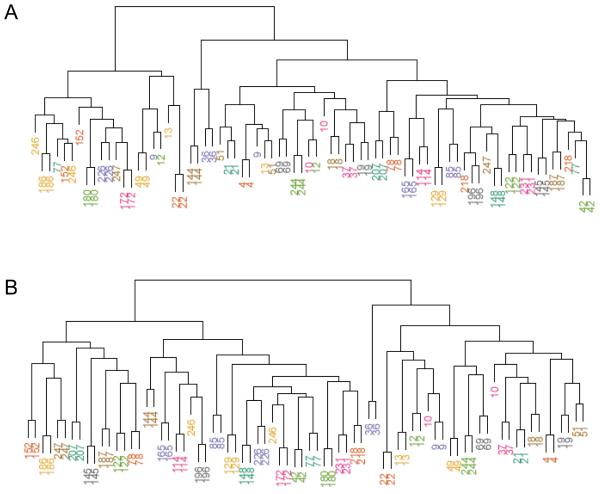
Clustering of the 227 VMR methylation profiles (Fig. 2A) revealed mixing of methylation profiles among the individuals, whereas use of only stable VMRs in the clustering algorithm uniquely identified each individual (Fig. 2B). These stable VMRs may represent polymorphic methylated regions that are not particularly susceptible to exposure modifications or that do not naturally change with age.
Fig. 2.
Similarity between individuals based on VMR methylation. (A) Dendogram based on clustering applied to methylation profiles at all 227 VMRs. (B) Dendogram based on clustering applied to methylation profiles using only the 119 stable VMRs. Numbers represent individual identification numbers.
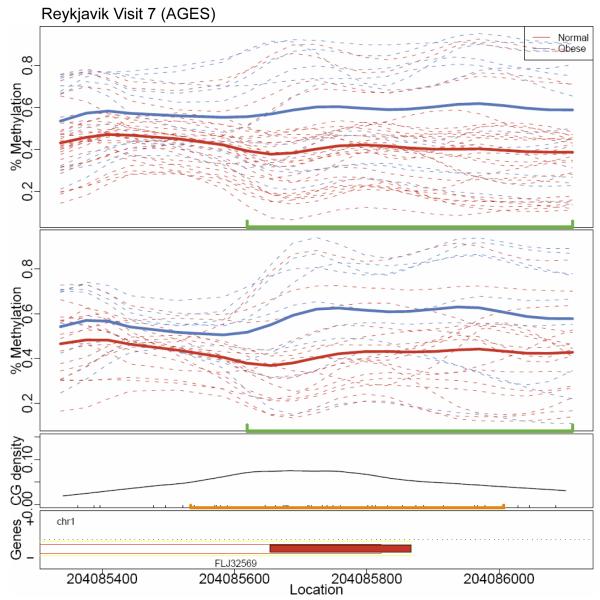
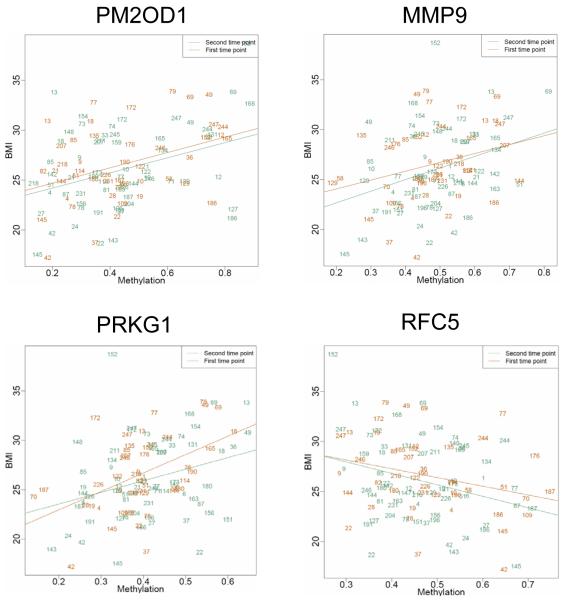
To explore how methylation of particular VMRs may play a role in disease risk, we determined the relationship between methylation and body mass index (BMI), an accessible and treatable phenotype that is known to have many disease correlates (15, 16). We identified 13 VMRs that met a false discovery rate (FDR) criterion of <25% in cross-sectional analyses of visit 7 (Table 3). Of these, four had a P value of <0.10 and the same strength and direction of correlation with BMI at the earlier visit 6 (Three of the 13 had conflicting direction of associations between visits.). These VMRs are in or near genes PM20D1, MMP9, PRKG1, and RFC5. The methylation curves among obese (BMI ≥30) and normal (BMI <25) subjects for the VMR at PM20D1 illustrate an ~20% increase in methylation that persists over time between the two visits (Fig. 3). Scatter plots for the relationship between methylation and BMI for all four VMRs exhibited significant correlations at both visits (Fig. 4).
