Abstract
Hedgehog was first described in Drosophila melanogaster by the Nobel laureates Eric Wieschaus and Christiane Nüsslein-Volhard. The hedgehog (Hh) pathway is a major regulator of cell differentiation, proliferation, tissue polarity, stem cell maintenance, and Carcinogenesis. The first link of Hh signaling to cancer was established through studies of a rare familial disease, Gorlin syndrome, in 1996. Follow-up studies revealed activation of this pathway in basal cell carcinoma, medulloblastoma and, leukemia as well as in gastrointestinal, lung, ovarian, breast, and prostate cancer. Targeted inhibition of Hh signaling is now believed to be effective in the treatment and prevention of human cancer. The discovery and synthesis of specific inhibitors for this pathway are even more exciting. In this review, we summarize major advances in the understanding of Hh signaling pathway activation in human cancer, mouse models for studying Hh-mediated Carcinogenesis, the roles of Hh signaling in tumor development and metastasis, antagonists for Hh signaling and their clinical implications.
Keywords: Hedgehog, Smoothened, PTCH1, cancer, signal transduction, clinical trials, animal model
Major advances in understanding the hedgehog (Hh) pathway have been made in the last 30 years. The Hh gene was identified in 1980 through genetic analyses of Drosophila fruit fly segmentation[1]. In early 1990's, three vertebrate homologues of the Hh gene were identified[2]–[6]. As an essential signaling pathway in embryonic development, the Hh pathway is critical for maintaining tissue polarity and stem cell population. In 1996, inactivation of this pathway was linked to the hereditary developmental disorder holoprosencephaly, whereas hyperactivation of this pathway was linked to human cancer[7]–[11]. More recently, an inhibitor of Hh signaling was successfully used in clinical trials of human cancer, further indicating the feasibility of Hh signaling inhibitors for cancer therapeutics. Figure 1 lists the major milestones of research on Hh signaling as related to cancer.
Figure 1. Major milestones in the studies of hedgehog signaling as related to human diseases, particularly cancer. For all references, please see the text for details.
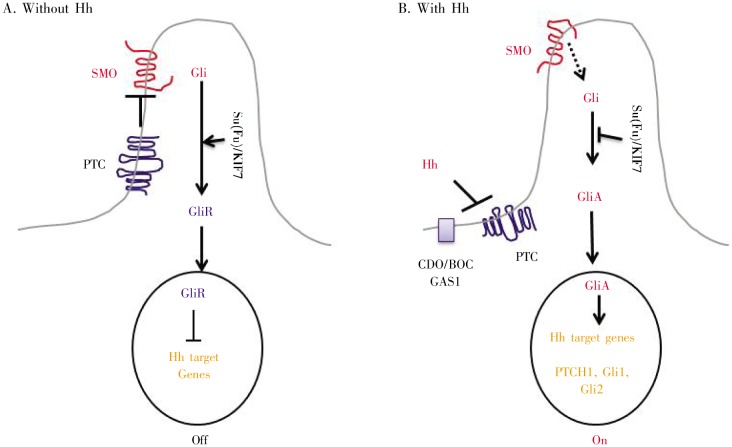
The general signaling mechanisms of the Hh pathway are conserved from flies to humans[12]. In the absence of Hh ligand, Smoothened (SMO), the seven transmembrane domain containing protein, serves as the key signal transducer, whose function is inhibited by another transmembrane protein Patched (PTC). An active Hh ligand (Shh, Ihh, Dhh, or the fly Hh homologue) binds to its receptor PTC and relieves this inhibition, allowing SMO to signal downstream, leading to the activation of Gli transcription factors. As a transcription factor, Gli protein associates with specific consensus sequences located in the promoter region of target genes, regulating target gene expression[13],[14]. Figure 2 shows a simplified diagram of the Hh signaling pathway.
Figure 2. A simplified model for Hh signaling in mammalian cells. SMO is the key signal transducer of the Hh pathway. A, in the absence of the Hh ligands, the Hh receptor PTC is thought to be localized in the cilium to inhibit SMO signaling (via an unknown mechanism). Gli molecules are processed with the help of Su(Fu)/KIF7 molecules into repressor forms, which disable the Hh signaling pathway. B, in the presence of Hh, PTC is thought to be shuttled out of cilium and is unable to inhibit SMO. Co-receptors of Hh ligands include CDO, BOC, and GAS1. Hh reception promotes SMO conformational change, facilitating Gli activation (GMA), stimulating Hh target gene expression. This process can be inhibited by KIF7 and Su (Fu). (Positive regulators are in red, negative regulators are in blue and target genes are in orange.).
Signal Transduction of the Hedgehog Pathway
Hh proteins [one Hh in Drosophila and three Hhs in mammals—Sonic Hedgehog (Shh), Indian Hedgehog (Ihh), and Desert Hedgehog (Dhh)] are secreted during development, functioning at short range to nearby cells and at long range to distant cells[15]–[17]. After translation, the Hh protein precursor undergoes auto-processing to release its N-terminal fragment (HhN), which is then covalently bound to a cholesterol moiety at the C-terminal end. Palmitoylation mediated by the Skinny Hedgehog acyltransferase occurs at the N-terminus of HhN[18]–[21]. Several molecules are involved in the movement, extracellular transport, and release of Hh proteins, including the transmembrane transporter-like protein Dispatched (Disp)[22]–[24], metalloproteases[25], the heparan sulfate proteoglycans Dally-like (Dlp) and Dally [26],[27] or their regulators[28], as well as enzymes such as Sulfateless and Tout velu[29]–[31].
Several molecules are engaged in the reception of Hh ligands, with PTC (one PTC in fly and two PTCs in vertebrates—PTCH1 and PTCH2) as the major receptor[32]. Studies from tissue cultured cells indicate that PTC inhibits SMO at a sub-stoichiometric concentration[33]. The Hh-interacting protein (HIP) can compete with PTC to bind Hh, resulting in the negative regulation of Hh signaling[34]. On the other hand, Ihog (or its vertebrate homologues CDO and BOC), GAS1, and Glypican-3 serve as co-receptors of Hh[35]–[42]. It is not clear how binding of Hh proteins results in the pathway activation. It is proposed that PTC limits SMO signaling by transporting small endogenous molecules specifically targeted to SMO. Candidates of these small molecules include PI4P, lipoproteins, and pro-vitamin D3[43]–[46]. However, how these molecules regulate SMO signaling is unknown.
Significant progress has been made toward our understanding of SMO signaling, with recent reports linking SMO to G protein coupling[47]–[50]. In particular, a study in Drosophila provides direct evidence for SMO-coupling to Gαi in the regulation of Hh pathway activation [48]. The physiological relevance of G protein coupling to SMO in Hh signaling during Carcinogenesis is unknown. In Drosophila, SMO function is stimulated through protein phosphorylation by PKA and Casein kinase I at the C-terminus [51],[52]. SMO mutants lacking these phosphorylation sites are defective in Hh signaling. However, these phosphorylation sites are not conserved in vertebrate SMO, indicating a different mechanism for SMO signaling in higher organisms[52]. There are two important events during mammalian SMO signaling. First, the SMO protein undergoes a conformational change favoring SMO signaling[53], although the regulatory mechanism underlying this conformational change is unclear. Second, the ciliary translocation of mammalian SMO protein is critical for Hh signaling (see below).
Accumulating evidence indicates that primary cilia play an important role in the Hh pathway[54]–[59]. The function of primary cilium is regulated by protein complexes involved in intra-flagellar transport (IFT), which functions in retrograde and anterograde movement of cargo within the primary cilia [60]. Mutations in IFT protein involved in predominantly primary cilium anterograde transportation are shown to result in mice with Hh loss of function phenotypes[55],[61]. Gli3 processing is the most significantly affected event in IFT mutants[56],[57],[61]. The presence of several Hh components upon Hh stimulation, including SMO and Gli molecules at the primary cilium, further supports the relevance of cilium in Hh signaling[62]–[65]. It has been shown that a SMO mutant lacking a ciliary translocation signal cannot mediate Hh signaling[54]. However, the translocation of SMO to cilium is not sufficient to activate Hh signaling [64],[65]. Using tissue-specific gene knockout, recent studies have revealed dual roles of cilium (via knocking out cilium component Kif3a) in Hh signaling-mediated Carcinogenesis in mice[66],[67]. Whereas the Kif3a gene is required for activated SMO-mediated tumor formation, and knocking out Kif3a accelerates Gli2-mediated Carcinogenesis. How SMO is translocated to the cilium in response to Hh signaling and how SMO activates downstream effectors are unclear. However, β-arrestin 2 can regulate ciliary localization of SMO[68]. The role of cilium for Hh signaling downstream of SMO is less clear, as not all of the signaling events occur in cilium. For example, cilium is not required for Su (Fu)-mediated regulation of Gli functions[69],[70].
