Abstract
Behaviors observed at the cellular level such as development and acquisition of effector functions by immune cells result from transcriptional changes. The biochemical mediators of transcription are sequence specific transcription factors (TFs), chromatin modifying enzymes, and chromatin, the complex of DNA and histone proteins. Covalent modification of DNA and histones, also termed epigenetic modification, influences the accessibility of target sequences for transcription factors on chromatin and the expression of linked genes required for immune functions. Genome-wide techniques such as ChIP-Seq have described the entire “cistrome” of transcription factors involved in specific developmental steps of B and T cells and started to define specific immune responses in terms of the binding profiles of critical effectors and epigenetic modification patterns. Current data suggest that both promoters and enhancers are prepared for action at different stages of activation by epigenetic modification through distinct transcription factors in different cells.
Introduction
There are numerous types of cells that comprise the immune system and many mechanisms that these cells use to rid the body of foreign invaders. In order to manipulate the immune response, it is necessary to understand the basic biochemical effectors of immunity. Recently, genome-scale measurements of transcription factor binding and histone modification in lymphocytes have extended our understanding of chromatin-based processes such as transcription, transcription factor binding, and histone modification, and synthesis of this data has led to the generation of an outline of genetic circuits that control immune function.
All genetic information is carried in the sequence of the DNA, but the complex of DNA and histone proteins, called chromatin, modulates interpretation of the sequence. The basic repeat unit of chromatin, the nucleosome, is formed by wrapping DNA around a histone octamer comprised of two copies of the histone proteins H2a, H2b, H3, and H4. Covalent modifications of DNA and histones influence the molecular processes that use chromatin as a substrate. In particular, it is well established that DNA methylation is involved in transcriptional repression, while post-translational modification on histones can be either activating or repressive depending on the nature and position of a particular modification. The location of nucleosomes along DNA also regulates the accessibility of critical cis- regulatory sequences (i.e. promoters and enhancers) to trans- regulatory transcription factors and has important roles in the immune function.
Previous studies have identified many DNA sequence-specific transcription factors that are required for the development and effector functions of B and T lymphocytes. Although many targets for each factor have been identified, it is necessary to identify all its target genes and the regulatory elements that mediate its function in order to comprehensively understand how each factor functions and how they interact and influence each other in the genome. Binding sites are typically identified by Chromatin immunoprecipitation (ChIP) followed by polymerase chain reaction (ChIP-PCR), however application of microarray (ChIP-chip) or next generation high throughput DNA sequencing (ChIP-Seq) makes it possible to enumerate target sites genome-wide. Because of advantages in cost, sensitivity, and speed, ChIP-Seq is the most versatile and only genome-scale technique for genomes larger than yeast of fly. In this review, we will first introduce recent progress in ChIP-Seq and related techniques, and discuss the application of these techniques to important immunological questions, emphasizing lymphocyte biology.
ChIP-Seq and related techniques
Both histone modification and transcription factor binding can be measured by the ChIP assay. The assay requires first cross-linking of chromatin to stabilize the interaction between protein factors and chromatin. Because histone-DNA contacts are very stable, histone modification can be measured with either cross-linked or native chromatin. To achieve high resolution, chromatin is broken to mononucleosome-sized fragments by sonication or micrococcal nuclease (MNase) digestion. Since MNase preferentially degrades nucleosome linker DNA, it results in uniform mono-nucleosome sized fragments and higher resolution of histone modifications. Sonication is the preferred method of chromatin fragmentation for mapping target sites of transcription factors in order to preserve their binding sites which are often located in the linker regions. Specific histone modifications or transcription factors are precipitated using antibodies and then DNA associated with the precipitated material is isolated. To identify all target sites the ChIP DNA is sequenced to saturation using next generation sequencing techniques (ChIP-Seq) (Schones and Zhao, 2008), which provides sufficient sequencing depth to recover potentially all target sites and provides a quantitative measurement of target distribution in the genome (Jothi et al., 2008; Zang et al., 2009; Zhang et al., 2008).
