Abstract
We evaluated the hypothesis that the N-terminal region of the replication control protein Cdc6 acts as an inhibitor of cyclin-dependent kinase (Cdk) activity, promoting mitotic exit. Cdc6 accumulation is restricted to the period from mid-cell cycle until the succeeding G1, due to proteolytic control that requires the Cdc6 N-terminal region. During late mitosis, Cdc6 is present at levels comparable with Sic1 and binds specifically to the mitotic cyclin Clb2. Moderate overexpression of Cdc6 promotes viability of CLB2Δdb strains, which otherwise arrest at mitotic exit, and rescue is dependent on the N-terminal putative Cdk-inhibitory domain. These observations support the potential for Cdc6 to inhibit Clb2-Cdk, thus promoting mitotic exit. Consistent with this idea, we observed a cytokinesis defect in cdh1Δ sic1Δ cdc6Δ2–49 triple mutants. However, we were able to construct viable strains, in three different backgrounds, containing neither SIC1 nor the Cdc6 Cdk-inhibitory domain, in contradiction to previous work. We conclude, therefore, that although both Cdc6 and Sic1 have the potential to facilitate mitotic exit by inhibiting Clb2-Cdk, mitotic exit nevertheless does not require any identified stoichiometric inhibitor of Cdk activity.
INTRODUCTION
Exit from mitosis is a critical cell cycle transition (Zachariae and Nasmyth, 1999). Although DNA replication and spindle morphogenesis are under positive control by cyclin-dependent kinase (Cdk), mitotic exit requires at least partial Cdk inactivation to occur. Thus, an oscillation in Cdk activity is obligatory for a complete cycle, because DNA replication and initiation of mitotic division are tied to high Cdk activity, but the replicated chromosomes assembled on the spindle will not complete division into two daughter cells until low Cdk activity is attained. Buildup of Cdk activity in the next cell cycle is then required before DNA replication can occur. This simple mechanism, combined with an oscillatory mechanism for Cdk activity, will thus reproduce much of the essential biology of the cell cycle (Nasmyth, 1996; Stern and Nurse, 1996; Zachariae and Nasmyth, 1999). Therefore, it is important to understand the mechanisms of Cdk inactivation.
A critical conserved mechanism for Cdk inactivation at exit from mitosis is cyclin proteolysis, under the control of the anaphase-promoting complex (APC) activated by Cdc20 and Cdh1 (Evans et al., 1983; Zachariae and Nasmyth, 1999; Morgan and Roberts, 2002; Wäsch and Cross, 2002). In budding yeast, two additional regulators have been proposed to inactivate Cdk to allow exit from mitosis: the Sic1 stoichiometric inhibitor, activated by the Cdc14 phosphatase (Visintin et al., 1998), and the N terminus of the Cdc6 replication protein (Calzada et al., 2001). This region of Cdc6 was identified as a Cdk-binding domain (Elsasser et al., 1996; Perkins et al., 2001; Weinreich et al., 2001). Calzada et al. (2001) reported that deletion of this region in the chromosomal copy of CDC6 (cdc6Δ2-47) caused a delay in mitotic exit, and remarkably, combination of the cdc6Δ2-47 mutation with deletion of SIC1 absolutely blocked mitotic exit. Cyclin proteolysis is known to be essential for mitotic exit (Wäsch and Cross, 2002), but the cdc6Δ2-47 sic1Δ mitotic exit block was shown to occur even in the presence of all cyclin proteolytic machinery. Thus, this result suggested coequal control of Cdk inactivation by cyclin proteolytic machinery and inhibition by stoichiometric binding proteins. This finding was surprising, because it was recently proposed that the primary biological role of Sic1 was in regulating replication origin reloading, not in regulating mitotic exit (Lengronne and Schwob, 2002). In addition, the Cdc6 N terminus is capable in some circumstances of inhibiting completion of mitosis (Weinreich et al., 2001), rather than promoting it as reported (Calzada et al., 2001). Thus, the block to mitotic exit in cdc6Δ2-47 sic1Δ strains was a striking and surprising observation.
The essential function of Cdc6 is to promote the assembly of prereplicative complexes (pre-RCs) at origins of DNA replication (Bell and Dutta, 2002). Assembled pre-RCs contain at least ORC, Cdc6, and Mcms and constitute a platform for the DNA replication machinery to come into place. Firing of DNA replication at these sites results in pre-RC disassembly, and reassembly of pre-RCs is prevented by redundant mechanisms resulting from Clb-Cdc28 actions that include phosphorylation of Cdc6 (Nguyen et al., 2001).
Transcription of CDC6 is most active in late mitosis/early G1 (Zhou and Jong, 1990; Zwerschke et al., 1994) via Swi5 (Piatti et al., 1995), a transcription factor that also activates the transcription of SIC1 (Knapp et al., 1996; Toyn et al., 1997). In cells that experience a prolonged G1, a second wave of CDC6 transcription occurs, which is dependent on SBF (Swi4/Swi6) or MBF (Mbp1/Swi6) (Piatti et al., 1995). Cdc6 proteolysis is differentially regulated by different cyclin-Cdc28 kinases (Piatti et al., 1995; Elsasser et al., 1999; Calzada et al., 2000; Drury et al., 2000). As a result of these controls, Cdc6 abundance reaches its maximum in late mitotic cells (Drury et al., 1997). This has the effect of positioning Cdc6 to promote pre-RC assembly after mitosis and also, hypothetically, to regulate mitotic exit.
Here, we evaluate Cdc6 as a Clb2-Cdc28 inhibitor in mitotic exit. We find that although biochemical and genetic experiments demonstrate that Cdc6 has the potential to act in this role, it is not required for this cell cycle step even in the absence of the other known Clb-Cdk inhibitor, Sic1.
MATERIALS AND METHODS
Yeast Strain Construction
Standard methods were used for mating, tetrad analysis, and transformations. A list of strains is provided in the supplementary information (Table SI). CLB2Δdb and CLB2Δdb,ken alleles were constructed as described previously (Wäsch and Cross, 2002). A two-copy integrant of GAL-SIC1 was required for construction of the CLB2Δdb,ken strain (our unpublished observations). swi5::kanMX was obtained from D. Stillman. CDC6 wild-type and mutant plasmids were obtained from M. Weinreich. The TUB1-GFP allele marked with HIS3 in the W303 strain background was obtained from A. Murray. Other plasmids and alleles were laboratory stocks. Protein A tagging of CDC6, cdc6Δ2-49, SIC1, and CLB2 at the C-terminal end was performed by genomic integration of a DNA sequence that encodes the IgG binding domains of protein A (PRA) from aureus and the HIS3MX selection marker from Schizosaccharomyces pombe (Wach et al., 1997; Rout et al., 2000). Tagging of CLN2, CLB2, CLB3, and CLB5 at the C-terminal end with nine repeats of the Myc epitope (MYC) was also done by genomic integration of a polymerase chain reaction (PCR) product that also contained the klTRP1 gene, as described previously (Knop et al., 1999).
Cell Cycle Blocks for Affinity Purifications
The CDC6-PRA and CDC6-PRA bar1 strains were grown in YPD to a density of ∼2.0 × 107 cells/ml at 30°C. The CDC6-PRA cdc20 GALL-CDC20 strain was grown at 30°C in YPG (YPGal) to a density of ∼2.0 × 107 cells/ml. CDC6-PRA bar1 strain were arrested by incubating for 2 h at 30°C in YPD (YPDex) containing 100 nM α-factor (synthesized by The Rockefeller University Protein Resource Center, New York, NY). CDC6-PRA cdc20 GALL-CDC20 cells were arrested at metaphase by adding 2% glucose to the YPG medium and incubating them for 3.5 h at 30°C. Cells were harvested by centrifugation and resuspended in 20 mM Na-HEPES (pH 7.4), 1.2% polyvinyl-pyrrolidone (Sigma-Aldrich, St. Louis, MO), 0.2 mg/ml phenylmethylsulfonyl fluoride (PMSF), and 4 μg/ml pepstatin A (1 ml of buffer/10-g cell pellet). The cell suspension was frozen in liquid nitrogen and lysed by grinding in a mortar grinder (RM100; Retsch, Newtown, PA) that was continuously cooled with liquid nitrogen (Schultz, 1999).