Several molecules have been identified to be genetically downstream of SMO signaling in Drosophila, including COS2 and Fused. How their vertebrate homologues function in Hh signaling is yet to be established. Recent in vivo studies support that a C0S2 homologue KIF7 functions in the Hh pathway, but no direct interaction between SMO and KIF7 is detected[71],[72], suggesting that the function of COS2 in vertebrates may be replaced by a few molecules. The phenotype of vertebrate Fused knockout mice is not similar to that observed in Shh null mice[73]–[75] and no changes of Hh signaling are observed in Fused null mice, suggesting that Fused is not critical for Hh signaling during early embryonic development of vertebrates.
In addition to the Drosophila homologues, mammalian cells have several novel cytoplasmic regulators of Hh signaling, including Rab23 [76] and tectonic[77]. Rab23 and tectonic are negative regulators downstream of SMO, but their exact mechanism of action remains to be elucidated. Unlike many Rab proteins, Rab23 is localized in the nucleus and cytoplasm[78], suggesting that Rab23 may have other uncharacterized functions apart from membrane trafficking. Through siRNA-based screenings, several additional molecules are identified to be involved in Hh signaling in mammalian cells[79],[80] but their exact functions are unclear.
Several evidences indicate that Suppressor of Fused [Su(Fu)] functions as a tumor suppressor gene in mammalian cells. Su(Fu) is originally identified genetically in Drosophila by its ability to suppress active fused mutations, but itself is not required for pathway activity. Unlike in Drosophila, Su (Fu) null mouse mutants fail to repress the pathway [81] and have some phenotypes similar to Ptch1 inactivation. Ptch1+/− mice are predisposed to developing medulloblastoma, rhabdomyosarcoma, and basal cell carcinomas [82]–[84], whereas Su(fu)+/− mice predominantly develop basaloid epidermal proliferation. Su (Fu) plays a central role in pathway repression, as indicated by data derived from Su(Fu) null MEFs and wild-type cells treated with Su(Fu) siRNA[81], where loss of Su(Fu) results in the activation of Hh signaling. At the molecular level, Su (Fu) associates with and inhibits Gli molecule function, and is required for Gli3 processing[85],[86]. One potential molecular basis by which Hh signaling releases the suppressing activity of Su(Fu) is the enhanced Su(Fu) protein degradation upon the activation of Hh signaling[87].
Hh signaling activation ultimately activates downstream Gli transcription factors, which can regulate target gene expression by directly binding to a consensus binding site (5′-TGGGTGGTC-3′) in the target gene promoter[13],[14],[88],[89]. The activity of Gli transcription factors can be regulated at several levels. First, the nuclear-cytoplasmic shuttling of Gli molecules is tightly regulated[85],[90]–[92]. Protein kinase A can retain Gli1 protein in the cytoplasm via a PKA site in the nuclear localization signal domain [90], whereas activated Ras signaling induces Gli nuclear localization[92]. Second, ubiquitination, acetylation, and protein degradation of Gli molecules is regulated by several distinct mechanisms, including β-TRCP–, cul3/BTB- and numb/ltch-mediated Gli ubiquitination[93]–[98]. In addition, Gli3 (Gli2 to a less extent) can be processed into transcriptional repressors, which may be mediated by the β-TRCP E3 ligase[95]. Defects in the retrograde motor for IFTs can affect Gli3 processing [99]. Furthermore, the transcriptional activity of Gli molecules is tightly regulated. Su(Fu) prevents nuclear translocation of Gli molecules and inhibits Gli1-mediated transcriptional activity[100].
Several feedback regulatory loops exist in this pathway, maintaining the level of Hh signaling in a given cell. PTC, HIP, GAS1, and Gli1 are components as well as target genes of this pathway. PTC and HIP provide negative feedback regulation, whereas Gli1 forms a positive regulatory loop. On the other hand, GAS1 is down-regulated by the Hh pathway but it is a positive regulator for Hh signaling. Alterations of these feedback loops would lead to abnormal signaling of this pathway, such as inactivation of PTCH1 in basal cell carcinoma (BCC).
The Link of Hh Signaling to Human Cancer
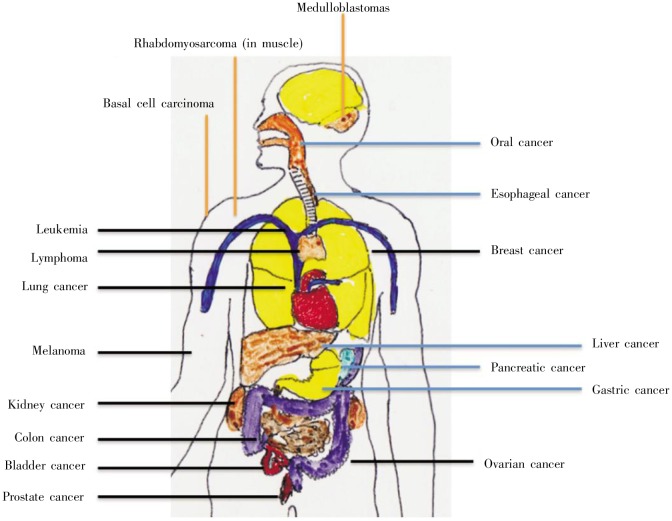
The initial link between Hh signaling and human cancers was made from the discovery that mutations of human PTCH1 are associated with a rare and hereditary form of BCC-basal cell nevus syndrome (also called Gorlin syndrome) [101]–[103]. Gorlin syndrome is a rare autosomal genetic disease with two distinct sets of phenotypes: a predisposition to develop cancer such as BCC and medulloblastoma, and developmental defects such as bifid ribs and ectopic calcification. The tumor suppressor role of PTCH1 is demonstrated in knockout mice, where Ptch1+/− mice develop tumors in addition to other features observed in patients with Gorlin syndrome, such as spina bifida occulta[83],[84],[104]. A variety of cancers are associated with the activation of hedgehog signaling (Figure 3 and Table 1)[105],[106].
Figure 3. Activation of Hh signaling in human cancer. Following the discovery of Hh signaling activation in Gorlin syndrome, increasing evidence suggests that Hh signaling is frequently activated in human cancer. Based on current findings, we group these cancers into three groups. Group one is associated with Gorlin syndrome, including basal cell carcinomas, medulloblastomas, and rhabdomyosarcomas (in muscle) (in red). Group two includes cancer types with reproducible data of Hh signaling activation from several groups, such as oral cancer and many gastrointestinal cancers (in blue). Group three includes cancer types with limited or variable results from different groups (in black). Several common cancer types are included in group three; further investigation will provide insight as to the significance of Hh signaling in these different types of cancer.
Table 1. Activation of hedgehog signaling in human cancer.
| Tumor type | Gene alteration | Mouse models | Functions | Hh-based clinical trial |
| BCCs | PTCH1, SMO, Su(Fu) PTCH1, | Ptch1+/−; R26-SmoM2/K14-cre; K5-SMO (or Gli2); K14-Shh Ptch1fl/fl/cre (K6a, K14 or Mx1) | Driver | Phase II/III |
| Medulloblastoma | SMO, Gli2, Su(Fu), Gli1, Ren | Ptch1+/−; SuFu+/−/p53+/−; R26-SmoM2/cre (Math1, hGFAP, Pax7); Ptch1fl/fl/cre (Math1, hGFAP, Olig2) | Driver | Phase II |
| Rhabdomyosarcoma | PTCH1, Gli1, Su(Fu) | Ptch1+/−; Su(Fu)+/−/p53−/−; SuFu+/−/Ptch1−/−; R26-SmoM2/CAG-CreER | Driver | |
| Esophageal cancer | Shh, Gli2 | Surgical rat models | Unknown | |
| Gastric cancer | Shh | Not available | Unknown | Phase II |
| Liver cancer | Shh | Not available | Unknown | |
| Pancreatic cancer | Shh, Ihh | R26-SmoM2/CAG-CreER; orthotopic | Metastasis | Phase II |
| Gliomas | Gli1, Shh | PDGF-B based mouse model; xenograft | Micro-environment | |
| Breast cancer | Ihh, Shh | Ptch1+/− (transient) | Unknown | |
| Prostate cancer | Shh | Not available | Unknown | |
| Lung cancer | Shh | Xenograft | Unknown | |
| Melanoma | Shh | Xenograft | Unknown | |
| Ovarian cancer | Shh | Xenograft | Unknown | Phase II (suspended) |
| Colon cancer | Shh | Xenograft | Unknown | Phase II (suspended) |
| Osteochondromas | Gli2 | Tg (Gli2;ColllAI); p53+/− | Unknown | |
| Kidney cancer | Shh | Not available | Unknown | |
| Endometrial cancer | Shh | Not available | Unknown | |
| Multiple myloma | Shh | Xenograft | Cancer stem cell | |
| Chronic myeloid leukemia | Shh | Not available | Unknown |
Activation of the Hedgehog Pathway in Human Cancer
BCC and medulloblastoma
Almost all BCCs have activated Hh signaling derived from PTCH1 (50%), SMO gene mutations (10%), or other genetic alterations[107]–[111]. Unlike wild-type SMO, expression of SmoM2, an activated SMO mutant identified in human BCCs, in mouse skin results in the formation of BCC-like tumors[107]. Su(Fu) is also mutated in a few BCCs[109]. From the compiled data, the genetic alteration of the Hh pathway is detected in about 70% of BCCs. Since most BCCs have activated Hh signaling, we predict that alterations in other Hh signaling molecules or related molecules may be responsible for Hh pathway activation in 30% of sporadic BCCs. At molecular level, activated Hh signaling in BCCs leads to cell proliferation through elevated expression of PDGFRα[112], whereas targeted inhibition of the pathway causes apoptosis via the induction of Fas[113].