The number of cells used for ChIP is critical for the success of the procedure. While the assay usually requires 1 to 10 million cells, recent progress has optimized ChIP conditions to significantly decrease starting cell number. So far this small cell technique is limited to histone modifications, and has not been reported for transcription factor binding (Adli et al., 2010).
In order to determine which regions of the genome are enriched and relate them to other known functional elements (i.e. other TF binding sites or genes), the short sequence reads (tags) generated are aligned to the reference genome. After conversion of the sequence data to position data, these short sequences (tags) are analyzed using various peak-calling algorithms to identify the ChIP-enriched regions as target sites of histone modifications or transcription factor binding. Although it is possible to visually identify or call binding sites from Genome Browser displays, application of the algorithms allows identification of all sites and permits statistical analysis of a binding event. Different algorithms are appropriate depending on the specific factor being analyzed. Several algorithms including MACS (Zhang et al., 2008) and SISSRs (Jothi et al., 2008) work well for identification of tightly localized signals such as transcription factor binding sites, H3K4me3, H3K9ac etc. Other algorithms, SICER (Zang et al., 2009) or ChromaBlocks (Hawkins et al., 2010), were specifically designed to identify chromatin domains of diffuse signals spread over a large genomic region such as H3K27 methylation and H4K16ac.
Another difficult, but key issue for ChIP-Seq, is the selection of proper controls for ChIP-Seq data analysis. The most common choices for control libraries are immunoprecipitates with total IgG or pre-enriched chromatin (input) used for ChIP. Most non-specific IgG preparations are usually not truly pre-immune IgGs (from the same animal before immunization), and don’t control for the non-specific and cross-reactivity of the affinity-purified antibody. Furthermore, non-specific IgGs usually pull down very little DNA and often lead to biased PCR amplification of sequences at only limited genomic loci. Consequently, this does not provide a good background model of the genome. For this reason, the chromatin input is a better control, because it generates an accurate estimation of biases that are introduced in ChIP assays due to sonication of chromatin and sequencing.
These technical caveats may be seen as trivial, but the size of genome wide data sets greatly increases the possibilities of false positives. Local differences in chromatin cause different sonication efficiencies, and a ChIP library will be enriched for these regions over unrelated and silent chromatin regions. In fact, active functional regions of the genome can be identified by their sensitivity to sonication just as they are sensitive to DNase (Teytelman et al., 2009). In addition, there are many data analysis steps between the raw data and the binary call of bound or unbound. Due to unfamiliarity and technical separation from these steps experimental biologists may be led to false conclusions.
DNA methylation, which mainly occurs within 5 -cytosine-guanine-3 dinucleotides (CpGs) in mammals, is perhaps the best understood epigenetic mechanism implicated in transcriptional repression. Methylated cytosine (MeC) can be detected by modification using sodium bisulfite, MeC binding proteins (Methyl-binding domain, MBDs), or antibodies against MeC. Bisulfite conversion-based methods provide single base pair resolution. Unfortunately, the scale needed to achieve meaningful coverage of a mammalian genome limits its broad application. To restrict the sequenced region, enrichment of genomic regions associated with methylated DNA with an antibody recognizing MeC (MeDIP) or MeC-binding proteins, followed by sequencing of the enriched DNA (MeDIP-Seq), provides a convenient and genome-wide strategy for detection of DNA methylaton (Bock et al., 2010). However, these affinity-based approaches suffer from low resolution and the bias that CpG-rich sequences are better enriched than equally methylated but CpG-poor regions. Thus, they are suited to compare the relative methylation levels of CpG-rich regions under different conditions but not for the quantitative measurement of each individual CpG base pair. Because of continuous decreases in cost and increases in sequencing capacity it is likely that bisulfite-conversion and sequencing will become routine. Further technical advances allow direct measurement of MeC, and new sequencing platforms with this capacity will soon be available to the community (Flusberg et al., 2010).