Table 3.
Stable VMRs associated with BMI. Bold values indicate confirmation in visit 6 analysis (P < 0.1 and consistent regression parameter estimates).
| Chromosome | Nearest gene | Visit 7 |
Visit 6 |
|||
|---|---|---|---|---|---|---|
| Q | P | Regression estimate | P | Regression estimate | ||
| Chromosome X | IL1RAPL2 | 0.114 | 0.00304 | −20.3 | 0.266 | −8.9 |
| Chromosome 1 | PM2OD1 | 0.114 | 0.00332 | 7.6 | 0.00824 | 7.7 |
| Chromosome 6 | NEDD9 | 0.114 | 0.00351 | 12.1 | 0.38 | 5.2 |
| Chromosome 20 | MMP9 | 0.160 | 0.00658 | 11.6 | 0.0605 | 8.9 |
| Chromosome 10 | SORCS1 | 0.215 | 0.0128 | −13.6 | 0.112 | −9.4 |
| Chromosome 10 | PRKG1 | 0.215 | 0.0132 | 11.8 | 0.000711 | 18.9 |
| Chromosome 12 | RFC5 | 0.243 | 0.0175 | −11.8 | 0.0653 | −8.8 |
| Chromosome 1 | TTC13 | 0.249 | 0.022 | 9.27 | 0.523 | 3.3 |
| Chromosome X | DACH2 | 0.249 | 0.0311 | −15.1 | 0.539 | 4.1 |
| Chromosome 5 | TRIM36 | 0.249 | 0.0326 | 11.3 | 0.0781 | −14.1 |
| Chromosome 14 | FLRT2 | 0.249 | 0.0278 | −9.5 | 0.19 | −5.8 |
| Chromosome 1 | C1orf57 | 0.249 | 0.0253 | −10.6 | 0.282 | −6.5 |
| Chromosome 18 | APCDD1 | 0.249 | 0.0332 | −10.7 | 0.901 | 0.7 |
Fig. 3.
Methylation curves at the gene PM20D1. Methylation curves for visit 7 and visit 6 data. Dashed lines are individual methylation curves. Solid lines are average curves by obese (blue) and normal (red) groups. The green line indicates the boundaries of the VMR. CpG density is shown in the third panel, with CpG islands marked in orange. Gene location is shown in the bottom panel.
Fig. 4.
Correlations between methylation and BMI at six BMI-related VMRs. Points are individual IDs. Blue indicates visit 7; red indicates visit 6.
DISCUSSION
We previously showed that global DNA methylation changes within individuals over time (11), and we have now confirmed and identified the locations of site-specific changes at dynamic VMRs with a genome-wide approach. In addition, we identified a separate set of stable VMRs that can be used to uniquely identify individuals in an epigenetic signature akin to genetic fingerprinting. This signature may be correlated with disease status, implying that an epigenetic signature can mark disease risk or disease states. In particular, we show stable VMRs that correlate with BMI at two separate visits a decade apart.