About one third of medulloblastomas have activated Hh signaling. Like BCCs, loss-of-function mutations of PTCH1 are often responsible for the pathway activation. Mutations in SMO and Su(Fu) are found only in a few cases. In addition, non-canonical activation of Gli2 via ATOH1 and Yap1 has been detected in medulloblastomas. Hh signaling is activated both in the desmoplastic form (more often) and the classic form of medulloblastomas.
Activation of Hh signaling in cancers not associated with Gorlin syndrome
Accumulating data supports the activation Hh signaling in many types of human cancer, including those associated or not associated with Gorlin syndrome. It is estimated that over 30% of human cancers demonstrate activated Hh signaling to a given extent, including brain tumors, melanomas, leukemias, lymphomas, gastrointestinal, prostate, lung, and breast cancers. Unlike the situation in BCCs and meduloblastomas, which are associated with Gorlin syndrome (type I cancer), gene mutation is not primarily responsible for activated Hh signaling in those cancers not associated with Gorlin syndrome (type II cancer)[114],[115]. The current understanding is that Hh signaling activation in type II cancers is caused by ligand-dependent mechanisms or non-canonical Hh signaling activation. The association of ligand-dependence (or ligand-independence) with a specific cancer type, tumor morphology or tumor stage has not yet been established.
The Role of Hh Signaling in Cancer Initiation, Progression, and Metastasis
Increasing evidence suggests that Hh signaling is involved in a specific stage of Carcinogenesis in a given cancer type. In Barrett's esophagus, an early precursor of esophageal adenocarcinomas, both Shh and Ihh are highly expressed in the epithelium, which is associated with stromal expression of Hh target genes Ptch1 and BMP4 [116]. Sox9, a target gene of BMP4, is highly expressed in the epithelial lesion [116]. These results indicate that Hh signaling plays an important role in the initiation of esophageal adenocarcinomas. In pancreatic cancer, activation of this pathway is found in (prostatic intraepithelial neoplasia, PIN) lesions as well in metastases [117]–[120], indicating that Hh signaling plays a significant role in pancreatic cancer. However, transgenic mice with pancreatic-specific expression of SHH or GLI2 develop undifferentiated pancreatic tumors which differ from pancreatic ductal adenocarcinomas (PDAC)[120]–[122], suggesting that sole activation of Hh signaling is not sufficient to drive PDAC development. In other tumors, such as gastric and prostate cancers, the activation of Hh signaling is associated with cancer progression[123]–[127]. Consistent with these findings, the inhibition of Hh signaling in prostate and gastric cancer cells reduces cell invasiveness[124],[127],[128] (our unpublished data). Reports also suggest that Hh signaling is required for development and progression of melanoma, glioma, breast cancer, ovarian cancer, leukemia, and B-cell lymphoma [129]–[134]. However, the role of Hh signaling in each cancer type has not been completely established. It is suggested that Hh signaling plays an important role for cancer stem cells in several cancer types, such as glioma, medulloblastoma, and possibly breast cancer (see more discussion below).
Increasing evidence indicates that Hh signaling is critical for cancer stem cell maintenance and function[135]–[137]. For example, leukemia stem cell maintenance and expansion is dependent on Hh signaling[135],[136]. Alteration of the Hh pathway is reported to affect the hematopoietic stem cell (HSC) population in some studies, but does not change HSC in other studies[136],[138]–[141]. Based on the cancer stem cell theory, it is anticipated that the activation of Hh signaling will exert resistance to cancer chemotherapy and radiotherapy [142]. Indeed, several studies have shown that the activation of Hh signaling is associated with resistance to chemotherapy and radiotherapy [143]–[145]. The Hh signaling inhibitor IPI-926 enhances the delivery of the chemotherapeutical drug gemcitabine in a mouse model of pancreatic cancer[144]. Further investigation is certainly warranted to determine the role of Hh signaling in the cancer stem cells of solid tumors.
Upon reviewing the literature on Hh signaling in human cancer, conflicting results concerning the activation of Hh signaling are often reported for the same cancer type. These discrepancies may arise due to the following reasons. First, the function of Hh signaling in human cancers may be context dependent, occurring in some tissues or cell lines but not in others. For example, accumulating data suggests that Hh signaling functions in maintaining cancer stem cell proliferation[135]–[137], but not the proliferation of all cancer cells. The percentage of cancer stem cells varies greatly among tumor types. Second, heterogeneity in tumor tissue often accounts for differences in the analysis of Hh target gene expression by real-time PCR. For example, prostate cancer specimens can be obtained from prostatectomy or transurethral resection of the prostate (TURP). Whereas the prostatectomy specimens contain only 5%–10% of tumor cells in the tissue, the TURP specimens generally have more than 70% of tumor cells. Thus, the data from these two types of specimens may differ due to the percentage of cancer cells in each tissue [127]. Laser microscope captured tissues will also have a significant amount of non-cancerous cells, with the percentage varying between operators. Third, a standard defining the activation of Hh signaling is required. Some investigators use the increased expression of Gli1 as the read-out[92],[130], whereas others examine the expression of several Hh target genes, such as Gli1, PTCH1, sFRP1, and HIp [120],[124],[125],[146],[147], or only use immunohistochemistry to
detect the activation of Hh signaling[133],[148]. In all, however, most studies use multiple approaches. Thus, the literature must be accepted with caution. Particular attention should be paid to the methodology used in the studies and the reproducibility of the results. In our view, employing immunohistochemistry to detect activation of the Hh signaling pathway for one Hh target gene is unreliable.
Animal Models for Hh-Mediated Carcinogenesis
It is widely accepted that correlation of Hh target gene expression with the tumor specimens is not sufficient to claim a role of Hh signaling in cancer. Establishing animal models using tissue-specific activation of Hh signaling is critical for understanding Hh signaling in Carcinogenesis. Currently, mouse models for BCC and medulloblastoma are well established, whereas mouse models for other Hh-signaling mediated types of Carcinogenesis need improvement. Table 1 summarizes the major mouse models for Hh signaling-mediated Carcinogenesis[105],[106].
Mouse models for BCCs
Wild-type mice never develop BCCs, even after treatment with carcinogen, UV or ionizing radiation. Ptch1+/− mice are susceptible to BCC development following UV irradiation or ionizing radiation[104]. The frequency of BCC development under these conditions is around 50% with 1 or 2 tumors per mouse[113],[149]. Due to the embryonic lethality of Ptch1+/−, tissue-specific knockout of Ptch1 has been generated[150]. By combining conditional gene knockout and the inducible activity of the keratin 6a promoter, Krt6a-cre:Ptch1neo/neo mice develop BCC following stimulation with retinoic acid[151]. In addition to the Ptch1 knockout mouse model, transgenic mice expressing Smo using Krt5 or Krt14 promoter also develop BCC-like tumors[107],[152]. However, these transgenic mice eventually lose the expression of Smo by an unknown mechanism. Using conditional knock-in technology, skin-specific SmoM2YFP (Krt14-creER: R26-SmoM2YFP or Krt14-cre: R26-SmoM2YFP) knock-in mice develop multiple microscopic BCCs at a very early age, providing an easy genetic assay for Hh signaling downstream of Smo[153]. Su(Fu)+/− mice develop skin lesions resembling skin hyperplasia but not BCC-like tumors[154]. Several transgenic mice have been developed using downstream transcriptional factors Gli1, Gli2, and Shh[155]–[157] The inducible expression of Gli2 in the skin results in BCCs after a few weeks. These mouse models provide a rich resource for furthering our understanding of Hh signaling-mediated BCC development.