Affinity Purification and Separation of Protein Complexes
Protein A affinity purification were performed based on a documented method (Aitchison et al., 1996; Rout et al., 1997). Ground cells (4 g) were resuspended in 20 ml of extraction buffer (20 mM Na-HEPES, pH 7.4, 300 mM NaCl, 0.1% Tween 20, 2 mM MgCl2, 0.002% DNase I, 0.2 mg/ml PMSF, 4 μg/ml pepstatin A) and incubated with gentle agitation at room temperature for 10 min to allow DNase I activity. The lysate was homogenized with a Polytron (PT 10/35; Brinkmann Instruments, Westbury, NY) for ∼20 s and then rotated for 1 h at 4°C. The soluble fraction was isolated by centrifugation at 3000 rpm (H-1000B; Sorvall, Newton, CT) for 10 min. IgG-DynaBeads were prepared by cross-linking rabbit IgG (ICN, Aurora, OH) to epoxy-activated DynaBeads (Dynal, Lake Success, NY). IgG-DynaBeads (15 μg) were added to each lysate, and the suspension was rotated for ∼18 h at 4°C. The IgGDynaBeads were collected with a magnet and washed five times with 1 ml of ice-cold extraction buffer. Isolated protein complexes were eluted from the IgG-DynaBeads in 0.5 M NH4OH 0.5 mM EDTA for 5 min at room temperature. The eluted proteins were frozen in liquid nitrogen and lyophilized in a SpeedVac (Thermo Savant, Holbrook, NY). Dried protein samples were dissolved in SDS-PAGE loading buffer [78 mM Tris-Cl, 0.003% bromphenol blue, 25 mM Tris base, 6.25 mM Tris (2-carboxyethyl) phosphine, 12.5% glycerol, 2.5% SDS], heated to 95°C for 5 min, and allowed to cool at room temperature for 10 min. Iodoacetamide (25 mM) was then added and samples were incubated for 30 min at room temperature to modify reduced cysteines. Samples were resolved by SDS-PAGE with a Novex 4-20% Tris-glycine polyacrylamide gel (Invitrogen, Carlsbad, CA). Proteins in the gel were visualized by Coomassie staining with GelCode Blue (Pierce Chemical, Rockford, IL).
Mass Spectrometric Identification of Protein Complex Components
Visible protein bands were excised from the Coomassie-stained gel and destained in 55% ammonium bicarbonate (100 mM)/45% acetonitrile with serial washes (4°C). Gel pieces were then dehydrated in acetonitrile, hydrated in 50 mM ammonium bicarbonate, and again dehydrated in acetonitrile. Proteins were digested in-gel with 75 ng of trypsin (Roche Diagnostics, Basel, Switzerland) per gel band in 50 mM ammonium bicarbonate for 6 h at 37°C. Tryptic peptides were extracted from the gel pieces with an 8-μl slurry of 1 volume of POROS R2 20 reverse-phase resin (Applied Biosystems, Foster City, CA) to 10 volumes of 5% formic acid/0.2% trifluoroacetic acid (TFA) at 4°C for ∼16 h. C18 Ziptips (Millipore, Billerica, MA) were conditioned with sequential washes of 0.1% TFA, 50% methanol/20% acetonitrile/0.1% TFA, and 0.1% TFA. POROS R2 20 resin was transferred to the conditioned Ziptips. The POROS R2 20 resin was washed in the Ziptip twice with 0.1% TFA. Tryptic peptides were eluted on the matrix-assisted laser desorption ionization (MALDI) target compact disk (Krutchinsky et al., 2001) with one-third saturated 2,5-dihydroxybenzoic acid (Lancaster Synthesis, Windham, NH) in 50% methanol:20% acetonitrile:0.1% TFA.
Tryptic peptides were identified by MALDI mass spectrometric analysis for protein identification (Beavis and Chait, 1996). Mass analysis of the tryptic peptides was performed with a MALDI-quadrupole-quadrupole-time-offlight mass spectrometer (Centaur Technology, Austin, TX; MDS Sciex, Concord, ON, Canada) modified with a compact disk sample stage as described previously (Krutchinsky et al., 2001). The highly accurate MALDI-mass spectrometry (MS) data was used to search the National Center for Biotechnology Center nonredundant protein database with the program ProFound to identify proteins from the tryptic peptide masses (Zhang and Chait, 2000). In addition to matching peptide masses to proteins, ProFound provides lists of peptide masses that can be used to confirm protein identifications by MS/MS analysis and lists of peptide masses that were not assigned to a protein in the MS analysis. To search for more proteins not identified in the MALDI-MS analysis and to confirm proteins identified in the single-stage MS analysis, amino acid sequence information for the tryptic peptides was obtained by MALDI-MS/MS fragmentation. Using the program M-IT sequencer, confirmation and unassigned masses obtained in the MALDI-quadrupole-quadrupole-time-of-flight analysis were used to prepare instrument files for MALDIMS/MS analysis (Krutchinsky et al., 2001). MALDI-MS/MS analysis of the same samples on the same MALDI target compact disk was performed using a modified LCQ Deca XP ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) as described previously (Krutchinsky et al., 2001). MALDI-MS/MS data was used to search the National Center for Biotechnology Information nonredundant protein database with the program Sonar (Genomic Solutions, Ann Arbor, MI) to identify proteins from the tryptic peptide fragmentation masses (Field et al., 2002).
Affinity Purifications and Western Blotting
For the experiments presented in Figure 1, B and C, a 50-ml culture of each strain was grown to mid-log phase. After measuring the OD600, cells were harvested from volumes around 50 ml that yielded similar cell masses. Cell pellets of ∼100 μl were washed in TE buffer and resuspended in 250 μl of extraction buffer (20 mM K-HEPES, pH 7.4, 110 mM K-acetate, 0.1% Tween 20, 1 mM dithiothreitol, 2 μg/ml DNaseI, 4 μg/ml pepstatin A, 0.2 mg/ml PMSF, 0.5% protease inhibitor cocktail (Sigma-Aldrich). Acid-washed glass beads (0.15 g) were added, and cell suspensions were vortexed for 5 min. Extraction buffer (500 μl) without DNaseI was added, and samples were vortexed for an additional 2 min. Samples were centrifuged for 30 s (tabletop Microfuge E; Beckman Coulter, Fullerton, CA), and the supernatants were removed. An additional 500-μl extraction buffer was added, and samples were vortexed for another 2 min. Samples were centrifuged for 30 s and the supernatants were combined. Pooled supernatants were centrifuged for 2 min. Of this final supernatant, 20 μl was kept for Western blotting analysis as “extract,” and the remainder was submitted to coaffinity purification. For this purpose, rabbit IgG-conjugated magnetic DynaBeads (Dynal) were added, and samples were incubated on a rotator at 4°C for 1 h. Beads were washed six times with 1 ml of extraction buffer without DNaseI. Proteins were eluted twice with 200 μl of 0.5 M NH4OH 0.5 mM EDTA for 5 min at room temperature. Eluates were pooled and lyophilized using a SpeedVac (Thermo Savant). Desiccated proteins were resuspended in 30 μl of SDS-PAGE sample buffer, and extract samples were dissolved in an equivalent volume of 2 × SDS-PAGE sample buffer. Samples were resolved by SDS-PAGE with a Novex 4-20% Tris-glycine polyacrylamide gel (Invitrogen, Carlsbad, CA) and transferred on nitrocellulose. Tenfold-diluted samples of the purification products were used for the gel destined to the anti-Protein A probing. Western blot analysis was performed, probing for the protein A moiety using a 1:1000 dilution of rabbit IgG (ICN) and probing for the Myc epitope using a 1:1000 dilution of a mouse monoclonal anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in Tris-buffered saline-Tween 20 5% milk (2-h incubation at room temperature). Secondary antibodies were horseradish peroxidase-coupled anti-rabbit and horseradish peroxidase-coupled anti-mouse, respectively, and both used as a 1:1000 dilution in Tris-buffered saline-Tween 20 5% milk (1-h incubation at room temperature). Chemiluminescence was used for the detection (Roche Diagnostics).
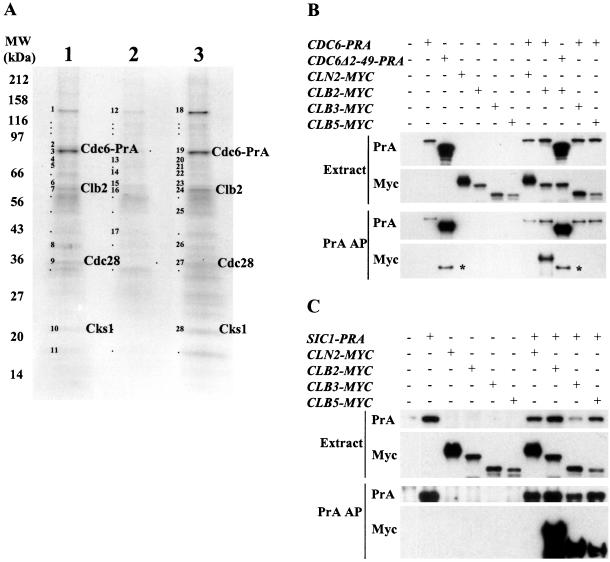
Figure 1.