Some have argued that DNA methylation changes over time and is an important biological mediator of environmental effects on human disease (17), whereas others support the concept of inherited DNA methylation patterns, implying that they are potentially variable across individuals but less likely to be dynamic over time (18). This has been a conundrum because these appear to be opposing ideas. However, we show that both ideas have merit. It is important to identify these regions in the context of disease consequences because those that are particularly labile may be the sites relevant when considering epigenetic marks as mediators of environmental effects, whereas those that are stable may be relevant as mediators or moderators of genetic effects. Further, thoseregions that do not change over time can be used as an epigenetic signature for an individual, similar to genotype. These regions can then be considered as candidates for assessment of methylation associations with disease or health-related phenotypes under specific risk models.
Our results help to focus the integration of methylation measurement into epidemiologic studies of disease risk by providing specific genomic sites for inquiry. Our exploration of possible correlations between methylation at these VMRs and an easily measured disease-related phenotype, BMI, identified 13 genes, 4 of which were consistently correlated with BMI across two separate study visits. Many of these 13 genes have been previously implicated in obesity or diabetes. MMP9, as well as another member of this family, MMP3, encodes a metallopeptidase that is up-regulated in obese individuals (19). Several matrix metalloproteinases (MMPs), including MMP9, are upregulated in human adipocytes (20). Matrix metallopeptidases have also been associated with obesity in rodent models (21, 22). PM20D1 is also a metalloproteinase and, although not yet well-characterized, may have similar implications for obesity. PRKG1, a guanosine 3′,5′-monophosphate (cGMP)–dependent protein kinase, plays an important role in foraging behavior, food acquisition, and energy balance (23). RFC5 is an intriguing gene because it encodes a metabolism-linked DNA replication complex loading protein, dysfunction of which leads to DNA repair defects. It might thus play a role in well-known but poorly understood DNA damage–related complications of diabetes.
In a mouse model of obesity, SORCS1 has been located at a type 2 diabetes quantitative trait locus (QTL) (24), and this finding has been confirmed in humans, where SORCS1 single-nucleotide polymorphisms (SNPs) and haplotypes were associated with fasting insulin secretion (25). IL1RAPL2 is located at a region on chromosome X that is associated with Prader-Willi–like syndrome, whereas DACH2 is also an X-linked gene associated with Wilson-Turner syndrome, both of which are Mendelian disorders with obesity features. TTC13 is part of a family containing another tetratricopeptide repeat gene, TTC8, which has been directly linked to Bardet-Biedl syndrome and includes obesity as a primary feature. APCDD1 is a positional candidate gene associated with a QTL that affects fat deposition in pigs (26) and is located at a region on chromosome 18 that is linked to percentage of body fat in men (27).
Our identification of VMRs is limited by the number of individuals contributing to this genome-wide CHARM analysis. It is likely that increased sample sizes will improve detection of additional VMRs. Further, the dynamic VMRs defined here are based on an 11-year window among elderly participants. It is important to also identify methylomic regions that show intraindividual changes during early segments of the life span and to connect these changes to particular environmental exposures. One potential caveat from these analyses is that the methylation patterns were obtained from DNA derived from blood and thus contain a mixture of cell types that could confound our results. However, in our previous study of global DNA methylation (that is, non–site-specific) in these samples, we found no relationship between lymphocyte count and methylation (11). A recent paper also showed that cellular heterogeneity was not associated with DNA methylation amounts for most sites they studied (18). Our use of blood as a DNA source may also limit the interpretations of these results, given the tissue specificity of DNA methylation. However, there is a growing precedent for lymphoid tissues serving as a good surrogate tissue for changes in other target tissues. For example, loss of imprinting of insulin-like growth factor 2 (IGF-2), one of the best-studied disease-related epigenetic mutations, is found in both lymphocytes and colon, and changes in either are associated with increased colorectal cancer risk (28). Finally, our exploration of the correlation between BMI and methylation was based on availability of quantitative data and relevance to human disease. We were unable to assess the relationship of VMRs to categorical outcomes in this sample. Although more comprehensive than previous genome-wide, site-specific methylation studies (7, 9), our analysis was limited by the sample number to the relationship between methylation and quantitative phenotype rather than categorical outcomes. This study supports further examination of other measures of obesity and disease outcomes such as diabetes and cardiovascular disease with respect to the particular VMRs identified here.