Mouse models for medulloblastomas
A small portion of Ptch1+/− mice (10%–30%) develop medulloblastomas and rhabdomyosarcomas [83],[84]. The synergy between the p53 pathway and Hh signaling is clearly shown in the medulloblastoma model. Whereas p53 null mice do not develop this type of tumor, all Ptch1+/−p53+/− mice develop medulloblastomas[158]. On the other hand, Ptch2+/− mice do not develop medulloblastoma per se, but Ptch1+/− mice have an increased incidence of medulloblastoma [159],[160]. Su (Fu)+/−mice develop skin phenotypes similar to Gorlin syndrome but are generally not tumor prone[154]. However, Su (Fu)+/− mice with a p53 null background frequently develop medulloblastomas characterized by Hh signaling alterations[161]. Although Ptch1+/−:Su(Fu)+/− mice are more likely to develop medulloblastoma than Ptch1+/− mice, the difference is not statistically significant[162]. In addition to the loss of tumor suppressor genes, transgenic mice expressing SmoM2 mutant under the control of neuroD2 promoter results in medulloblastoma [163]. Tissue-specific activation of Hh signaling via Ptch1 knockout or SmoM2 expression using granule neuron precursor lineage specific promoters (Math1, GFAP, Oligo-2, TLx3), but not the purkinje neuron specific promoter, leads to the formation of medulloblastoma[164],[165], indicating that granule neuron precursors are the source for the development of medulloblastoma. Further analysis shows that CD15 is the medulloblastoma stem cell marker[137],[166].
Mouse models for Hh signaling-mediated Carcinogenesis in other organ sites
Postnatal induction of an activated allele of Smoothened (R26-SmoM2) using a ubiquitously expressed, inducible Cre transgene (CAGGS-CreER) has been used to explore the role of Hh signaling-mediated Carcinogenesis in mice [153]. In this model, all mice developed rhabdomyosarcoma and basal cell carcinoma, with 40% also developed medulloblastoma. In addition, pancreatic lesions resembling low-grade mucinous cystic neoplasms in humans and diverticular harmartomatous lesions in both the intestine and stomach are observed. However, no other tumor types are observed in this mouse model, suggesting that activation of Hh signaling is not sufficient to initiate tumor development in the prostate, lung, breast and gastrointestinal tract.
Similar data are observed in other studies. For example, it is shown in orthotopic mouse models that Hh signaling is necessary for pancreatic cancer metastasis[167] (also our unpublished data). Pancreatic tissue-specific deletion of Smo, on the other hand, did not affect the formation of pancreatic ductal adenocarcinomas (PDAC), whereas GLI2 expression (CLEG2:Pdx1-cre mice) or Shh expression only lead to formation of undifferentiated pancreatic tumors [120]–[122],[168]. These results indicate that activation of Hh signaling alone is not sufficient to drive PDAC formation, but is essential for tumor progression and metastasis. In consistent with this theory, PDAC development in Kras+/G12D:Pdx1-cre mice is not affected by the removal of the Smo gene, and Pdx-1-driven expression of SmoM2 does not result in PIN lesions despite that paracrine Hh signaling is observed in the pancreatic tissue[122],[168].
A recent study indicates that Shh expression in the epithelium of Barrett's esophagus can lead to stromal expression of Hh signaling target genes[116]. Using a Shh transgenic mouse model, it is shown that epithelial expression of Shh can lead to stromal expression of the Hh target gene BMP4, its target gene Sox9 in the epithelium and a columnar phenotype of mouse esophageal epithelium, resembling a feature in human Barrett's esophagus. These data suggest that the activation of Hh signaling can drive the formation of some features resembling Barrett's esophagus in mice.
For investigating the role of Hh signaling in other cancer types, the major models are based on xenografts in immunodeficient mice (nude or SCID mice)[106]. With potential implications of Hh signaling inhibitors for clinical cancer treatment, more established mouse models will be needed. Because modeling cancer metastasis is a challenge, we anticipate an increase in the use of orthotopic mouse models for studying Hh signaling in cancer progression and metastasis in the foreseeable future.
Small Molecule Modulators of Hedgehog Signaling
More than 50 compounds have been identified to have inhibitory effects on Hh signaling. Of these, 4 are being used in clinical trials. There are 3 major targeting sites for Hh signaling inhibitors identified so far: Hh molecules (Shh neutralizing antibodies, small molecule Robotnikinin); SMO protein (cyclopamine and its derivatives IPI-926, Cyc-T, and synthetic compounds GDC-0449, Cur61414, XL-139, and LDE-225); and Gli inhibitors (HPI-1, HPI-2, GANT-56, and GANT-61)[105]. We can divide Hh signaling inhibitors into three groups: natural products (cyclopamine and its derivatives, and other natural products), synthetic small molecules, and Hh signaling modulators. Table 2 lists major current Hh signaling inhibitors[46],[113],[125],[169]–[186].
Table 2. Hedgehog signaling inhibitors.
| Inhibitor | Other name | 50% inhibition concentration (IC50) | In vitro/in vivo studies | References |
| Cyclopamine | 300 nmol/L | In vivo & in vitro | [111],[171],[172] | |
| KAAD-cyclopamine | 20 nmol/L | In vitro cultured cells | [123],[169],[182] | |
| Jervine | 500 nmol/L | In vitro and cultured embryos | [183]–[185] | |
| Cyc-T | Cyclopamine tartrate salt | 20 nmol/L | In vitro & in vivo studies | [174] |
| Cur-61414 | 200 nmol/L | Phase I clinical trial | [186] | |
| Sant-1, 2, 3, 4 | 20–200 nmol/L | In vitro studies | [187] | |
| Compound 5 | <100 nmol/L | In vitro studies | [188] | |
| Compound Z | <1 nmol/L | In vitro studies | [189] | |
| 2-amino-thiazole | 30 nmol/L | In vitro studies | [189] | |
| Gant-58, 61 | 5 µmol/L | In vitro & in vivo studies | [190] | |
| IPI-926 | <20 nmol/L | Phase I clinical trial | [191] | |
| GDC-0449 | Vismodegib | <20 nmol/L | Phase I/II/III clinical trials | [175] |
| BMS-833923 (XL139) | XL139 | <20 nmol/L | Phase I clinical trial | NCI clinical trial database |
| LDE-225 | <20 nmol/L | Phase I clinical trial | NCI clinical trial database | |
| Vitamin D3 | 100 µmol/L | In vitro | [46] | |
| Robotnikinin | >10 µmol/L | In vitro | [192] | |
| HPI-1, 2, 3, 4 | <10 µmol/L | In vitro | [176] | |
| Itraconazole | <1.5 µmol/L | In vitro and xenograft | [181] |
Natural products (cyclopamine, its derivatives, and others)
Cyclopamine, a plant-derived steroidal alkaloid, inhibits Hh signaling through direct binding to the transmembrane helices of SMO[171]. Identification of specific, small molecule antagonists of SMO has revealed exciting new prospects for the targeted therapy of human cancers associated with Hh signaling.
The specificity of cyclopamine varies depending on the concentration used. While cyclopamine at a low concentration (< 10 µmol/L) has specific inhibitory effects on Hh signaling, high doses of cyclopamine can result in cell death without affecting Hh target gene expression[187]. In several mouse models, the in vivo effect of cyclopamine on tumor shrinkage has been demonstrated. Oral delivery of cyclopamine blocks the growth of UV-induced BCCs in Ptch1+/− mice by 50%[113]. The treatment in this model also prevents development of additional microscopic BCCs, implying a cancer prevention potential of cyclopamine. Similarly, cyclopamine is shown to be effective in reducing the development of medulloblastoma in Ptch1+/− mice[169] and tumor growth of cancer cell lines in nu/nu mice[92],[120],[124],[170]. Additional modifications on cyclopamine aiming at increasing acid stability and aqueous solubility are now available, such as IPI-926 and Cyc-T[176],[188]. IPI-926 is now at use in a Phase I clinical trial.
Synthetic Hh signaling antagonists
Increasing synthetic Hh antagonists are being reported in the literature, with most compounds targeting SMO. Four of these compounds are now in clinical trials (Table 2) [182],[183], including GDC-0449. The successful clinical trials with GDC-0449 against human BCCs further encourage the translational investigation in this area [183]. The clinical trial of the same compound in a medulloblastoma patient led to rapid tumor shrinkage, but was complicated by drug resistance due to a SMO mutation, disabling the binding of GDC-0449 to SMO. This case report implies a need for novel alternative strategies for treatment of cancers associated with Hh signaling. There are also several small molecules targeting at Shh or Gli [185],[189],[190]. Due to the wide spread existence of non-conical regulation of Gli transcription factors and the potential resistance to SMO inhibitors, antagonists targeting downstream effectors of the Hh pathway constitute a valuable resource for developing chemotherapeutic strategies against Hh pathway-related cancers.