Cdc6 associates with Clb2. (A) Cdc6 forms a complex with Clb2, Cdc28, and Cks1. Asynchronous CDC6-PRA cells (lane 1), α-factor–arrested CDC6-PRA bar1 cells (lane2) or glucose-arrested CDC6-PRA cdc20 GALL-CDC20 cells (lane 3) were submitted to a protein A affinity purification. Bands marked with either a dot or number were excised and analyzed by MALDI-mass spectrometry to identify proteins. Bands containing proteins identified with high confidence are numbered and the identifications are listed in the supplementary Table SI. Bands identified with a dot did not give a high-confidence identification. Proteins that seem to be specific to the Cdc6-PrA pullout are labeled. (B) Cdc6 associates with Clb2 and not with other cyclins and its N terminus is required for this association. CDC6-PRA or cdc6Δ2-49-PRA or CDC6-wt (untagged) cells also expressing CLN2-MYC, CLB2-MYC, CLB3-MYC, or CLB5-MYC were submitted to protein A affinity purification. Top, whole cell soluble extracts probed by Western blot for protein A and Myc. Bottom, purified proteins probed by Western blot for protein A and Myc. In the anti-MYC Western blot, the bands indicated by the asterisk (*) corresponded to the protein A tag from Cdc6Δ2-49-PrA that was recognized by the anti-MYC antibody due to its high abundance. (C) Sic1 associates with Clb2, Clb3, and Clb5 but not Cln2 (methods as in B).
Microscopy and Fluorescence-activated Cell Sorting (FACS)
Immunofluorescence directed against tubulin and 4,6-diamidino-2-phenyindole (DAPI) DNA staining were carried out as described previously (Jacobson et al., 2000). Fluorescence and differential interference contrast images were acquired on an Axioplan 2 microscope (Carl Zeiss, Thornwood, NY) with a 100 × 1.4 numerical aperture Planapochromat objective (Vermont Optechs, Charlotte, VT), fitted with a Hamamatsu Orca ER cooled charge-coupled device camera (Sciscope Instrument, Iowa City, IA) controlled by MetaVue software (Universal Imaging, Downingtown, PA). DNA content analysis by FACS was performed as described previously (Epstein and Cross, 1992).
RESULTS
Cdc6 Associates Specifically and Stably with Clb2
Cdc6 has been reported to associate with Cdc28 in vitro and in vivo through its amino terminus (Elsasser et al., 1996, 1999; Calzada et al., 2000). Association between Cdc6 and the Clb5 B-type cyclin has been reported in vitro (Elsasser et al., 1996). To explore potential associations of Cdc6 with cyclins in vivo and at endogenous expression levels, we used strains encoding a protein A fusions of Cdc6 expressed from the endogenous promoter. The Cdc6-PrA fusion protein was functional since CDC6-PRA cells grew normally. We affinity-purified Cdc6-PrA and analyzed the composition of the complex by SDS-PAGE and mass spectrometry (Figure 1A). We found that Cdc6 was degraded in an α-factor arrest, consistent with previous observations (Drury et al., 1997), and we were able to use the band pattern obtained in this condition as an indicator of which bands were contributed from contaminating proteins. All detectable bands were analyzed (supplementary information; Table SII). Strong bands that were observed from the purification done on asynchronous CDC6-PRA cells or metaphase-blocked CDC6-PRA cdc20 GALL-CDC20 cells and not observed from the purifi-cation done on α-factor–arrested CDC6-PRA bar1 cells were identified as Cdc6-PrA, Clb2, Cdc28, and Cks1. The similar staining intensity of the bands corresponding to the latter three proteins combined with the absence of other strong bands specific to this purification point at the existence of a complex between Cdc6 and a stoichiometric Clb2–Cdc28–Cks1 complex in vivo. All other proteins identified were common contaminants in this procedure, such as abundant ribosomal and metabolic proteins and chaperones (supplementary information; Table SII). Surprisingly, Cdc6-PrA did not copurify with significant amounts of other components of the pre-RC, such as MCM, Cdt1, and ORC complexes, even though these complexes were all extracted and effectively purified using the protein A tag under the same conditions (our unpublished observations).
Cyclins other than Clb2 could have been present in low amounts and could have eluded our mass spectrometric analysis. Therefore, we conducted coaffinity purifications by using strains that expressed Myc-tagged versions of four cyclins or protein A fusions of Cdc6 and Cdc6Δ2-49, or both, all expressed under the control of their endogenous promoters (Figure 1B). We used Cln2-Myc, Clb2-Myc, Clb3-Myc, and Clb5-Myc as a representative set, due to their functional significance (Cross et al., 2002). After protein A affinity purification, purified proteins were resolved on SDS-PAGE and probed for the Myc moiety by Western blotting. Of the four cyclins tested, only Clb2-Myc copurified with Cdc6-PrA, and this association was not observed when Cdc6Δ2-49-PrA was used as a bait, despite the higher abundance of this stabilized mutant protein. Therefore, Cdc6 associates specifically with Clb2 but does not stably associate with Cln2, Clb3, or Clb5 ex vivo, and this association is dependent on the amino terminus of Cdc6. For comparison, we performed the same analysis on cells coexpressing Sic1-PrA and the Myc-fusion cyclins (Figure 1C). We verified that Sic1-PrA could interact very strongly with all B-type cyclins tested (Clb2-Myc, Clb3-Myc, and Clb5-Myc) but not with the G1 cyclin Cln2-Myc. Therefore, the specificity of the cyclin–Cdc6 interaction was higher than the Sic1–cyclin interactions. These results contrast with those of Elsasser et al. (1996) who proposed that Clb5 could interact with Cdc6 based on in vitro binding data.
Cdc6 Abundance and Clb2 Binding through the Cell Cycle
Cdc6 should be present at a concentration similar to Clb2 in mitotic cells if it were to contribute significantly to mitotic exit through stoichiometric inhibition of Clb2–Cdk complexes. To test this idea, we used strains encoding protein A fusions of both Cdc6 and Clb2 expressed from their endogenous promoters. Using a strategy described elsewhere (Cross et al., 2002), we derived estimates of copy numbers per diploid cell in asynchronous cultures (Table 1). This analysis revealed that Cdc6 has an abundance greater than Sic1 and about two-thirds that of Clb2. Thus, Cdc6 could potentially contribute significant stoichiometric inhibition of Clb/Cdc28 kinase activity.
Table 1.
Quantitation of Cdc6-PrA, C1b2-PrA, and Sic1-PrA
| Copies/diploid cell (asynchronous)
|
|||
|---|---|---|---|
| Strain | Protein A-tagged protein | This study | Cross et al. (2002) |
| AL4 | Cdc6 | 624 ± 7 (3) | NA |
| AL4 | Sic1 | 195 ± 29 (3) | NA |
| MK3 | Clb2 | 1,006 ± 125 (3) | 1,128 ± 231 (4) |
| MK3 | Sic1 | 265 ± 29 (3) | 214 ± 42 (5) |
AL4, MAT a/α CDC6-PRA::HIS3MX/CDC6 SIC1-PRA::HIS3MX/SIC1; MK3, MAT a/α CLB2-PRA::HIS3MX/CLB2 SIC1-PRA::HIS3MX/SIC1.
Quantitations were performed on these heterozygous diploid strains as described in text; MK3 was one of the strains tested previously (Cross et al., 2002). Numbers are in estimated copies per cell in asynchronous diploid cells. Published numbers are provided for comparison. The numbers of independent cultures tested are in parentheses.
To act as an inhibitor of Clb2/Cdc28, Cdc6 must also be present at the same time in the cell cycle. Cdc6 has been reported to be present in late mitosis and G1 (Piatti et al., 1995; Drury et al., 1997). Clb2 levels rise in mitotic cells and decrease rapidly before mitotic exit. We examined the abundance patterns of Cdc6 and Clb2 through the cell cycle. We used CDC6-PRA cdc20 GALL-CDC20 cells that we arrested in metaphase by adding glucose. We released the cells by transfer to galactose medium and assessed the fluctuations of Cdc6-PrA and Clb2 by Western blotting (Figure 2A). Arrested cells displayed high levels of Clb2 and moderate levels of Cdc6-PrA. Cdc6-PrA reached a peak of abundance 20 min after release in galactose, whereas Clb2 disappeared quickly and was almost absent by 20 min. Cdc6-PrA persisted after cell division, reaching its lowest level after 70 min, in freshly budded cells, and then rose. Clb2 stayed at low levels until it reappeared at 70 min as cells budded. These results confirm that periods with high levels of Cdc6 overlap with the period during which Clb2 levels fall, consistent with a role of Cdc6 in Clb2-Cdk1 inhibition.