The position and occupancy of nucleosomes can be determined by sequencing the ends of mono-nucleosome associated DNA generated by micrococcal nuclease (MNase) (Schones et al., 2008). Because MNase preferentially degrades linker DNA, sequencing of the ends of DNA fragments generated by MNase digestion provides nucleosome position information. One issue for mapping nucleosome distribution is whether the chromatin should be cross-linked with formaldehyde before MNase digestion. In general, cross-linking is not required since the nucleosome structure is relatively stable under the MNase-Seq conditions and the results with or without prior cross-linking are fairly similar (unpublished information, K. Cui). In some cases, cross-linking may further stabilize fragile nucleosomes at promoter and enhancer regions. Unfortunately, cross-linking with HCHO may also stabilize non-nucleosome protein complexes on chromatin, and protect DNA from the MNase digestion. This can result in some protected regions not related to nucleosome structure, complicating interpretation.
Traditionally, functional enhancers and promoters were predicted by the presence of DNase I hypersensitive (HS) sites on Southern blot assays. Application of the next generation sequencing technique to detect the DNase I HS sites reveal the genome-wide distribution of HS and their association with histone modifications (Boyle et al., 2008). These data are extremely valuable as a gold standard for enhancer identification. Similarly, the accessibility of chromatin to restriction enzymes has been combined with next generation sequencing (NA-Seq), which provides an alternative method to monitor genome-wide the status of chromatin during differentiation of immune cells (Gargiulo et al., 2009).
General features of T cell epigenomes
The most comprehensively characterized epigenome is of the human CD4+ T cells, with data of genome-wide distribution of more than 20 histone methylation, 18 histone acetylation, histone variant H2A.Z, nucleosome positions, RNA Polymerases II and III and various transcription factors and co-factors (Barski et al., 2010; Barski et al., 2007; Schones et al., 2008; Wang et al., 2008). These datasets confirmed and extend previous observations that histone acetylation and many methylation events are positively correlated with gene activation. H3K27me2, H3K27me3, H3K9me2, H3K9me3, and H4K20me3 are exceptions that are negatively correlated with gene expression.
Furthermore, active genes tend to be associated with multiple “active” modifications, whereas silent genes are associated with only a few “repressive” modifications or are not associated with any modification.
The general correlation between histone modification and gene activity had already been established, but the length of modification islands is best observed by studying genome-wide modification patterns. This provides insight into the mechanisms of deposition of a given post-translational modification (PTM). For example, H3K27me3 marks are spread broadly over repressed genes, due to the self-propagation of this mark that is added and recognized by distinct subunits of the polycomb repressor complexes (Xu et al., 2010). H3K4me3, associated with active promoters, is highly localized to a few nucleosomes around promoter regions, likely deposited by complexes that directly interact with sequence specific factors. Other activating modifications, H3K4me1 for example, are also broadly spread at many genomic regions (Barski et al., 2007), but the mechanisms that give rise to these large islands are not known.
Different functional genomic regions are associated with distinct sets of histone modifications. The transcribed regions of active genes are usually associated with histone H4 acetylation, mono-methylation of H2BK5, H3K9, H3K27 and H4K20, H3K36me3, and mono-, di-, or trimethylation of H3K79. Active promoters are enriched with multiple modifications including H3K4me1, H3K4me2, H3K4me3, H3K9ac and histone variant H2A.Z. More than 25% of all promoters are associated with a common set of 17 modifications in addition to other modifications (Wang et al., 2008). Transcriptional enhancers, defined as regions of DNA that activate transcription but are not located at the transcriptional start site (TSS), are marked by histone modifications such as H3K4me1 (Heintzman et al., 2009). Although H3K4me1 is detected at most enhancers, it is not the best mark to pinpoint functional enhancer elements because of its broad distribution in the genome. In contrast, H3K4me2, H3K9ac, H3K27ac and H3K18ac are better markers of enhancers because of their sharp signals (Creyghton et al.; He et al., 2010; Rada-Iglesias et al.; Roh et al., 2007; Wang et al., 2008). p300, the histone acetyltransferase (HAT) that acetylates H3 on K18 and 27 is also a good enhancer marker (Jin et al., 2011; Visel et al., 2009). Differential marking of enhancers by H3K27me3 or H3K27ac may indicate alternative status of enhancer activity as demonstrated in embryonic stem (ES) cells (Rada-Iglesias et al., 2011).