The implications of these results are wide-ranging. An individual epigenetic signature that is stable over time has not previously been described. Such a signature could be driven by underlying sequence variation, early environmental exposure (for example, prenatally), or both. These stable VMRs would likely complement genotype because they would also reflect early exposure. In addition, we have proposed that some genetic variants would drive increasing site-specific stochastic epigenetic variation, and thus, the variance of methylation in a population could be predicted by genotype but the methylation level in an individual would not be predictable from genotype and would require direct measurement (14).
Whether in part or completely genetically driven, this epigenotype may be more proximate to the ultimate phenotype, in this case BMI, and thus have considerable value for disease risk assessment. Although the sample size is larger than previous genome-scale, gene-specific methylation studies, it is still relatively small compared to classical sequence-driven approaches such as genome-wide association studies (GWAS). Even so, the data suggest that this epigenomic approach to disease phenotype will be an important complement to such studies. Even with the restraint of relatively small numbers of samples, we could identify four genes with VMRs related to BMI. In addition, the identification of stable VMRs may have long-term consequences for developing personalized epigenomics in medicine, with the hope of forging a connection between personal genomes and early (for example, in utero) environmental influences.
MATERIALS AND METHODS
Samples
Nonimmortalized lymphocyte samples were taken from participants of the AGES Reykjavik Study (29). Seventy-four samples contributed to these analyses. These samples meet our high-quality array data criteria and were from a randomly chosen set of 100 samples from the 638 AGES participants that had ample DNA from two visits. CHARM data were only considered in analyses if they passed our internal quality assessment. For cross-sectional analyses of the most recent collection (visit 7), 64 samples contributed data, whereas 48 samples contributed to cross-sectional analyses of the earlier visit 6 data. For identification of dynamic VMRs, a subset of 38 samples had quality CHARM data at both time points. For the analyses with BMI presented here, BMI was calculated as the body weight in kilograms divided by the height in meters squared.
Genome-wide methylation assay
We used CHARM analysis, which is a microarray-based method agnostic to preconceptions about methylation, including location relative to genes and CpG content (30, 31). The resulting quantitative measurements of methylation, denoted with M, are log ratios of intensities from total (Cy3) and McrBC-fractionated (Cy5) DNA: Positive and negative M values are quantitatively associated with methylated and unmethylated sites, respectively. For each sample, we analyzed ~4.5 million CpG sites across the genome with a custom-designed NimbleGen HD2 microarray, including all of the classically defined CpG islands and nonrepetitive progressively lower CpG density genomic regions of the genome until the array is saturated. We include 4500 control probes to standardize these M values so that unmethylated regions are associated, on average, with values of 0. CHARM is 100% specific at 90% sensitivity for known methylation marks identified by other methods (for example, in promoters) while including more than half of the genomes not identified by conventional region before selection. The CHARM results have also been extensively corroborated by quantitative bisulfite pyrosequencing analysis (30).
Identification of VMRs
We first screened the methylome for regions where methylation varied substantially across individuals. We termed these VMRs to distinguish them from regions identified for their discrimination of groups, such as tissue types or cases versus controls, which we and others have previously called differentially methylated regions (DMRs). Our use of the term VMR can be considered a specific type of metastable epiallele introduced by Rakyan et al.(32) to denote variable expression of imprinted loci or variable methylation of an agouti methylation variant.
To identify VMRs from our data, we first processed the raw CHARM data with the statistical procedure described (33). This statistical procedure produced quality metrics (between 0 and 100%) for each sample and, for those that passed our quality test (>80%), a vector of methylation percentage estimates for each feature on the array. These were then smoothed with the standard CHARM approach to reduce measurement error (30, 31). We denote the resulting methylation percentages for subject i at microarray feature j for time t as Mijt.