Figure 2.
Cdc6 temporally overlaps and associates with Clb2 in mitotic cells. (A) Timing of expression. CDC6-PRA (or cdc6Δ2-49-PRA, or SIC1-PRA) cdc20 GALL-CDC20 cells were grown in YPG, arrested in YPD for 3 h and released in YPG. Top, budding index (squares, CDC6-PRA cells; diamonds, cdc6Δ2-49-PRA cells; triangles, SIC1-PRA cells). Bottom, Western blot analysis of the cultures probing for protein A and Clb2. In cdc6Δ2-49-PRA cells, a breakdown fragment of Cdc6Δ2-49-PrA migrated at a molecular weight identical to Clb2, possibly accounting for the low signal observed for unbudded cells in the Clb2 blot because the protein A could be detected by the Clb2 antibody. (B) Cdc6 associates with Clb2 in mitotic cells. Right, a time course of protein A affinity purification was performed on CDC6-PRA cdc20 GALL-CDC20 cells grown in YPG, arrested in YPD for 3 h, and released in YPG. Samples were taken every 10 min after release for affinity purification. Left, controls for the affinity purification; all asynchronous cultures. Lane a, CDC6-PRA cells; lane b, CLB2-MYC cells; lane c, CDC6-PRA cells mixed with CLB2-MYC cells before cell breakage (control for postlysis binding); lane d, CDC6-PRA cdc20 GALL-CDC20 cells; and lane e, CDC6-PRA CLB2-MYC cdc20 GALL-CDC20 cells. Top, budding index; middle, whole cell soluble extracts probed by Western blot for protein A and Myc; bottom, purified proteins probed by Western blot for protein A and Myc.
We were interested to observe the accumulation of Cdc6 in the cdc20-blocked cultures, in which active Clb–Cdk1 complexes accumulate; in comparison, Sic1-PrA failed to accumulate detectably at this block (Figure 2A, t = 0). This observation suggests that “mode 3” proteolysis (Drury et al., 2000), described as controlling Cdc6 levels in cells expressing high Clb levels, is rather inefficient at removing Cdc6 at endogenous expression levels. Mode 3 proteolysis is considered not to require the Cdc6 N terminus, so the persistence of constitutive levels of Cdc6Δ2-49 throughout the cell cycle (Figure 2A) is also consistent with relatively low effectiveness of mode 3 in removing Cdc6 at endogenous expression levels.
To visualize the timing of occurrence of the Cdc6–Clb2 complex, we synchronized CDC6-PRA CLB2-MYC cdc20 GALL-CDC20 cells in metaphase by arresting them in glucose, releasing them in galactose, and then performing protein A affinity purifications on culture samples taken at different time points (Figure 2B). Cdc6-PrA associated with Clb2-Myc at essentially all times when Clb2-Myc was present. We did not attempt to calculate the proportion of total Clb2-Myc that remained stably bound to Cdc6-PrA through this procedure, although it was clearly only a minor fraction based on relative amounts loaded and exposure times used (our unpublished observations). It is hard to interpret this because of possible losses of bound Clb2-Myc during the extensive washes of Cdc6-PrA during the purification.
Moderate Overexpression of CDC6 Helps Cells Attain Mitotic Exit
Strongly overexpressed Cdc6 from the GAL1 promoter was shown previously to act genetically to inhibit Clb-Cdc28 function and delay entry in mitosis (Bueno and Russell, 1992; Basco et al., 1995; Perkins et al., 2001). Because we observed specific binding of Cdc6 to Clb2, we wanted to see whether a mitotic exit defect specifically due to Clb2 could be overcome by moderate Cdc6 overexpression. To do this, we examined the ability of low- and high-copy plasmids (∼1–3 copies and ∼10 copies/cell, respectively) containing CDC6 with its own promoter to rescue the lethal mitotic exit defect associated with removal of the Clb2 destruction box (CLB2Δdb) (Wäsch and Cross, 2002). CDC6 and SIC1 could both carry out significant rescue of CLB2Δdb lethality even at low copy, and rescued very efficiently at high copy (supplementary information; Figure S1). To increase the stringency of the assay, we also tested CLB2Δdb,ken, in which the KEN boxes of Clb2, recognized specifically by Cdh1, were also mutated. Rescue of this mutant required higher dosage of both CDC6 and SIC1, consistent with the idea that the low-copy rescue required activation of Cdh1-dependent proteolysis of Clb2Δdb. Notably, CDC6 was at least as efficient as SIC1 in these assays.
Rescue by CDC6 in these assays required the N-terminal 49 amino acids (Figure 3). The N-terminal phosphorylation sites were important for the rescue of CLB2Δdb,ken, but not CLB2Δdb. Removal of the candidate cyclin interaction sequence in amino acids 8–17 (Weinreich et al., 2001) had only moderate effects on rescue.
Figure 3.
Mutants expressing stabilized forms of Clb2 are rescued by CDC6 in an N terminus-dependent manner. CLB2Δdb GALSIC1(1 ×) or CLB2Δdb,ken GAL-SIC1 (2 ×) cells were transformed with plasmids expressing CDC6, cdc6Δ2-49, cdc6-3P-, and cdc6Δ8-17. Galactose-grown cultures were serially diluted (10-fold dilutions) on galactose- or glucose-containing rich medium and incubated for 2 d at 30°C.
Surprisingly, rescue was severely impaired by the K114E mutation, which blocks Cdc6 replicative function (Weinreich et al., 1999) but is outside the Cdk-interacting domain as defined previously (Weinreich et al., 2001) (Figure S2). This suggests that rescue of CLB2Δdb lethality may involve the replicative function of Cdc6 to some extent, as well as its Cdk-inhibitory ability. Perhaps replication origin licensing is somewhat inefficient under these conditions due to residual Clb activity during mitotic exit (Nguyen et al., 2001), yielding an increased requirement for Cdc6 replicative activity. In any case, these results confirm the idea that the Cdc6 Nterminus can contribute to negative regulation of Clb2-Cdk in mitotic exit.
Viability of sic1 Null Strains Lacking the Cdc6 Cdk Inhibitory Domain in the W303, S288C, and BF264-15D Strain Backgrounds
Based on their observation that combining deletion of SIC1 with deletion of the N terminus of Cdc6 was synthetic lethal, Calzada et al. 2001) made the important suggestion that Cdk inhibition by stoichiometric binding inhibitors was required for mitotic exit. However, we found that it was possible to construct this double mutant. In three different genetic backgrounds, viable sic1Δ cdc6Δ2-49 cells could be generated (Figure 4).
Figure 4.
cdc6Δ2-49 sic1 mutants are viable. (A) In the W303 background, a deletion of coding sequence for amino acids 2–49 was introduced in the chromosome with no associated marker, as described in the text and Figure S3. Left, sic1::HIS3 cdc6Δ2-49 pURA3-SIC1 strains were constructed by tetrad analysis, and loss of the pSIC1-URA3 plasmid selected on 5-FOA. Strains with and without the plasmid were tested for the CDC6 N-terminal deletion and for the sic1::HIS3 mutation by PCR, and for growth rate by serial dilution at 30 and 37°. Right, segregants from a cross with cdh1::LEU2, sic1::HIS3, and cdc6Δ2-49 segregating were tested by PCR. The second PCR analysis tests specifically for the presence of the coding sequence for amino acids 2-49. The strains were tested for growth rate as in the left panel. (B) In the S288C background (Figure 4B), we made a strain that was cdc6Δ::kanMX sic1Δ::kanMX pURA3-CDC6, and a control cdc6Δ::kanMX SIC1 pURA3-CDC6 strain (complete coding sequence deletions). All kanMX disruptions were confirmed by PCR. We transformed these strains with lowcopy (CEN) plasmids encoding wild-type and the three mutant CDC6 genes, expressed from the CDC6 promoter (Weinreich et al., 2001). All transformants, except for the vector controls could readily lose the URA3/CDC6 plasmid. (C) BF264-15D-congenic background: We backcrossed cdc6Δ2-49 from W303 (see A) into the BF264-15D congenic strain background (see text). Isogenic pairs of sic1Δ pURA3-SIC1 segregants (cdc6Δ2-49 or CDC6) from the fifth backcross were selected for pURA3-SIC1 plasmid loss by FOA and tested for growth rate by serial dilution at 30 and 37°. PCR characterization of the strains was performed as in A.