Hh signaling modulators
Recent studies indicate that vitamin D3, whose secretion can be facilitated by PTCH1, can inhibit SMO signaling through direct binding to SMO. This finding raises a possibility to treat BCCs with nutritional supplements[46]. Promising data show that the effect of tazarotene, a retinoid with retinoic acid receptor (RAR) beta/gamma specificity against BCC Carcinogenesis is sustained after its withdrawal[191]. The common cooking ingredient curcumin has also been shown to block Hh signaling-mediated Carcinogenesis. Several natural products, including genistein, EGCG and resveratrol, are also shown to affect Hh signaling in a mouse model of prostate cancer[192]. A commonly used antifungal agent itraconazole, is shown to affect Hh signaling [186]. The detailed molecular mechanisms of action for these signaling modulators remain elusive.
Summary
The linkage of Hh signaling activation to a variety of human cancers and the discovery of novel Hh signaling inhibitors provides opportunities for developing novel cancer therapeutic strategies. However, several major challenges must be overcome before Hh signaling inhibitors are thrust into clinical application. These include a lack understanding of the molecular mechanisms for Hh signaling-mediated Carcinogenesis, identifying correct tumor types for therapeutic application, the need for reliable and reproducible mouse models for testing and optimizing drug dosages to minimize side effects, and novel stra tegies to mitigate drug resistance.
Acknowledgments
This work was supported by grants from the National Cancer Institute CA94160 and Wells Center for Pediatric Research. Due to a large number of publications in hedgehog signaling activation in human cancer, this manuscript only cited those with the most relevance. The authors wished to acknowledge the contribution of all the laboratories to this area of research.
References
- 1.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in drosophila [J] Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos [J] Cell. 1993;75(7):1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 3.Echelard Y, Epstein DJ, St-Jacques B, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of cns polarity [J] Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 4.Riddle RD, Johnson RL, Laufer E, et al. Sonic hedgehog mediates the polarizing activity of the zpa [J] Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 5.Chang DT, Lopez A, von Kessler DP, et al. Products, genetic linkage and limb patterning activity of a murine hedgehog gene [J] Development. 1994;120(11):3339–3353. doi: 10.1242/dev.120.11.3339. [DOI] [PubMed] [Google Scholar]
- 6.Roelink H, Augsburger A, Heemskerk J, et al. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord [J] Cell. 1994;76(4):761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 7.Epstein EH. Basal cell carcinomas: Attack of the hedgehog [J] Nat Rev Cancer. 2008;8(10):743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J. Hedgehog signaling in prostate cancer [J] Future Oncol. 2005;1(3):331–338. doi: 10.1517/14796694.1.3.331. [DOI] [PubMed] [Google Scholar]
- 9.Xie J. Hedgehog signaling pathway: Development of antagonists or cancer therapy [J] Curr Oncol Rep. 2008;10(2):107–113. doi: 10.1007/s11912-008-0018-7. [DOI] [PubMed] [Google Scholar]
- 10.Xie J. Molecular biology of basal and squamous cell carcinomas [J] Adv Exp Med Biol. 2008;624:241–251. doi: 10.1007/978-0-387-77574-6_19. [DOI] [PubMed] [Google Scholar]
- 11.Jiang J, Hui CC. Hedgehog signaling in development and cancer [J] Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog [J] Nat Rev Genet. 2006;7(11):841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki H, Hui C, Nakafuku M, et al. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro [J] Development. 1997;124(7):1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 14.Kinzler KW, Vogelstein B. The Gli gene encodes a nuclear protein which binds specific sequences in the human genome [J] Mol Cell Biol. 1990;10(2):634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling [J] Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 16.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles [J] Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 17.Taipale J, Beachy PA. The hedgehog and wnt signalling pathways in cancer [J] Nature. 2001;411(6835):349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 18.Lee JJ, Ekker SC, von Kessler DP, et al. Autoproteolysis in hedgehog protein biogenesis [J] Science. 1994;266(5190):1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 19.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development [J] Science. 1996;274(5285):255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 20.Porter JA, von Kessler DP, Ekker SC, et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling [J] Nature. 1995;374(6520):363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 21.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for n-palmitoylation of sonic hedgehog [J] J Biol Chem. 2008;283(32):22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami T, Kawcak T, Li YJ, et al. Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling [J] Development. 2002;129(24):5753–5765. doi: 10.1242/dev.00178. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Erkner A, Gong R, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched [J] Cell. 2002;111(1):63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 24.Caspary T, Garcia-Garcia MJ, Huangfu D, et al. Mouse dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling [J] Curr Biol. 2002;12(18):1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- 25.Dierker T, Dreier R, Petersen A, et al. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells [J] J Biol Chem. 2009;284(12):8013–8022. doi: 10.1074/jbc.M806838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckett K, Franch-Marro X, Vincent JP. Glypican-mediated endocytosis of hedgehog has opposite effects in flies and mice [J] Trends Cell Biol. 2008;18(8):360–363. doi: 10.1016/j.tcb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Lum L, Yao S, Mozer B, et al. Identification of hedgehog pathway components by RNAi in drosophila cultured cells [J] Science. 2003;299(5615):2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 28.Baena-Lopez LA, Rodriguez I, Baonza A. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like [J] Proc Natl Acad Sci USA. 2008;105(28):9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toyoda H, Kinoshita-Toyoda A, Fox B, et al. Structural analysis of glycosaminoglycans in animals bearing mutations in sugarless, sulfateless, and tout-velu. Drosophila homologues of vertebrate genes encoding glycosaminoglycan biosynthetic enzymes [J] J Biol Chem. 2000;275(29):21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- 30.Bellaiche Y, The I, Perrimon N. Tout-velu is a drosophila homologue of the putative tumour suppressor EXT-1 and is needed for hh diffusion [J] Nature. 1998;394(6688):85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 31.Koziel L, Kunath M, Kelly OG, et al. Ext1-dependent heparan sulfate regulates the range of ihh signaling during endochondral ossification [J] Dev Cell. 2004;6(6):801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for sonic hedgehog [J] Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 33.Taipale J, Cooper MK, Maiti T, et al. Patched acts catalytically to suppress the activity of smoothened [J] Nature. 2002;418(6900):892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 34.Chuang PT, McMahon AP. Vertebrate hedgehog signalling modulated by induction of a hedgehog-binding protein [J] Nature. 1999;397(6720):617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 35.Martinelli DC, Fan CM. Gas1 extends the range of hedgehog action by facilitating its signaling [J] Genes Dev. 2007;21(10):1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seppala M, Depew MJ, Martinelli DC, et al. Gas1 is a modifier for holoprosencephaly and genetically interacts with sonic hedgehog [J] J Clin Invest. 2007;117(6):1575–1584. doi: 10.1172/JCI32032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen BL, Tenzen T, McMahon AP. The hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development [J] Genes Dev. 2007;21(10):1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada A, Charron F, Morin S, et al. Boc is a receptor for sonic hedgehog in the guidance of commissural axons [J] Nature. 2006;444(7117):369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- 39.Tenzen T, Allen BL, Cole F, et al. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice [J] Dev Cell. 2006;10(5):647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, Kang JS, Cole F, et al. Cdo functions at multiple points in the sonic hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly [J] Dev Cell. 2006;10(5):657–665. doi: 10.1016/j.devcel.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Yao S, Lum L, Beachy P. The ihog cell-surface proteins bind hedgehog and mediate pathway activation [J] Cell. 2006;125(2):343–357. doi: 10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 42.Capurro Ml, Xu P, Shi W, et al. Glypican-3 inhibits hedgehog signaling during development by competing with patched for hedgehog binding [J] Dev Cell. 2008;14(5):700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Yavari A, Nagaraj R, Owusu-Ansah E, et al. Role of lipid metabolism in smoothened derepression in hedgehog signaling [J] Dev Cell. 2010;19(1):54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khaliullina H, Panakova D, Eugster C, et al. Patched regulates smoothened trafficking using lipoprotein-derived lipids [J] Development. 2009;136(24):4111–4121. doi: 10.1242/dev.041392. [DOI] [PubMed] [Google Scholar]