We used cross-sectional analysis of visit 7 data to identify polymorphic VMRs on the basis of extreme interindividual variance across consecutive probes. Specifically, we estimated between-subject variability with the median absolute deviation, a robust estimate of the SD (14). We computed the median of ∣Mijt – mjt∣ across subjects, with mjt, the median Mijt across subjects i, and referred to it as sjt (14). To avoid false positives in subsequent analysis of correlations with covariates, we required a stringent definition for designating a polymorphic VMR: a region of 10 or more consecutive probes attaining values of sjt above the 99th percentile of all the sjt and an average sjt of >0.125. We chose these cutoff values with permutation tests. Specifically, we randomized the genomic order of the CHARM probes and applied the above algorithm to find VMRs (including the smoothing step) for each permuted data set. Using our criteria, we obtained 0 false positives. Lower-ing either the number of consecutive probes or the average sjt thresholds produced false positives.
These VMRs were then annotated for genomic location and gene proximity. Genes within 3 kb of VMRs were considered in a Gene Ontology (GO) analysis of biological process categories. For each GO category, we performed a hypergeometric test (34), with corresponding nominal P value, to determine enrichment of genes near VMRs. We also calculated the FDR for each category statistic to account for the multiple comparisons.
Identification of stable versus dynamic VMRs
Using the average Mijt within the range of each VMR, we generated methylation profiles for each sample. This included a vector of k VMR values for each subject i and time point t. We calculated Dik, the median absolute within-person difference between methylation profiles from visit 6 to visit 7 for each VMR k. We then fit a two-component Gaussian mixture model (35) to these values and used the resulting estimated posterior distributions to classify VMRs into three groups: “stable” (those with posterior probability of membership in the lower distribution >0.99, reflecting little intraindividual change over time); “dynamic” (those with posterior probability of membership in the higher distribution >0.99, reflecting those with high intraindividual change over time); and “ambiguous” (those not meeting either criteria, and thus in the overlap between the two distributions). (Among the stable VMRs, there is some change over time observed in both directions, and when one takes the absolute value of this difference, the result is a small positive number, and thus the central tendency of Dk for stable VMRs is not 0.)
To evaluate discrimination of individuals based on methylation patterns, we applied hierarchical clustering to the vectors of methylation values for the VMRs and graphed individuals into a dendrogram based on similarity of VMRs. We then selected only those VMRs designated as stable in the analysis above and repeated the hierarchical clustering and dendrogram graphic.
Identification of BMI-related methylated regions
We performed cross-sectional analyses for data at each visit separately. For each stable VMR, we fit a linear regression model to summarize the relationship between BMI and methylation. Specifically, for each VMR k, we fit the following model:
where Yi is the BMI for individual i, Mik is the methylation level for individual i in the kth VMR, and e represents unexplained variability. Here, bk represents the parameter of interest that summarizes the correlation between BMI and methylation. This produced one Wald statistic for each VMR. We fit this model to the data from visit 7, and to account for the multiple comparisons due to multiple VMRs, we reported a list of regions with an FDR of 0.30. To confirm these results, we independently applied the same regression approach to visit 6 and obtained estimates of b along with P values.
Supplementary Material
36. Acknowledgments
We thank E. Briem for CHARM hybridization. Funding: Supported by NIH grants 1R01ES015211 (D.F.) and 2P50HG003323 (A.P.F.). The AGES Reykjavik Study is funded by NIH contract N01-AG-12100, Hjartavernd (the Icelandic Heart Association), and Althingi (the Icelandic Parliament). This research was funded in part by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Competing interests: Johns Hopkins School of Medicine is filing a provisional patent based on these data.
Accession numbers: The CHARM data can be found in GEO GSE23858.
SUPPLEMENTARY MATERIAL www.sciencetranslationalmedicine.org/cgi/content/full/2/49/49ra67/DC1 Table S1. Variably methylated regions across individuals.