In the W303 background (Figure 4A), we made precise gene replacements of CDC6 with cdc6Δ2-49, cdc6-3P- (lacking three Cdk sites in the N terminal 50 amino acids of Cdc6) and cdc6Δ8-17, all of which were reported to be defective in Cdk binding and mitotic regulation (Weinreich et al., 2001). The allele construction is described in the supplementary information (Figure S3). These CDC6 alleles are exact chromosomal gene replacements without associated marker sequences, unlike the cdc6Δ2-47 allele constructed in Calzada et al. (2001), in which a URA3 gene was inserted 3′ to CDC6, disrupting the neighboring YJL193W open reading frame (Calzada et al., 2001).
We could readily combine all the cdc6 alleles (cdc6Δ2-49, cdc6-3P-, cdc6Δ8-17, and cdc6Δ2-47::loxP) with a sic1::HIS3 deletion allele, in standard tetrad analysis. In most experiments, we had a CEN-SIC1/URA3 plasmid segregating, so that we could recover all genotypes (even if SIC1-requiring) and test for a SIC1 requirement by 5-fluoroorotic acid (5-FOA) resistance (selecting plasmid loss events). In 4:0 Ura+: Ura- tetrads from crosses with sic1::HIS3 homozygous and cdc6Δ2-49 segregating, FOA-resistance segregated 4:0 (i.e., cdc6Δ2-49 sic1Δ strains did not require the SIC1 plasmid for viability). In crosses where cdh1::LEU2 was also segregating, we observed 2:2 FOA-S Leu+:FOA-R Leu- segregation, independent of cdc6 genotype (i.e., we could detect a SIC1 requirement in cdh1Δ strains, in the same experiment, providing an internal control). Identical results were obtained using the cdc6-3P-, cdc6Δ8-17, and cdc6Δ2-47::loxP alleles (our unpublished observations). We carried out this entire analysis in W303 with independent duplicates of all gene replacements, starting from independent cdc6Δ2-47::loxP-URA3-loxP insertions, with identical results.
We could also recover sic1::HIS3 cdc6Δ2-49 segregants without the SIC1/URA3 plasmid, thus without the use of 5-FOA selection. Among such Ura- segregants, 9 of 22 sic1::HIS3 CDH1, 6 of 10 SIC1 cdh1::LEU2, and 7 of 12 SIC1 CDH1 segregants contained the cdc6Δ2-49 mutation. sic1::HIS3 cdc6Δ2-49 segregants were viable, and displayed only a slight growth defect at 37°C. In cultures grown at 30°C, we observed no excess accumulation of budded binucleate cells with long spindles (the reported lethal phenotype for sic1Δ cdc6Δ2-47) (Table 2; our unpublished observations). Accumulation of long spindles monitored using green fluorescent protein (GFP)-labeled tubulin was similar in these two backgrounds (in one experiment, 9% of a cycling log phase sic1Δ CDC6 strain and 12% of a cycling sic1Δ cdc6Δ2-49 strain had long spindles). Similar results were obtained with sic1Δ CDC6 GAL-SIC1 or sic1Δ cdc6Δ2-49 GAL-SIC1 strains shifted to glucose medium to deplete Sic1 (our unpublished observations). These results contrast with the 100% block as binucleate cells with long spindles reported for sic1Δ cdc6Δ2-47 (Calzada et al., 2001).
Table 2.
Test of sic1Δ cdc6Δ2-49 strains for delay in exit from mitosis
| Strain | Medium | % Unbudded cells | % Binucleated cells |
|---|---|---|---|
| sic1Δ GAL-SIC1 | Gal | 31 | 17 |
| sic1Δ cdc6Δ2-49 GAL-SIC1 | Gal | 22 | 16 |
| sic1Δ | Gal | 31 | 17 |
| sic1Δ cdc6Δ2-49 | Gal | 27 | 19 |
| sic1Δ GAL-SIC1 | Gal → Glu (2.5 h) | 26 | 16 |
| sic1Δ cdc6Δ2-49 GAL-SIC1 | Gal → Glu (2.5 h) | 28 | 21 |
| sic1Δ | Gal → Glu (2.5 h) | 11 | 13 |
| sic1Δ cdc6Δ2-49 | Gal → Glu (2.5 h) | 13 | 19 |
Strains of the indicated genotype were grown in the indicated medium and shifted from YEPGal into YEPD (Gal → Glu) for 2.5 h (long enough to quantitatively deplete Sic1 in GAL-SIC1 strains; Wäsch and Cross, 2002). Note: the reported phenotype of mitotic exit defect in sic1Δ cdc6Δ2-47 strains was ∼100% budded binucleate cells (Calzada et al., 2001).
In the S288C background (Figure 4B), we used a plasmidshuffle strategy to establish that cdc6Δ::kanMX sic1Δ::kanMX strains were viable with the three mutant CDC6 genes, expressed from the CDC6 promoter (Weinreich et al., 2001) as their sole source of CDC6. The mutant transformants grew well, and did not accumulate binucleate cells (our unpublished observations); thus, they did not exhibit a significant delay in mitotic exit. This result extends the lack of requirement for Sic1 or the Cdc6 N terminus to another strain background.
We performed six generations of backcross of cdc6Δ2-49 from W303 into the BF264-15D strain background (Figure 4C). In each generation, the cdc6Δ2-49 strain was either SIC1 or else carried pURA3-SIC1, so that selection of modifiers affecting viability or vigor of sic1Δ cdc6Δ2-49 strains should be avoided. In most backcrosses, the backcrossed cdc6Δ2-49 strain was also cdh1Δ, allowing an internal control for detection of a Sic1 requirement due to sic1Δ cdh1Δ spore inviability, as described above. In every generation of backcross, we could readily recover viable cdc6Δ2-49 sic1Δ segregants, whereas viable sic1Δ cdh1Δ segregants were not recovered. In third, forth, fifth, and sixth generation backcrosses to BF264-15D, sic1Δ segregants lacking the pURA3-SIC1 plasmid were tested by PCR, and 5 of 7, 6 of 16, 3 of 3, and 4 of 9 were cdc6Δ2-49. Note that these segregants were haploids that lacked the pURA3/SIC1 plasmid from their initial meiotic generation, thus this was an unbiased and unselected sample. These results rule out an unlinked modifier from W303, allowing viability of the double mutant (P << 0.01), and show that even among strains 98% genetically identical to BF264-15D, cdc6Δ2-49 sic1Δ segregants were easily recovered. A W303-specific modifier linked to SIC1 is ruled out by the backcrossing strategy, because in some crosses the sic1 allele was from a BF264-15D parent. A W303-specific modifier linked to CDC6 remains a formal possibility to explain the discrepancy with results in Calzada et al. (2001). After the fifth backcross, we compared isogenic pairs of sic1Δ pURA3-SIC1 segregants (cdc6Δ2-49 or CDC6) before and after selection of loss of the pURA3-SIC1 plasmid by 5-FOA selection. We observed no significant growth defect for any genotype.
The strains described in Calzada et al. (2001) all contained hemagglutinin (HA)-epitope-tagged CLB2, to facilitate biochemical analysis. We investigated the possibility that the CLB2-HA allele could cause the discrepancy between our results. In the W303 background, we were able to recover viable CLB2-HA cdc6Δ2-49 sic1Δ spores, at the expected Mendelian frequency, that generated normal-size colonies. Of 32 sic1::HIS3 segregants tested by PCR, eight were cdc6Δ2-49 CLB2-HA, nine were CDC6 CLB2-HA, seven were cdc6Δ2-49 CLB2, and eight were CDC6 CLB2. A sic1Δ cdc6Δ2-49 CLB2-HA strain was backcrossed twice in two independent lines to a sic1Δ BF264-15D strain. The results provided no evidence for inviability or any significant partial mitotic exit defect of sic1Δ cdc6Δ2-49, with either CLB2-HA or CLB2-wt (our unpublished observations). The hypothesis that a single-gene suppressor specific to BF264-15D provided inviability of cdc6Δ2-49 sic1Δ CLB2-HA was not supported (P << 0.05; our unpublished observations). Therefore, it is unlikely that the inclusion of HA-tagged Clb2 in the experiments of Calzada et al. (2001) are responsible the discrepancy between their results and ours. This discrepancy remains unexplained at present.