In addition to histone modification, histone position is also dynamically regulated. Active genes with elongating Pol II and genes with paused Pol II exhibited several highly positioned and phased nucleosomes relative to their TSSs (Schones et al., 2008). These results suggest that a nucleosome structure at the transcription initiation site is not compatible with Pol II binding and has to be relocated to allow binding of Pol II. Accordingly, there is no characteristic nucleosome pattern in genes when they are not associated with Pol II. Enhancer function also involves nucleosome reorganization that is regulated by T cell receptor (TCR) signaling (Schones et al., 2008). Since the nucleosome structure is generally inhibitory to binding of transcription factors, enhancer nucleosomes are reorganized such that target motifs are located in the linker region (He et al., 2010). This feature has been successfully used for identification of functional enhancers.
Changes in epigenetic markings are observed during differentiation, cytokine expression, and trans-differentiation of helper T cells
Naïve CD4+ T cells can be differentiated into different T helper cells including Th1, Th2 and Th17 cells characterized by expression of specific cytokines and transcription factors. Several studies characterized the epigenetic pattern associated with these genes in T helper lineages (Agarwal and Rao, 1998; Akimzhanov et al., 2008; Avni et al., 2002; Chen et al., 2005; Fields et al., 2004; Hatton et al., 2006; Santangelo et al., 2002; Schoenborn et al., 2007). A genome-wide data set for H3K4me3 and H3K27me3 in naïve, Treg, and T helper cells demonstrates that the signature cytokine gene loci are associated with the epigenetic modification corresponding to their expression pattern. The Th1-specific Ifng gene (coding the cytokine interferon-gamma) is associated with H3K4me3 in Th1 cells and H3K27me3 in all other cells, whereas the Th2-specific Il4 gene (coding the cytokine interleukin 4) is associated with H3K4me3 only in Th2 cells and H3K27me3 in other cells, consistent with these cells being epigenetically stable lineages (Wei et al., 2009).
In contrast to these expected patterns, key transcription factors required for Th cell differentiation have different patterns of modification. The Tbx21 gene, which encodes T-bet, a key transcription factor for Th1 cell differentiation, is associated with both H3K4me3 and H3K27me3 in naïve cells. After differentiation, the promoter resolves to only H3K4me3 in Th1 cells, consistent with its active expression, while it remains associated with both H3K4me3 and H3K27me3 in other Th cells.
The co-existence of H3K4me3 and H3K27me3, termed bivalent modification, was proposed to play critical roles unique to ES differentiation (Bernstein et al., 2006); however, it exists at a large number of genes in adult tissue and is probably a general mechanism to keep genes in a silent, but inducible state (Cui et al., 2009; Mohn et al., 2008; Roh et al., 2006). T helper cells are considered terminally differentiated cells, but the unresolved bivalency at the promoters of key transcription factor genes in the non-expressing cells suggested that their expression can be induced under appropriate conditions, leading to alternate cell fate. Indeed, the expression of Tbx21 is induced in natural T regulatory cells under Th1 cell culture conditions, which is correlated with expression of IFN-γ, suggesting that the cells can assume a Th1 cell fate. The fate of Th17 cells could also be changed during late development (Lee et al., 2009), which is attributed to the epigenetic instability of the IL-17 locus (Mukasa et al., 2010). These results collectively support the model that monovalent modification (H3K4me3 alone for an active locus or H3K27me3 alone for an inactive locus) confers epigenetic and phenotypic stability. The opposite of this, epigenetic indecision (lack of any modification or both active and repressive modifications at inactive locus) allows T helper cell plasticity.