- 45.Callejo A, Culi J, Guerrero I. Patched, the receptor of hedgehog, is a lipoprotein receptor [J] Proc Natl Acad Sci USA. 2008;105(3):912–917. doi: 10.1073/pnas.0705603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bijlsma MF, Spek CA, Zivkovic D, et al. Repression of smoothened by patched-dependent (pro-)vitamin d3 secretion [J] PLoS Biol. 2006;4(8):e232. doi: 10.1371/journal.pbio.0040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philipp M, Fralish GB, Meloni AR, et al. Smoothened signaling in vertebrates is facilitated by a g protein-coupled receptor kinase [J] Mol Biol Cell. 2008;19(12):5478–5489. doi: 10.1091/mbc.E08-05-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogden SK, Fei DL, Schilling NS, et al. G protein galphai functions immediately downstream of smoothened in hedgehog signalling [J] Nature. 2008;456(7224):967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molnar C, Holguin H, Mayor F, Jr, et al. The G protein-coupled receptor regulatory kinase GPRK2 participates in Hedgehog signaling in Drosophila [J] Proc Natl Acad Sci USA. 2007;104(19):7963–7968. doi: 10.1073/pnas.0702374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riobo NA, Saucy B, Dilizio C, et al. Activation of heterotrimeric g proteins by smoothened [J] Proc Natl Acad Sci USA. 2006;103(33):12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia J, Tong C, Wang B, et al. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I [J] Nature. 2004;432(7020):1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 52.Zhang C, Williams EH, Guo Y, et al. Extensive phosphorylation of smoothened in hedgehog pathway activation [J] Proc Natl Acad Sci USA. 2004;101(52):17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch [J] Nature. 2007;450(7167):252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 54.Corbit KC, Aanstad P, Singla V, et al. Vertebrate smoothened functions at the primary cilium [J] Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 55.Huangfu D, Liu A, Rakeman AS, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins [J] Nature. 2003;426(6962):83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 56.May SR, Ashique AM, Karlen M, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli [J] Dev Biol. 2005;287(2):378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 57.Huangfu D, Anderson KV. Cilia and hedgehog responsiveness in the mouse [J] Proc Natl Acad Sci USA. 2005;102(32):11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Davenport JR, Croyle MJ, et al. Disruption of IFT results in both exocrine and endocrine abnormalities in the pancreas of Tg737(orpk) mutant mice [J] Lab Invest. 2005;85(1):45–64. doi: 10.1038/labinvest.3700207. [DOI] [PubMed] [Google Scholar]
- 59.Hoover AN, Wynkoop A, Zeng H, et al. C2cd3 is required for cilia formation and hedgehog signaling in mouse [J] Development. 2008;135(24):4049–4058. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling [J] Cell. 2006;125(3):439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Cortellino S, Wang C, Wang B, et al. Defective ciliogenesis, embryonic lethality and severe impairment of the sonic hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene ift122/wdr10, partially overlapping with the DNA repair gene med1/mbd4 [J] Dev Biol. 2009;325(1):225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haycraft CJ, Banizs B, Aydin-Son Y, et al. Gli2 and gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function [J] PLoS Genet. 2005;1(4):e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Zhou Z, Walsh CT, et al. Selective translocation of intracellular smoothened to the primary cilium in response to hedgehog pathway modulation [J] Proc Natl Acad Sci USA. 2009;106(8):2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium [J] PLoS One. 2009;4(4):e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohatgi R, Milenkovic L, Corcoran RB, et al. Hedgehog signal transduction by smoothened: Pharmacologic evidence for a 2-step activation process [J] Proc Natl Acad Sci USA. 2009;106(9):3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han YG, Kim HJ, Dlugosz AA, et al. Dual and opposing roles of primary cilia in medulloblastoma development [J] Nat Med. 2009;15(9):1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong SY, Seol AD, So PL, et al. Primary cilia can both mediate and suppress hedgehog pathway-dependent tumorigenesis [J] Nat Med. 2009;15(9):1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovacs JJ, Whalen EJ, Liu R, et al. Beta-arrestin-mediated localization of smoothened to the primary cilium [J] Science. 2008;320(5884):1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen MH, Wilson CW, Li YJ, et al. Cilium-independent regulation of gli protein function by sufu in hedgehog signaling is evolutionarily conserved [J] Genes Dev. 2009;23(16):1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia J, Kolterud A, Zeng H, et al. Suppressor of fused inhibits mammalian hedgehog signaling in the absence of cilia [J] Dev Biol. 2009;330(2):452–460. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheung HO, Zhang X, Ribeiro A, et al. The kinesin protein kif7 is a critical regulator of gli transcription factors in mammalian hedgehog signaling [J] Sci Signal. 2009;2(76):ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 72.Endoh-Yamagami S, Evangelista M, Wilson D, et al. The mammalian cos2 homolog kif7 plays an essential role in modulating hh signal transduction during development [J] Curr Biol. 2009;19(15):1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 73.Wilson CW, Nguyen CT, Chen MH, et al. Fused has evolved divergent roles in vertebrate hedgehog signalling and motile ciliogenesis [J] Nature. 2009;459(7243):98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merchant M, Evangelista M, Luoh SM, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus [J] Mol Cell Biol. 2005;25(16):7054–7068. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen MH, Gao N, Kawakami T, et al. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development [J] Mol Cell Biol. 2005;25(16):7042–7053. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse sonic hedgehog signalling pathway [J] Nature. 2001;412(6843):194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 77.Reiter JF, Skarnes WC. Tectonic, a novel regulator of the hedgehog pathway required for both activation and inhibition [J] Genes Dev. 2006;20(1):22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang S, Yang L, An Y, et al. Expression of hedgehog signaling molecules in lung cancer [J] Acta Histochem. 2010 Jul 23; doi: 10.1016/j.acthis.2010.06.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 79.Varjosalo M, Bjorklund M, Cheng F, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling [J] Cell. 2008;133(3):537–548. doi: 10.1016/j.cell.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 80.Evangelista M, Lim TY, Lee J, et al. Kinome sirna screen identifies regulators of ciliogenesis and hedgehog signal transduction [J] Sci Signal. 2008;1(39):ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- 81.Svard J, Henricson KH, Persson-Lek M, et al. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian hedgehog signaling pathway [J] Dev Cell. 2006;10(2):187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 82.Aszterbaum M, Epstein J, Oro A, et al. Ultraviolet and ionizing radiation enhance the growth of bccs and trichoblastomas in patched heterozygous knockout mice [J] Nat Med. 1999;5(11):1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- 83.Hahn H, Wojnowski L, Zimmer AM, et al. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of gorlin syndrome [J] Nat Med. 1998;4(5):619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 84.Goodrich LV, Milenkovic L, Higgins KM, et al. Altered neural cell fates and medulloblastoma in mouse patched mutants [J] Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 85.Barnfield PC, Zhang X, Thanabalasingham V, et al. Negative regulation of gli1 and gli2 activator function by suppressor of fused through multiple mechanisms [J] Differentiation. 2005;73(8):397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 86.Kise Y, Morinaka A, Teglund S, et al. Sufu recruits gsk3beta for efficient processing of gli3 [J] Biochem Biophys Res Commun. 2009;387(3):569–574. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 87.Yue S, Chen Y, Cheng SY. Hedgehog signaling promotes the degradation of tumor suppressor sufu through the ubiquitin-proteasome pathway [J] Oncogene. 2009;28(4):492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- 88.Kinzler KW, Ruppert JM, Bigner SH, et al. The gli gene is a member of the kruppel family of zinc finger proteins [J] Nature. 1988;332(6162):371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 89.Ruppert JM, Kinzler KW, Wong AJ, et al. The gli-kruppel family of human genes [J] Mol Cell Biol. 1988;8(8):3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheng T, Chi S, Zhang X, et al. Regulation of gli1 localization by the camp/protein kinase a signaling axis through a site near the nuclear localization signal [J] J Biol Chem. 2006;281(1):9–12. doi: 10.1074/jbc.C500300200. [DOI] [PubMed] [Google Scholar]