Removal of the Cdc6 N Terminus Confers a Cytokinesis Defect in sic1Δ cdh1Δ Cells
Despite the fact that the sic1Δ cdc6Δ2-49 mutant was viable, in contradiction to the results of Calzada et al. (2001), the ability of Cdc6 to rescue CLB2Δdb mitotic exit defects suggest that Cdc6 has the potential to help mitotic exit. We therefore examined more closely the possibility that the Cdc6 N terminus might cooperate with Sic1 under some circumstances. Sic1 is thought to cooperate with Cdh1 in regulating Clb-Cdk activity (see INTRODUCTION), so we asked what would happen in the absence of all three potential regulators.
sic1Δ cdh1Δ GAL-SIC1 strains proliferate slowly on glucose medium, forming small colonies with >10% plating efficiency (Wäsch and Cross, 2002); this limited proliferation is dependent on the Cdc6 N terminus (Figure 5; Cross, 2003). As shown above, cdh1Δ cdc6Δ2-49 and sic1Δ cdc6Δ2-49 double mutants have no significant proliferation defect, so the strong inviability of the triple mutant represents functional overlap among all three genes.
Figure 5.
Removal of the Cdc6 N terminus reduces viability of sic1Δ cdh1Δ cells. Galactose-grown cultures were serially diluted (10-fold dilutions) on galactose- or glucose-containing rich medium and incubated for 3 d at 30°C. All strains contained the TUB1-GFP allele (used in Figure 6).
To investigate the basis for the enhanced lethality of this triple mutant, we examined the morphology of sic1Δ cdh1Δ with or without the cdc6Δ2-49 mutation (Figure 6). We used GAL-SIC1 to allow the culture of these cells in galactose and their subsequent arrest in glucose. Phase contrast microscopy images, Tub1-GFP fluorescence images, and FACS measurements allowed the visualization of cell morphology, spindle morphology, and DNA content of various mutants cultured in galactose or glucose (Figure 6A). In galactose, only minor differences were observed among the GAL-SIC1–containing strains (Figure 6A, left).
Figure 6.
Removal of the Cdc6 N terminus confers a cytokinesis defect in sic1Δ cdh1Δ cells. (A) Cells with the indicated genotypes (the same strains as in Figure 5) were cultured in galactose-containing medium (left) and then transferred to glucose-containing medium for 4 h (right). Differential interference contrast (DIC) pictures, Tub1-GFP fluorescence pictures, and FACS profiles are shown for each condition. Brackets in the lower DIC picture indicate the four-cell body objects that were resistant to sonication (see text). (B) Cells with the indicated genotypes (wild-type TUB1) were cultured in galactose-containing medium and then transferred to glucose-containing medium. FACS profiles were taken after 2.5 and 5 h. Cells taken at 2.5 h were fixed and immunofluorescence was used to visualize spindles in DAPI-stained cells; long spindles with separated nuclei (anaphase spindles) were counted.
sic1Δ cells showed no clear peak of cells with 1C DNA content, consistent with the role of SIC1 in preventing DNA replication in early G1 cells by inhibiting Cdk activity (Lengronne and Schwob, 2002). sic1Δ cdc6Δ2-49 GAL-SIC1 cells (Figure 6A, fourth row) grew well in glucose and seemed identical to sic1Δ cells (Figure 6A, first row). sic1Δ cdh1Δ GAL-SIC1 cells (Figure 6A, fifth row) proliferated in galactose with a somewhat reduced G1 population in comparison to the sic1Δ GAL-SIC1 cells (Figure 6A, first row). However, in glucose, sic1Δ cdh1Δ GAL-SIC1 cells (Figure 6A, fifth row) arrested mostly as large-budded cells with replicated DNA and generally short spindles, consistent with previous observations (Wäsch and Cross, 2002). The additional removal of the Cdc6 N terminus had a dramatic effect in this context. In glucose, sic1Δ cdh1Δ cdc6Δ2-49 GAL-SIC1 cells (Figure 6A, sixth row) reproducibly accumulated as “four-cell body objects” that were resistant to sonication. These objects always had two spindles, one spreading across “cell body 1” and “cell body 2” and one spreading across “cell body 3” and “cell body 4.” As was observed with the sic1Δ cdh1Δ GALSIC1 cells in glucose, the spindles were generally short. Consistent with these four-cell body objects, we observed a marked increase in frequency of cells with 4C DNA content.
We used immunofluorescence directed to tubulin and DAPI staining to visualize the nuclei (Figure 6B). For both sic1Δ cdh1Δ cdc6Δ2-49 GAL-SIC1 cells and sic1Δ cdh1Δ GALSIC1 cells transferred to glucose medium both generally had one short spindle per nucleus. In both cases, this is inconsistent with a mitotic exit defect, because cells did not accumulate as binucleates with long spindles. These results are consistent with previous findings on sic1Δ cdh1Δ cells (Wäsch and Cross, 2002). The simplest explanation of these observations is that the additional removal of the Cdc6 N terminus confers a cytokinesis defect to cells lacking SIC1 and CDH1. This defective cytokinesis could account for the enhancement of lethality of sic1Δ cdh1Δ by cdc6Δ2-49 (Figure 5). The accumulation of 4C DNA content cells suggests that even when cytokinesis is defective, DNA replication in the next cell cycle is not significantly impaired. This is surprising, considering the known role of Clb activity in blocking origin licensing (Nguyen et al., 2001).
CDC6 Overexpression Rescues sic1Δ cdh1Δ Cells
The apparent overlap in function between SIC1, CDH1, and CDC6 led us to test whether increased CDC6 gene dosage could bypass the requirement for either CDH1 or SIC1 for viability. We found that extra CDC6 rescued the reduced viability of sic1Δ cdh1Δ cells (Figure 7). Deletion of Cdc6 amino acids 2–49 or mutation of the three Cdk phosphorylation sites within amino acids 2–49 severely reduced the ability of CDC6 to rescue sic1Δ cdh1Δ cells. Deletion of amino acids 8–17 had a lesser but detectable effect (our unpublished observations). These results are consistent with the idea that Sic1, Cdh1, and the Cdk inhibitory domain in the Cdc6 N terminus have overlapping functions.
Figure 7.
CDC6 rescues sic1Δ cdh1Δ cells. A cdh1Δ sic1Δ strain carrying a pSIC1-URA3 plasmid was transformed with LEU2 plasmids, either vector (RS415, low-copy, or RS425, high-copy), RS415 containing CDH1 as a positive control, or RS425 carrying wt and mutant CDC6. (A) Transformants were patched on -leu and then replica-plated to -ura (indicating retention of the pSIC1-URA3 plasmid) or FOA (indicating ability to lose this plasmid). Note that one RS425-CDC6 transformant spontaneously lost the pSIC1-URA3 plasmid. (B) Serial dilutions of transformants on -leu or FOA.
swi5Δ cdh1Δ Inviability and Its Rescue by CDC6
The results mentioned above demonstrate that Cdc6 has the potential to act as a coinhibitor of Clb-Cdc28 with Sic1. It is interesting that both these proteins are controlled by the Swi5 transcription factor, resulting in a burst of transcription for both at the M/G1 border (Piatti et al., 1995; Knapp et al., 1996; Toyn et al., 1997). However, Swi5 is not essential, suggesting that other mechanisms may provide sufficient Clb-Cdc28 inactivation in its absence. We tested whether Cdh1 could be the relevant activity, and we found that indeed, swi5Δ cdh1Δ double mutants were inviable in tetrad analysis (our unpublished observations), whereas both single mutants were fully viable. Lethality could be rescued by SIC1 overexpression from the GAL1 promoter (Figure 8A). We used this to determine the lethal phenotype of swi5Δ cdh1Δ GAL-SIC1 by turning off the GAL1 promoter with glucose medium. This mutant did not display a rapid specific cell cycle arrest; rather, cells went through a variable number of divisions before ultimately becoming inviable (that is, unable to form colonies even when plated on galactose medium) with mostly replicated DNA (Figure 8B, second row).
Figure 8.
CDH1, SWI5, and CDC6 interact genetically. (A) cdh1Δ swi5Δ mutants are inviable and can be rescued by SIC1 and by CDC6. Rescue by CDC6 depends on both its N terminus and its DNA-replication activity. cdh1Δ swi5Δ GAL-SIC1 cells were transformed with plasmids expressing SIC1, CDC6, cdc6Δ2-49, cdc6-3P-, cdc6Δ8-17, and cdc6-K114E. Galactose-grown cultures were serially diluted (10-fold dilutions) on galactose-or glucose-containing rich medium and incubated for 2 d at 30°C. (B) Cells with the indicated genotype were grown in galactose-containing medium and transferred to glucose-containing medium for 3 h. FACS profiles are shown, with the percentage of binucleated cells in the top right corner.