How does bivalent modification prime genes for induction in response to TCR or differentiation signals? Histone methylation provides recognition signals for various cofactors of transcription and thereby modulates transcription. H3K27me3 is added by the Enhancer of Zeste homolog 2 (Ezh2) containing PRC2 complex and recognized by the PRC1 repressive complex that maintains a repressive chromatin environment for transcription (Margueron and Reinberg, 2011). The function of H3K4me3 in regulating gene activation is mediated by a number of mechanisms. First, it interacts with ATP-dependent chromatin remodeling complexes Chd1 and NURF (Pray-Grant et al., 2005; Wysocka et al., 2006), which open chromatin to allow binding of other transcription regulators. Second, H3K4me3 signals mediate binding of the NuA3 histone acetyltransferase (Martin et al., 2006). Third, the MLL and hSET1 complexes, which are responsible for adding the H3K4 methylation mark, are associated with the MOF acetylase (Dou et al., 2005; Wysocka et al., 2005) and Sin3 deacetylase (Wysocka et al., 2003), respectively. These data suggest that H3K4 methylation modulates the histone acetylation status of a gene by recruiting HATs and histone deacetylases (HDACs). Indeed, promoters marked by H3K27me3 alone are not be acetylated even in the presence of HDAC inhibitors, but bivalent promoters are rapidly acetylated by HDAC inhibitor treatment (Wang et al., 2009). Inhibition of WDR5, an essential subunit of the MLL complexes that are responsible for H3K4me3, compromise the histone acetylation induced by HDAC inhibitors. These results support the model that H3K4 methylation facilitates the dynamic cycle of acetylation and deacetylation by transient binding of HATs and HDACs at bivalent promoters. This maintains a silent state, but at the same time primes them for induction in response to appropriate environmental cues (Wang et al., 2009).
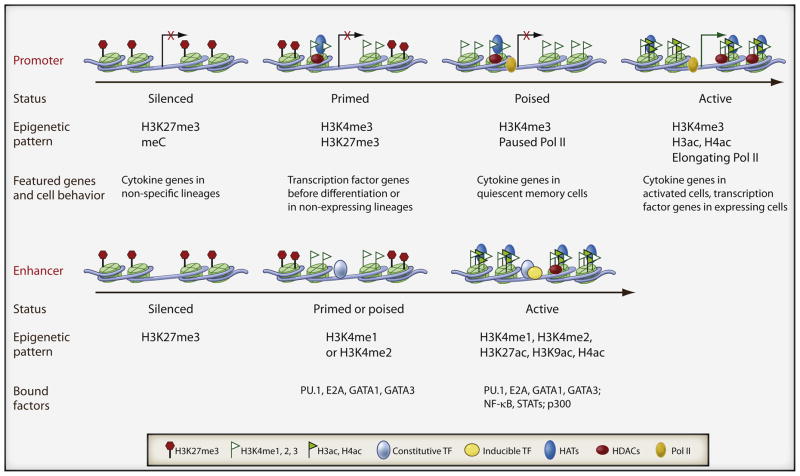
The epigenetic marks associated with gene promoters that are turned on very quickly after a stimulus are nearly identical to that of active genes, and little if any epigenetic change is associated with these genes during a short-term expression change. Therefore, active histone modifications provide a chromatin environment to support on-going transcription, and to poise genes for rapid induction after a stimulus (Figure 1, top panel). Among genes induced in human resting CD4+ T cell by T cell receptor signaling after 18 hours, most are associated with prior H3K4 methylation, H2A.Z, and Pol II, and very little H3K27me3; this pattern does not change much during the course of gene induction (Barski et al., 2009). In contrast, developmental genes are primed by the active mark, H3K4 methylation, and the repressive mark, H3K27me3; additionally, they are associated with only very low levels of histone acetylation and Pol II. Thus, activation of these genes is much slower than that of the fully poised genes.