REFERENCES AND NOTES
- 1.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 6.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 7.Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr. Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- 11.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. U.S.A. 2010;107(Suppl. 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams KF, Schatzkin A, Harris TB, Kipnis V, Mouw T, Ballard-Barbash R, Hollenbeck A, Leitzmann MF. Overweight, obesity, and mortality in a large prospective cohort of persons 50 to 71 years old. N. Engl. J. Med. 2006;355:763–778. doi: 10.1056/NEJMoa055643. [DOI] [PubMed] [Google Scholar]
- 16.Flegal KM, Graubard BI, Williamson DF, Gail MH. Cause-specific excess deaths associated with underweight, overweight, and obesity. JAMA. 2007;298:2028–2037. doi: 10.1001/jama.298.17.2028. [DOI] [PubMed] [Google Scholar]
- 17.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, Putter H, Slagboom PE, Heijmans BT. Variation, patterns, and temporal stability of DNA methylation: Considerations for epigenetic epidemiology. FASEB J. 2010 doi: 10.1096/fj.09-150490. 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- 19.Nair S, Lee YH, Rousseau E, Cam M, Tataranni PA, Baier LJ, Bogardus C, Permana PA. Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia. 2005;48:1784–1788. doi: 10.1007/s00125-005-1868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara A, Lim FL, Mazzatti DJ, Trayhurn P. Microarray analysis identifies matrix metalloproteinases (MMPs) as key genes whose expression is up-regulated in human adipocytes by macrophage-conditioned medium. Pflugers Arch. 2009;458:1103–1114. doi: 10.1007/s00424-009-0693-8. [DOI] [PubMed] [Google Scholar]
- 21.Uemura S, Matsushita H, Li W, Glassford AJ, Asagami T, Lee KH, Harrison DG, Tsao PS. Diabetes mellitus enhances vascular matrix metalloproteinase activity: Role of oxidative stress. Circ. Res. 2001;88:1291–1298. doi: 10.1161/hh1201.092042. [DOI] [PubMed] [Google Scholar]
- 22.Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J. Biol. Chem. 2003;278:11888–11896. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 23.Kaun KR, Sokolowski MB. cGMP-dependent protein kinase: Linking foraging to energy homeostasis. Genome. 2009;52:1–7. doi: 10.1139/G08-090. [DOI] [PubMed] [Google Scholar]
- 24.Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET, Stapleton DS, Gray-Keller MP, Young MB, Stoehr JP, Lan H, Boronenkov I, Raess PW, Flowers MT, Attie AD. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat. Genet. 2006;38:688–693. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- 25.Goodarzi MO, Lehman DM, Taylor KD, Guo X, Cui J, Quiñones MJ, Clee SM, Yandell BS, Blangero J, Hsueh WA, Attie AD, Stern MP, Rotter JI. SORCS1: A novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes. 2007;56:1922–1929. doi: 10.2337/db06-1677. [DOI] [PubMed] [Google Scholar]
- 26.Lee KT, Park EW, Moon S, Park HS, Kim HY, Jang GW, Choi BH, Chung HY, Lee JW, Cheong IC, Oh SJ, Kim H, Suh DS, Kim TH. Genomic sequence analysis of a potential QTL region for fat trait on pig chromosome 6. Genomics. 2006;87:218–224. doi: 10.1016/j.ygeno.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Lewis CE, North KE, Arnett D, Borecki IB, Coon H, Ellison RC, Hunt SC, Oberman A, Rich SS, Province MA, Miller MB. Sex-specific findings from a genome-wide linkage analysis of human fatness in non-Hispanic whites and African Americans: The HyperGEN study. Int. J. Obes. 2005;29:639–649. doi: 10.1038/sj.ijo.0802916. [DOI] [PubMed] [Google Scholar]
- 28.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 29.Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, Thorgeirsson G, Aspelund T, Garcia ME, Cotch MF, Hoffman HJ, Gudnason V. Age, Gene/Environment Susceptibility–Reykjavik Study: Multidisciplinary applied phenomics. Am. J. Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 33.Ayree MJ, Wu Z, Ladd-Acosta C, Herb B, Feinberg AP, Yegnasubramanian S, Irizarry RA. Biostatistics. Department of Biostatistics, Johns Hopkins University; 2010. Accurate genome-scale percentage DNA methylation estimates from microarray data. Working Paper 208, http://www.bepress.com/jhubiostat/paper208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 35.Banfield JD, Raftery AE. Model-based Gaussian and non-Gaussian clustering. Biometrics. 1993;49:803–821. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.