This non-Cdc phenotype is difficult to analyze in detail, but the lethality of the double mutant was nevertheless tight enough for us to test the ability of wild-type and mutant CDC6 and SIC1 to rescue at varying copy number (Figure 8A). Rescue was quite efficient even with low-copy-number plasmids containing CDC6 or SIC1. Rescue required the Cdc6 N terminus; unlike the results with CLB2Δdb (see above), the N-terminal phosphorylation sites and the candidate cyclin-binding sequence (amino acids 8–17) were both strongly required for rescue. In addition, the K114E mutation strongly reduced rescue, suggesting that both the Cdk inhibition and the DNA replication licensing function of Cdc6 were involved. As with rescue of CLB2Δdb lethality (see above), this could suggest that inefficient replication origin licensing under these conditions results in an increased requirement for Cdc6 replicative activity.
The swi5Δ cdh1Δ cells accumulated as large-budded cells with a short spindle (our unpublished observations), therefore closely resembling sic1Δ cdh1Δ cells (Wäsch and Cross, 2002). We tested the effect of removing the Cdc6 N terminus on the swi5Δ cdh1Δ mutant. cdc6Δ2-49 swi5Δ cdh1Δ GALSIC1 cells were viable in galactose but arrested in glucose with a marked increase in 4C DNA (Figure 8B, third row). Like cdc6Δ2-49 sic1Δ cdh1Δ GAL-SIC1 cells, cdc6Δ2-49 swi5Δ cdh1Δ GAL-SIC1 cells transferred to glucose medium accumulated as four-cell body objects with generally short spindles (our unpublished observations). Thus, although diffi-cult to interpret in terms of a specific cell cycle requirement, the results with swi5Δ cdh1Δ lethality support the idea that Cdc6, Sic1 and Cdh1 can cooperate to regulate Clb-Cdc28 activity and the effect of the loss of all three activities may be a failure of cytokinesis.
Functional Overlap between the Cdc6 N Terminus, Sic1, and the Mitotic Exit Network
Cdc14 and Cdc15 are part of the mitotic exit network (Bardin and Amon, 2001). Suggestive of some degree of involvement of the Cdc6 N terminus in mitotic exit, we found that cdc6Δ2-49 acts as a weak enhancer of the cdc14-1 (Figure 9A) or cdc15-2 (Figure 9B) temperature-sensitive mutations, although sic1Δ acts as a considerably stronger enhancer. Reciprocally, high-copy CDC6 but not cdc6Δ2-49 weakly suppresses cdc14-1 at semipermissive temperature (Figure 9C). This suppression is much weaker than that by high-copy SIC1. It is also notable that some of the enhancing effects of cdc6Δ2-49 on cdc14-1 occur even under conditions of SIC1 overexpression (Figure 9A, see cdc6Δ2-49 strains on YEPGal plates, at 30°C), suggesting that the Cdc6 N terminus could have a role in this context that cannot be provided by extra-Sic1. Thus, the Cdc6 N terminus may have the potential to cooperate with the mitotic exit network or with Sic1 or with both, but this is only detectable in highly compromised backgrounds (in the absence of Cdh1, or upon partial inactivation of Cdc14 or Cdc15).
Figure 9.
Genetic interaction of cdc6Δ2-49 and sic1Δ with the mitotic exit network. (A) Interaction with cdc14-1. A W303-congenic cdc14-1 strain was constructed by repeated backcrossing and tetrad analysis, and a cdc6Δ2-49/+ sic1Δ/+ cdc14-1/+ GAL-SIC1::TRP1/trp1 diploid was constructed. Haploid segregants of the indicated genotype (all GAL-SIC1 cdc14-1) were tested for growth by serial dilution and incubation at the indicated temperature, on galactose (GAL-SIC1 on) or glucose (GAL-SIC1 off). (B) Interaction with cdc15-2. Methods as in A. (C) The indicated plasmids were introduced into a cdc14-1 strain and replica-plated to plates that were incubated at the indicated temperatures.
DISCUSSION
The possibility of the involvement of Cdc6 in mitotic exit through Cdk inhibition intrigued us for two main reasons. First, it would mean that a protein required for DNA replication would find a second function in another major cellular event, distant both in nature and in time. Second, it would reinforce the idea that Cdk inactivation, through stoichiometric inhibition, is used by the cell as a significant mechanism to achieve mitotic exit. If true, this would contrast with other eukaryotes, for which APC-dependent degradation of B-type cyclins accounts for most or all Cdk inactivation at mitotic exit.
In this work, we show that Cdc6 fills a number of biochemical features consistent with its ability to act as a Clb2-Cdc28 inhibitor contributing to mitotic exit. We also show functional evidence for this role in crippled genetic backgrounds. However, we prove that the function of Cdc6 as Cdk inhibitor is dispensable, even in the absence of Sic1, contradicting a previous report (Calzada et al., 2001).
Are the Amounts of Cdc6 Enough to Inhibit Clb2?
Our biochemical observations clearly show that Cdc6 interacts with Clb2 in vivo. This is the first set of observations on Cdc6–cyclin interactions made under conditions in which no protein is overexpressed. Quantitative measurements showed that Cdc6 and Clb2 had similar abundances in asynchronous cultures, with Clb2 being in slight excess over Cdc6. We also observed that Clb2 oscillates with higher amplitude than Cdc6 and peaks in budded cells. Therefore, it is reasonable to conclude that Clb2 alone (without counting other mitotic Clbs) exceeds Cdc6 in abundance for most of the time when cells are budded. But if Cdc6 exceeded mitotic Clbs in abundance, sufficient inhibition could potentially be achieved to block mitosis. This is likely to occur when Cdc6 is overexpressed (Bueno and Russell, 1992; Basco et al., 1995; Perkins et al., 2001).
It has been suggested that Cdc6 at normal expression levels is required to prevent mitosis if DNA has not replicated (“reductional mitosis”) (Piatti et al., 1995; Weinreich et al., 2001). A novel G1/M checkpoint pathway has been suggested based on the observation that cdc7 and dbf4 temperature-sensitive mutants undergo reductional mitosis at the restrictive temperature in some genetic backgrounds, in a way that is independent of the known checkpoint pathways (Toyn et al., 1995). Such a pathway could require Cdc6 (Weinreich et al., 2001). However, it is unclear, based on our quantitations, whether there is enough Cdc6 to account for preventing reductional mitosis if Cdc6 acts a simple stoichiometric inhibitor.
Importantly, at endogenous expression levels, as Clb2 levels fall due to APC activation during mitosis, our quantitation and determination of cell cycle patterns of accumulation (Table 1 and Figure 2) clearly imply that Cdc6 levels will exceed Clb2 levels, allowing the potential for effective inhibition contributing to mitotic exit. In contrast, Sic1 may not accumulate to significant levels until Clb2 degradation is almost complete (Figure 2), so the primary biological role of Sic1 may be in Clb inhibition in G1, as suggested previously (Wäsch and Cross, 2002; Lengronne and Schwob, 2002).
Although we observe specific Cdc6–Clb2 interaction, the primary function of this interaction may not be inhibition of Clb2-associated kinase activity. It is possible that this interaction is involved in other processes, such as inhibition of prereplicative complex formation by Clb2 (Nguyen et al., 2001). Further work will be required to evaluate these possibilities.
How Much Cdc6 Is Needed for Replication?
Our quantitation of Cdc6 is relevant to the molecular mechanisms responsible for licensing DNA replication. A recent estimate for the number of functional origins of DNA replication gave a number ∼300–400 in haploids (Wyrick et al., 2001). Our estimate of 600–650 for the number of molecules of Cdc6 per diploid cell is in the same narrow range. Therefore, it is possible that only one molecule of Cdc6 is used at each functional origin.
Possible Role for Cdk Inhibition by Cdc6 in Mitotic Exit
Overexpression of the well-characterized Clb-Cdk inhibitor SIC1 can rescue mutants defective in mitotic completion such as temperature-sensitive mutants of the DBF2 mitotic kinase (Donovan et al., 1994) and mutants of CLB2 that fail to degrade Clb2 due to resistance to the APC-proteasome pathway (Wäsch and Cross, 2002). These results suggested that SIC1 has the potential to promote mitotic exit by lowering mitotic Clb-Cdk activity. In this study, we tested CDC6 for a similar functional potential using the CLB2Δdb and CLB2Δdb,ken strains that produce Clb2 proteins that are refractory to APC-mediated degradation. We found that CDC6, like SIC1, could rescue these mutants, implying that these two Cdk inhibitors have the potential to help mitotic exit through Cdk inhibition.