- 91.Kogerman P, Grimm T, Kogerman L, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of gli-1 [J] Nat Cell Biol. 1999;1(5):312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 92.Stecca B, Mas C, Clement V, et al. Melanomas require hedgehog-gli signaling regulated by interactions between gli1 and the ras-mek/akt pathways [J] Proc Natl Acad Sci USA. 2007;104(14):5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan Y, Bai CB, Joyner AL, et al. Sonic hedgehog signaling regulates gli2 transcriptional activity by suppressing its processing and degradation [J] Mol Cell Biol. 2006;26(9):3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huntzicker EG, Estay IS, Zhen H, et al. Dual degradation signals control gli protein stability and tumor formation [J] Genes Dev. 2006;20(3):276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang B, Li Y. Evidence for the direct involvement of {beta}trcp in gli3 protein processing [J] Proc Natl Acad Sci USA. 2006;103(1):33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Marcotullio L, Ferretti E, Greco A, et al. Numb is a suppressor of hedgehog signalling and targets gli1 for itch-dependent ubiquitination [J] Nat Cell Biol. 2006;8(12):1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 97.Jiang J. Regulation of hh/gli signaling by dual Ubiquitin pathways [J] Cell Cycle. 2006;5(21):2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- 98.Canettieri G, Di Marcotullio L, Greco A, et al. Histone deacetylase and cullin3-ren (kctd11) Ubiquitin ligase interplay regulates hedgehog signalling through gli acetylation [J] Nat Cell Biol. 2010;12(2):132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 99.Huangfu D, Anderson KV. Signaling from smo to ci/gli: Conservation and divergence of hedgehog pathways from drosophila to vertebrates [J] Development. 2006;133(1):3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 100.Cheng SY, Bishop JM. Suppressor of fused represses glimediated transcription by recruiting the sap18-msin3 corepressor complex [J] Proc Natl Acad Sci USA. 2002;99(8):5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of drosophila patched in the nevoid basal cell carcinoma syndrome [J] Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 102.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome [J] Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 103.Epstein E., Jr Genetic determinants of basal cell carcinoma risk [J] Med Pediatr Oncol. 2001;36(5):555–558. doi: 10.1002/mpo.1129. [DOI] [PubMed] [Google Scholar]
- 104.Aszterbaum M, Beech J, Epstein EH., Jr Ultraviolet radiation mutagenesis of hedgehog pathway genes in basal cell carcinomas [J] J Investig Dermatol Symp Proc. 1999;4(1):41–45. doi: 10.1038/sj.jidsp.5640179. [DOI] [PubMed] [Google Scholar]
- 105.Yang L, Xie G, Fan Q, et al. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications [J] Oncogene. 2010;29(4):469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 106.Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma [J] Biochim Biophys Acta. 2010;1805(2):181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 107.Xie J, Murone M, Luoh SM, et al. Activating smoothened mutations in sporadic basal-cell carcinoma [J] Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 108.Lam CW, Xie J, To KF, et al. A frequent activated smoothened mutation in sporadic basal cell carcinomas [J] Oncogene. 1999;18(3):833–836. doi: 10.1038/sj.onc.1202360. [DOI] [PubMed] [Google Scholar]
- 109.Reifenberger J, Wolter M, Knobbe CB, et al. Somatic mutations in the ptch, smoh, sufuh and tp53 genes in sporadic basal cell carcinomas [J] Br J Dermatol. 2005;152(1):43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 110.Reifenberger J, Wolter M, Weber RG, et al. Missense mutations in smoh in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system [J] Cancer Res. 1998;58(9):1798–1803. [PubMed] [Google Scholar]
- 111.Couve-Privat S, Bouadjar B, Avril MF, et al. Significantly high levels of ultraviolet-specific mutations in the smoothened gene in basal cell carcinomas from DNA repair-deficient xeroderma pigmentosum patients [J] Cancer Res. 2002;62(24):7186–7189. [PubMed] [Google Scholar]
- 112.Xie J, Aszterbaum M, Zhang X, et al. A role of pdgfralpha in basal cell carcinoma proliferation [J] Proc Natl Acad Sci USA. 2001;98(16):9255–9259. doi: 10.1073/pnas.151173398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Athar M, Li C, Tang X, et al. Inhibition of smoothened signaling prevents ultraviolet b-induced basal cell carcinomas through regulation of fas expression and apoptosis [J] Cancer Res. 2004;64(20):7545–7552. doi: 10.1158/0008-5472.CAN-04-1393. [DOI] [PubMed] [Google Scholar]
- 114.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for hedgehog ligand stimulation in growth of digestive tract tumours [J] Nature. 2003;425(6960):846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 115.Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer [J] Nature. 2003;422(6929):313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 116.Wang DH, Clemons NJ, Miyashita T, et al. Aberrant epithelial-mesenchymal hedgehog signaling characterizes barren's metaplasia [J] Gastroenterology. 2010;138(5):1810–1822. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer [J] Oncogene. 2009;28(40):3513–3525. doi: 10.1038/onc.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feldmann G, Fendrich V, McGovern K, et al. An orally bioavailable small-molecule inhibitor of hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer [J] Mol Cancer Ther. 2008;7(9):2725–2735. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma X, Sheng T, Zhang Y, et al. Hedgehog signaling is activated in subsets of esophageal cancers [J] Int J Cancer. 2006;118(1):139–148. doi: 10.1002/ijc.21295. [DOI] [PubMed] [Google Scholar]
- 120.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis [J] Nature. 2003;425(6960):851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pasca di Magliano M, Sekine S, Ermilov A, et al. Hedgehog/ras interactions regulate early stages of pancreatic cancer [J] Genes Dev. 2006;20(22):3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nolan-Stevaux O, Lau J, Truitt ML, et al. Gli1 is regulated through smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates pdac cell survival and transformation [J] Genes Dev. 2009;23(1):24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fan L, Pepicelli CV, Dibble CC, et al. Hedgehog signaling promotes prostate xenograft tumor growth [J] Endocrinology. 2004;145(8):3961–3970. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- 124.Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis [J] Nature. 2004;431(7009):707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 125.Ma X, Chen K, Huang S, et al. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas [J] Carcinogenesis. 2005;26(10):1698–1705. doi: 10.1093/carcin/bgi130. [DOI] [PubMed] [Google Scholar]
- 126.Sanchez P, Hernandez AM, Stecca B, et al. Inhibition of prostate cancer proliferation by interference with sonic hedgehog-gli1 signaling [J] Proc Natl Acad Sci USA. 2004;101(34):12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sheng T, Li C, Zhang X, et al. Activation of the hedgehog pathway in advanced prostate cancer [J] Mol Cancer. 2004;3:29. doi: 10.1186/1476-4598-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He J, Sheng T, Stelter AA, et al. Suppressing wnt signaling by the hedgehog pathway through sfrp-1 [J] J Biol Chem. 2006;281(47):35598–35602. doi: 10.1074/jbc.C600200200. [DOI] [PubMed] [Google Scholar]
- 129.Ehtesham M, Sarangi A, Valadez JG, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells [J] Oncogene. 2007;26(39):5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 130.Bhattacharya R, Kwon J, Ali B, et al. Role of hedgehog signaling in ovarian cancer [J] Clin Cancer Res. 2008;14(23):7659–7666. doi: 10.1158/1078-0432.CCR-08-1414. [DOI] [PubMed] [Google Scholar]
- 131.Fiaschi M, Rozell B, Bergstrom A, et al. Development of mammary tumors by conditional expression of gli1 [J] Cancer Res. 2009;69(11):4810–4817. doi: 10.1158/0008-5472.CAN-08-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kasper M, Jaks V, Fiaschi M, et al. Hedgehog signalling in breast cancer [J] Carcinogenesis. 2009;30(6):903–911. doi: 10.1093/carcin/bgp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liao X, Siu MK, Au CW, et al. Aberrant activation of hedgehog signaling pathway in ovarian cancers: Effect on prognosis, cell invasion and differentiation [J] Carcinogenesis. 2009;30(1):131–140. doi: 10.1093/carcin/bgn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lindemann RK. Stroma-initiated hedgehog signaling takes center stage in b-cell lymphoma [J] Cancer Res. 2008;68(4):961–964. doi: 10.1158/0008-5472.CAN-07-5500. [DOI] [PubMed] [Google Scholar]
- 135.Dierks C, Beigi R, Guo GR, et al. Expansion of bcr-abl-positive leukemic stem cells is dependent on hedgehog pathway activation [J] Cancer Cell. 2008;14(3):238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 136.Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia [J] Nature. 2009;458(7239):776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Read TA, Fogarty MP, Markant SL, et al. Identification of cd15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma [J] Cancer Cell. 2009;15(2):135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hofmann I, Stover EH, Cullen DE, et al. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis [J] Cell Stem Cell. 2009;4(6):559–567. doi: 10.1016/j.stem.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gao J, Graves S, Koch U, et al. Hedgehog signaling is dispensable for adult hematopoietic stem cell function [J] Cell Stem Cell. 2009;4(6):548–558. doi: 10.1016/j.stem.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Siggins SL, Nguyen NY, McCormack MP, et al. The hedgehog receptor patched1 regulates myeloid and lymphoid progenitors by distinct cell-extrinsic mechanisms [J] Blood. 2009;114(5):995–1004. doi: 10.1182/blood-2009-03-208330. [DOI] [PubMed] [Google Scholar]
- 141.Merchant A, Joseph G, Wang Q, et al. Gli1 regulates the proliferation and differentiation of hscs and myeloid progenitors [J] Blood. 2010;115(12):2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells [J] Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 143.Sims-Mourtada J, Izzo JG, Apisarnthanarax S, et al. Hedgehog: An attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response [J] Clin Cancer Res. 2006;12(21):6565–6572. doi: 10.1158/1078-0432.CCR-06-0176. [DOI] [PubMed] [Google Scholar]