Figure 1. Epigenetic memories of different activation stages of promoters and enhancers.
The upper panel shows the different status of promoter activation indicated with different colors below the cartoon. The distinctive epigenetic modifications associated with the promoter status and classes of genes that exhibit the epigenetic pattern in particular lineages or cell activation states are also indicated.
The lower panel shows the different status of enhancer activation and the associated epigenetic modification with each status. The bound transcription factors and co-factors with each status are also indicated.
Establishment of new histone modification patterns is observed at cytokine genes after extended TCR signaling (Araki et al., 2009; Aune et al., 2009), which is the result of the differentiation process associated with the extended TCR signaling. Cytokine genes in quiescent memory cells are associated with this signal-dependent poised promoter configuration. Examples of this are the Ifng gene in resting Th1 cells and Il4 gene in resting Th2 cells. Both so can be rapidly transcribed upon antigen engagement, suggesting that this kind of special chromatin configuration at key cytokine and effector genes forms the basis of immune memory (Barski et al., 2009).
Analogous to promoters, enhancers are associated with distinct epigenetic modification patterns when they are silent, active, or poised (Figure 1, bottom panel). Silent enhancers are associated with H3K27me3 and no active modification. Much like the case of promoters, enhancers are often developmentally poised with histone methylation marks, poised enhancers may also have H3K4me1 and H3K27me3 when they are inactive, but remain accessible. Signal dependent poised enhancers are marked by H3K4me1,2, which have lost the silencing marks, but remain inactive. Full activity is regulated by histone acetylation and is acutely induced at binding sites of inducible transcription factors and deposited by p300 (Ghisletti et al., 2010). Accordingly the correlation between enhancer activity and p300 or H3K27ac, the mark deposited by p300, is high; both p300 binding and H3K27ac have been successfully used as signals to predict functional enhancers (Creyghton et al., 2010). Modulation of the acetylation status is a critical step for initiating and terminating enhancer activity.
Prevention of DNA methylation is another mechanism to prime enhancer activity. For example, DNA in the enhancer of the T cell-specific ptcra gene must be demethylated at the earliest stages of development, using machinery that is only present in ES cells. If methylated transgenes are introduced to T cell lines, the transgene is not demethylated, the enhancer doesn’t acquire active histone modifications, and finally the cells are no longer able to express this gene. In contrast if the transgene is not methylated, or if it is introduced to ES cells, where demethylation occurs, full activity is observed (Xu et al., 2007; Xu et al., 2009). This mechanism of enhancer preparation will be more difficult to explore at a global level, but it is important to consider.
The “Average” Epigenome
The general arrangement and features of epigenomes of are the same in all tissues that have been measured including ES cells, hematopoietic stem cells, neurons , and T cells. From the epigenetic perspective, all genes and enhancers conform to the same pattern of histone modifications, and can be classified to a small number of states: silent, developmentally- or signal dependent-poised, or active. The correlation between these states and functional activity is so high that a statistical analysis of histone modifications can identify genes, enhancers, repeat elements, silent chromatin, or other functional elements without prior knowledge of the genome (Ernst and Kellis, 2010). Each state is defined by the presence of several histone modifications that always co-exist, as is the case with a core set of 17 modifications previously detected at promoters (Wang et al., 2008).
The functional relevance of having many histone modifications together is difficult to know. Histone modifying enzymes typically exist in large multi-protein complexes that contain several enzymatic subunits, and therefore may simply travel together, and in many cases the mark may not have a function. More likely different modifications have different functions and work together in an additive or collaborative fashion to maintain a fully active configuration of chromatin. Still another possibility is that different active modifications function redundantly in order to maintain a robust chromatin structure. In this case, an occasional absence of one modification will not have deleterious effects on the chromatin structure. In addition to the inter-locus comparison of different modification patterns, inhibition of enzymes that are responsible for the modification, either by SiRNA mediated reduction in protein levels in vitro or targeted deletion in animals, can provide valuable information on the function of a modification. Direct mechanistic insight may be derived from experiments using chemically synthesized and modified chromatin as has recently been proposed, but these are beyond our current technical abilities (Allis and Muir, 2011).