In addition to testing the ability of CDC6 to rescue mutants that are defective in mitotic exit, we used a complementary strategy to test the involvement of CDC6 in mitotic exit. This strategy consisted in testing the effect of deleting the N terminus of Cdc6 in a mutant that barely reaches low enough Cdk activity to exit mitosis, the cdh1Δ sic1Δ mutant (Wäsch and Cross, 2002). The inviability of the resulting triple mutant could be explained by a failure in cytokinesis; surprisingly, DNA replication in the succeeding cell cycle was not significantly defective. Consistent with these observations, the reverse experiment showed that extracopies of CDC6 but not cdc6Δ2-49 completely rescued the inviability of cdh1Δ sic1Δ cells. We also found that the double mutant cdh1Δ swi5Δ showed a striking reduction in viability, which is also rescued by moderate Sic1 or Cdc6 overexpression. This is consistent with the known role of Swi5 as a transcription factor that activates the expression of both SIC1 and CDC6 (see INTRODUCTION). In addition, we observed that deletion of the Cdc6 N terminus caused a defect in cytokinesis in the cdh1Δ swi5Δ mutant, as with the cdh1Δ sic1Δ cdc6Δ2-49 mutant.
Another line of evidence pointed at a possible involvement of Cdc6 in mitotic exit, in the context of mutations in the mitotic exit network. We observed an enhanced lethality of cdc14-1 and cdc15-2 temperature-sensitive mutants by deletion of the Cdc6 N terminus and a weak suppression of cdc14-1 by high-copy CDC6 but not cdc6Δ2-49 at a semipermissive temperature.
Overall, these experiments support an ancillary role for the Cdc6 N terminus in regulating mitotic exit.
Stoichiometric Cdk Inhibitors Are Not Required for Mitotic Exit
Despite the clear potential for the Cdc6 N terminus and Sic1 to function in mitotic exit, we found that the Cdk-inhibitory functions of these genes could be readily removed alone or in combination with no obvious defect in mitotic exit, in three different genetic backgrounds. This clearly contradicts the result of Calzada et al. (2001) and demonstrates that Clb-Cdk stoichiometric inhibition by the known inhibitors is not essential for mitotic exit. Because detecting an effect on mitotic exit of removing the Cdc6 N terminus requires highly compromised genetic backgrounds (cdh1Δ sic1Δ, cdh1Δ swi5Δ, cdc14ts, and cdc15ts), we conclude that this function of Cdc6 does not contribute significantly to mitotic exit in a wild-type background. The proposal of Calzada et al. (2001) represented the first example of essential Cdk control by stoichiometrically binding inhibitors. Because inhibition by Sic1 and Cdc6 is dispensable, as we demonstrated in this study, cyclin proteolysis dependent on Cdc20 and Cdh1 may be the central and evolutionarily conserved Cdk-inactivating event regulating mitotic exit (Wäsch and Cross, 2002).
Supplementary Material
Acknowledgments
We thank Lea Schroeder for technica1 assistance in performing the experiment in Table 2 and to Alison North for help with microscopy. We also thank A. Murray, E. Schiebel, S. Oliver, D. Stillman, and M. Weinreich for strains and plasmids; and A. Bueno, the members of the Rout laboratory, and the Chait laboratory for useful discussions. Funding was provided by Public Health Service grants GM-47238 and CA-89810, and by Defense Advanced Research Projects Agency F30602-01-2-0572.
Online version of this article contains supplemental data for some figures. Online version is available at www.molbiolcell.org.
References
- Aitchison, J.D., Blobel, G., and Rout, M.P. (1996). Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274, 624-627. [DOI] [PubMed] [Google Scholar]
- Bardin, A.J., and Amon, A. (2001). Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2, 815-826. [DOI] [PubMed] [Google Scholar]
- Basco, R.D., Segal, M.D., and Reed, S.I. (1995). Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol. Cell Biol. 15, 5030-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis, R.C., and Chait, B.T. (1996). Matrix-assisted laser desorption ionization mass-spectrometry of proteins. Methods Enzymol. 270, 519-551. [DOI] [PubMed] [Google Scholar]
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- Bueno, A., and Russell, P. (1992). Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11, 2167-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada, A., Sacristan, M., Sanchez, E., and Bueno, A. (2001). Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature 412, 355-358. [DOI] [PubMed] [Google Scholar]
- Calzada, A., Sanchez, M., Sanchez, E., and Bueno, A. (2000). The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275, 9734-9741. [DOI] [PubMed] [Google Scholar]
- Cross, F.R. (2003). Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell 4, 741-752. [DOI] [PubMed] [Google Scholar]
- Cross, F.R., Archambault, V., Miller, M., and Klovstad, M. (2002). Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 13, 52-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, J.D., Toyn, J.H., Johnson, A.L., and Johnston, L.H. (1994). P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 8, 1640-1653. [DOI] [PubMed] [Google Scholar]
- Drury, L.S., Perkins, G., and Diffley, J.F. (1997). The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L.S., Perkins, G., and Diffley, J.F. (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231-240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Chi, Y., Yang, P., and Campbell, J.L. (1999). Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser, S., Lou, F., Wang, B., Campbell, J.L., and Jong, A. (1996). Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell 7, 1723-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, C.B., and Cross, F.R. (1992). CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6, 1695-1706. [DOI] [PubMed] [Google Scholar]
- Evans, T., Rosenthal, E.T., Youngblom, J., Distel, D., and Hunt, T. (1983). Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33, 389-396. [DOI] [PubMed] [Google Scholar]
- Field, H.I., Fenyo, D., and Beavis, R.C. (2002). RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimises protein identification, and archives data in a relational database. Proteomics 2, 36-47. [PubMed] [Google Scholar]
- Jacobson, M.D., Gray, S., Yuste-Rojas, M., and Cross, F.R. (2000). Testing cyclin specificity in the exit from mitosis. Mol. Cell Biol. 20, 4483-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, D., Bhoite, L., Stillman, D.J., and Nasmyth, K. (1996). The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell Biol. 16, 5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Krutchinsky, A.N., Kalkum, M., and Chait, B.T. (2001). Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal. Chem. 73, 5066-5077. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., and Schwob, E. (2002). The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9, 1067-1078. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O., and Roberts, J.M. (2002). Oscillation sensation. Nature 418, 495-496. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (1996). At the heart of the budding yeast cell cycle. Trends Genet. 12, 405-412. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.Q., Co, C., and Li, J.J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068-1073. [DOI] [PubMed] [Google Scholar]
- Perkins, G., Drury, L.S., and Diffley, J.F. (2001). Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20, 4836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, S., Lengauer, C., and Nasmyth, K. (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a 'reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Blobel, G., and Aitchison, J.D. (1997). A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715-725. [DOI] [PubMed] [Google Scholar]
- Schultz, M.C. (1999). Chromatin assembly in yeast cell-free extracts. Methods 17, 161-172. [DOI] [PubMed] [Google Scholar]
- Stern, B., and Nurse, P. (1996). A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12, 345-350. [PubMed] [Google Scholar]
- Toyn, J.H., Johnson, A.L., Donovan, J.D., Toone, W.M., and Johnston, L.H. (1997). The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics 145, 85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn, J.H., Johnson, A.L., and Johnston, L.H. (1995). Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol. Cell Biol. 15, 5312-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., Craig, K., Hwang, E.S., Prinz, S., Tyers, M., and Amon, A. (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2, 709-718. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065-1075. [DOI] [PubMed] [Google Scholar]
- Wäsch, R., and Cross, F.R. (2002). APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418, 556-562. [DOI] [PubMed] [Google Scholar]
- Weinreich, M., Liang, C., Chen, H.H., and Stillman, B. (2001). Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98, 11211-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, M., Liang, C., and Stillman, B. (1999). The Cdc6p nucleotide-binding motif is required for loading MCM proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96, 441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J.J., Aparicio, J.G., Chen, T., Barnett, J.D., Jennings, E.G., Young, R.A., Bell, S.P., and Aparicio, O.M. (2001). Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., and Nasmyth, K. (1999). Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13, 2039-2058. [DOI] [PubMed] [Google Scholar]
- Zhang, W., and Chait, B.T. (2000). ProFound: an expert system for protein identifi-cation using mass spectrometric peptide mapping information. Anal. Chem. 72, 2482-2489. [DOI] [PubMed] [Google Scholar]
- Zhou, C., and Jong, A. (1990). CDC6 mRNA fluctuates periodically in the yeast cell cycle. J. Biol. Chem. 265, 19904-19909. [PubMed] [Google Scholar]
- Zwerschke, W., Rottjakob, H.W., and Kuntzel, H. (1994). The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem. 269, 23351-23356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.