- 144.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer [J] Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yoshikawa R, Nakano Y, Tao L, et al. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy [J] Br J Cancer. 2008;98(10):1670–1674. doi: 10.1038/sj.bjc.6604361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang S, He J, Zhang X, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas [J] Carcinogenesis. 2006;27(7):1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 147.Lee Y, Kawagoe R, Sasai K, et al. Loss of suppressor-of-fused function promotes tumorigenesis [J] Oncogene. 2007;26(44):6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 148.Kubo M, Nakamura M, Tasaki A, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer [J] Cancer Res. 2004;64(17):6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 149.Mancuso M, Pazzaglia S, Tanori M, et al. Basal cell carcinoma and its development: Insights from radiation-induced tumors in ptch1-deficient mice [J] Cancer Res. 2004;64(3):934–941. doi: 10.1158/0008-5472.can-03-2460. [DOI] [PubMed] [Google Scholar]
- 150.Ellis T, Smyth I, Riley E, et al. Patched 1 conditional null allele in mice [J] Genesis. 2003;36(3):158–161. doi: 10.1002/gene.10208. [DOI] [PubMed] [Google Scholar]
- 151.Adolphe C, Hetherington R, Ellis T, et al. Patched1 functions as a gatekeeper by promoting cell cycle progression [J] Cancer Res. 2006;66(4):2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 152.Grachtchouk V, Grachtchouk M, Lowe L, et al. The magnitude of hedgehog signaling activity defines skin tumor phenotype [J] EMBO J. 2003;22(11):2741–2751. doi: 10.1093/emboj/cdg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mao J, Ligon KL, Rakhlin EY, et al. A novel somatic mouse model to survey tumorigenic potential applied to the hedgehog pathway [J] Cancer Res. 2006;66(20):10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Svard J, Heby-Henricson K, Persson-Lek M, et al. Genetic elimination of suppressor of fused reveals an essential repressor function in the mammalian hedgehog signaling pathway [J] Dev Cell. 2006;10(2):187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 155.Grachtchouk M, Mo R, Yu S, et al. Basal cell carcinomas in mice overexpressing gli2 in skin [J] Nat Genet. 2000;24(3):216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 156.Nilsson M, Unden AB, Krause D, et al. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing gli-1 [J] Proc Natl Acad Sci USA. 2000;97(7):3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Oro AE, Higgins KM, Hu Z, et al. Basal cell carcinomas in mice overexpressing sonic hedgehog [J] Science. 1997;276(5313):817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 158.Romer JT, Kimura H, Magdaleno S, et al. Suppression of the shh pathway using a small molecule inhibitor eliminates medulloblastoma in ptc1 (+/-)p53 (-/-) mice [J] Cancer Cell. 2004;6(3):229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 159.Nieuwenhuis E, Motoyama J, Barnfield PC, et al. Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia [J] Mol Cell Biol. 2006;26(17):6609–6622. doi: 10.1128/MCB.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee Y, Miller HL, Russell HR, et al. Patched2 modulates tumorigenesis in patched1 heterozygous mice [J] Cancer Res. 2006;66(14):6964–6971. doi: 10.1158/0008-5472.CAN-06-0505. [DOI] [PubMed] [Google Scholar]
- 161.Lee Y, Kawagoe R, Sasai K, et al. Loss of suppressor-of-fused function promotes tumorigenesis [J] Oncogene. 2007;26(44):6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 162.Svard J, Rozell B, Toftgard R, et al. Tumor suppressor gene co-operativity in compound patched1 and suppressor of fused heterozygous mutant mice [J] Mol Carcinog. 2009;48(5):408–419. doi: 10.1002/mc.20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hallahan AR, Pritchard Jl, Hansen S, et al. The smoa1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas [J] Cancer Res. 2004;64(21):7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 164.Schuller U, Heine VM, Mao J, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form shh-induced medulloblastoma [J] Cancer Cell. 2008;14(2):123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang ZJ, Ellis T, Markant SL, et al. Medulloblastoma can be initiated by deletion of patched in lineage-restricted progenitors or stem cells [J] Cancer Cell. 2008;14(2):135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ward RJ, Lee L, Graham K, et al. Multipotent cd15+ cancer stem cells in patched-1-deficient mouse medulloblastoma [J] Cancer Res. 2009;69(11):4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- 167.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: A new paradigm for combination therapy in solid cancers [J] Cancer Res. 2007;67(5):2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tian H, Callahan CA, DuPree KJ, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic Carcinogenesis [J] Proc Natl Acad Sci USA. 2009;106(11):4254–4259. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanchez P, Ruiz i Altaba A. In vivo inhibition of endogenous brain tumors through systemic interference of hedgehog signaling in mice [J] Mech Dev. 2005;122(2):223–230. doi: 10.1016/j.mod.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 170.Berman DM, Karhadkar SS, Hallahan AR, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade [J] Science. 2002;297(5586):1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 171.Chen JK, Taipale J, Cooper MK, et al. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened [J] Genes Dev. 2002;16(21):2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huang S, He J, Zhang X, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas [J] Carcinogenesis. 2006;27(7):1334–1340. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 173.Cooper MK, Porter JA, Young KE, et al. Teratogen-mediated inhibition of target tissue response to shh signaling [J] Science. 1998;280(5369):1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- 174.Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis [J] Development. 1999;126(21):4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- 175.Mistretta CM, Liu HX, Gaffield W, et al. Cyclopamine and jervine in embryonic rat tongue cultures demonstrate a role for shh signaling in taste papilla development and patterning: Fungiform papillae double in number and form in novel locations in dorsal lingual epithelium [J] Dev Biol. 2003;254(1):1–18. doi: 10.1016/s0012-1606(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 176.Xie J, Garrossian M. The Board of Regents of the University of Texas System, Assignee. Cyclopamine tartrate salt and uses thereof [P]. US patent application number WO 2009099625. 2009.
- 177.Frank-Kamenetsky M, Zhang XM, Bottega S, et al. Small-molecule modulators of hedgehog signaling: Identification and characterization of smoothened agonists and antagonists [J] J Biol. 2002;1(2):10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen JK, Taipale J, Young KE, et al. Small molecule modulation of smoothened activity [J] Proc Natl Acad Sci USA. 2002;99(22):14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Borzillo GV, Lippa B. The hedgehog signaling pathway as a target for anticancer drug discovery [J] Curr Top Med Chem. 2005;5(2):147–157. doi: 10.2174/1568026053507732. [DOI] [PubMed] [Google Scholar]
- 180.Rubin LL, de Sauvage FJ. Targeting the hedgehog pathway in cancer [J] Nat Rev Drug Discov. 2006;5(12):1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 181.Lauth M, Bergstrom A, Shimokawa T, et al. Inhibition of gli-mediated transcription and tumor cell growth by small-molecule antagonists [J] Proc Natl Acad Sci USA. 2007;104(20):8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer [J] Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma [J] N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 184.Stanton BZ, Peng LF, Maloof N, et al. A small molecule that binds hedgehog and blocks its signaling in human cells [J] Nat Chem Biol. 2009;5(3):154–156. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hyman JM, Firestone AJ, Heine VM, et al. Small-molecule inhibitors reveal multiple strategies for hedgehog pathway blockade [J] Proc Natl Acad Sci USA. 2009;106(33):14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kim J, Tang JY, Gong R, et al. Itraconazole, a commonly used antifungal that inhibits hedgehog pathway activity and cancer growth [J] Cancer Cell. 2010;17(4):388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yauch RL, Gould SE, Scales SJ, et al. A paracrine requirement for hedgehog signalling in cancer [J] Nature. 2008;455(7211):406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 188.Tremblay MR, Lescarbeau A, Grogan MJ, et al. Discovery of a potent and orally active hedgehog pathway antagonist (ipi-926) [J] J Med Chem. 2009;52(14):4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- 189.Von Hoff DD, Lorusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma [J] N Engl J Med. 2009;361(12):1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 190.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor gdc-0449 [J] N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.So PL, Fujimoto MA, Epstein EH., Jr Pharmacologic retinoid signaling and physiologic retinoic acid receptor signaling inhibit basal cell carcinoma tumorigenesis [J] Mol Cancer Ther. 2008;7(5):1275–1284. doi: 10.1158/1535-7163.MCT-07-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Slusarz A, Shenouda NS, Sakla MS, et al. Common botanical compounds inhibit the hedgehog signaling pathway in prostate cancer [J] Cancer Res. 2010;70(8):3382–3390. doi: 10.1158/0008-5472.CAN-09-3012. [DOI] [PubMed] [Google Scholar]