As noted above established histone modification patterns self-propagate, but changes in epigenetic patterns are mediated by sequence specific transcription factors. One of the best-studied examples is the Cd4 enhancer that is required to establish epigenetic patterns during development, but becomes dispensable in mature cells (Chong et al., 2010). Sequence specific transcription factors bind to enhancers and recruit epigenetic enzymes to effect their gene expression changes. Below we focus on the transcription factors that mediate T helper differentiation and B cell commitment. These are chosen because they are paradigms for rapid induction of gene expression after extracellular signaling in the case of T cells, the B cell transcriptional cascade is well known, and currently both systems have been well studied by genome wide techniques.
Identification of direct target genes of key transcription factors during T helper cell differentiation
Because of the different life cycles of pathogens such as viruses, extracellular bacteria, or parasites different clearing mechanisms must be employed. The immune system has several distinct batteries of immune effectors that are suited for different pathogens so that Th1 cells are tuned toward viruses and promote cell lysis, while Th2 cell responses are more appropriate to clear extracellular pathogens, and lead to humoral responses. Th17 and Treg cells are implicated in controlling inflammation and suppressive function, respectively. Key transcription factors regulating these various Th cells have been well characterized. Tbx21, coding T-bet, is critically for the Th1 cell fate (Afkarian et al., 2002; Kaplan et al., 1996b; Szabo et al., 2000; Thierfelder et al., 1996), while GATA3 is essential for the Th2 cell differentiation (Kaplan et al., 1996a; Takeda et al., 1996; Zheng and Flavell, 1997). Th17 cell commitment requires the TGF-β signaling pathway and a key transcription factor, RORγt (Chen et al., 2006; Dong, 2008; Korn et al., 2007; Nurieva et al., 2007; Wei et al., 2007; Yang et al., 2008; Zhou et al., 2007). Meanwhile, Foxp3 is a key transcription factor required for Treg cell differentiation and function (Elias et al., 2008; Lohr et al., 2006; Miyara and Sakaguchi, 2007; Zheng and Rudensky, 2007).
Various members of the Signal transducer and activator of transcription proteins (STATs) function to either enhance or inhibit T helper cell differentiation and function (Adamson et al., 2009). STAT4 and STAT6 reciprocally regulate the differentiation of Th1 and Th2 cells, respectively. In response to cytokine signaling including IL-12 and IL-23, STAT4 directly activates the expression of the Th1 cell cytokine IFN-γ (Thierfelder et al., 1996), and indirectly regulates the Th1 cell transcription factor T-bet via the kinase MAP3K8 through a positive feedback loop (Watford et al., 2008). STAT6 promotes Th2 cell differentiation by activating the Il4r gene, and is required for expression of the Th2 cell transcription factor GATA3 (Kurata et al., 1999; Liao et al., 2008; Zheng and Flavell, 1997). In other contexts, STAT5 promotes Treg cell differentiation by directly binding to the Foxp3 promoter and activating its expression (Burchill et al., 2007). Meanwhile, STAT3 is a positive regulator of Th17 differentiation by directly or indirectly activating the Rorc gene that encodes RORγt (Laurence et al., 2007).
STAT proteins also act as negative regulators of alternative T helper fates. For example, STAT5 inhibits Th17 cell differentiation(Laurence et al., 2007), whereas STAT3 inhibits Foxp3 expression(Zhou et al., 2007). In other contexts STAT3 physically interacts with Foxp3 and is required for the immunosuppressive function of Treg cells (Chaudhry et al., 2009). Meanwhile, STAT6, which is critically required for Th2 cells, also inhibits Foxp3 expression (Takaki et al., 2008).
Numerous genes are known to be affected by deletion of these key transcription factors in various Th cells. However, in order to gain full understanding of the global regulatory networks controlled by these factors, all targets must be determined. While a few studies have been published on identification of Foxp3 and GATA3 targets using DNA microarrays (Jenner et al., 2009; Yagi et al., 2010; Zheng et al., 2007), more genome wide studies have focused on various STATs as reviewed below.
Wei and colleagues (Wei et al., 2010), identified genome-wide target sites of STAT4 under Th1 cell conditions and target sites of STAT6 under Th2 cell conditions. About 4000 genes were bound by each transcription factor in these cells; and about 63% of the binding occurred in intergenic regions and 31% localized to promoters. About 50% of the STAT4 and STAT6 target genes overlapped. A smaller set of genes (508) was found bound by STAT6 in another study (Elo et al., 2010), which identified the primary and secondary direct target genes during a time course experiment. Recent data indicated that STAT5 is also critically involved in the differentiation of Th2 cells by regulating the expression of Il4r. STAT5 becomes bound to their target sites at the beginning of Th2 cell differentiation and the binding is sustained or enhanced after two rounds of Th2 cell polarization at many critical target genes (Liao et al., 2008). Comparison of the STAT5 and STAT6 target genes reveal a statistically significant overlap (Elo et al., 2010), suggesting a similar targeting mechanism. These data demonstrate that STAT5, STAT4 and STAT6 have broad effects in Th1 and Th2 cell differentiation and provide a comprehensive view of their target genes and potential function in T helper cells. Despite the well-established function of STAT proteins as activators of transcription, these genome-wide studies identified hundreds of genes that are directly repressed by STAT binding. Interestingly, these factors bind to a subset of Th1- and Th2 cell-specific genes and act in an opposing manner to modulate epigenetic modifications and gene expression (Wei et al., 2010). However, it is not clear how STAT proteins mediate transcriptional repression of target genes and oppose the function of the other STATs. One possibility is that these STATs, which recognize similar binding motifs, could compete for the same binding sites and recruit different co-factors for either transcriptional activation or repression.
The balance of Th17 and Treg cells is critical for host immunity, which is reciprocally regulated by STAT5 and STAT3. Using a colitis model, a recent study showed that STAT3 has an essential role in driving both colitis and systemic inflammation by promoting Th17 cell differentiation and inhibiting Treg cell conversion. Among the 3000 genes bound by STAT3 as identified by ChIP-Seq, are most of the genes known to be involved in Th17 cell differentiation (Durant et al., 2010). Following stimulation of CD4+ T cells with IL-21, STAT3 binds its target sites, the majority overlapped with the binding sites of IRF4. Interestingly, deletion of IRF4 resulted in a global loss of STAT3 binding as compared to the wild type cells (Kwon et al., 2009). The dependence of STAT3 binding to target sites on the presence of IRF4 is consistent with a critical role of IRF4 in the development of inflammatory Th17 cells (Brustle et al., 2007).
Characterization of global regulatory networks in B cell development
The transcriptional regulatory network critical for B cell development has been extensively studied. The self-reinforcing, core set of transcription factors that limit alternative lineage potential and specify the B cell fate is the sequentially expressed E2A, EBF1, and Pax5 proteins (Ramirez et al., 2010). In brief, E2A expression is induced as cells enter the lymphoid lineages and helps to restrict erythroid and myeloid potential. Evidence for this includes single cell cultures of E2A deleted cells (Dias et al., 2008), and an E2A deficient progenitor cell line. Although this cell line resembles and has the growth requirements of a population of cells that is normally B cell restricted, it maintains multi-lineage potential (Ikawa et al., 2004). E2A collaborates with PU.1, another important transcriptional regulator, in several hematopoietic lineages. The IL-7R is an important target of Pu.1 at this stage (DeKoter et al., 2002). A combination of E2A, Pu.1, and Stat5 induce expression of EBF1 in early B cell progenitors (Roessler et al., 2007).
Within the blood lineages EBF1 expression is limited to B cells, and a central role of EBF1 in B cell development is highlighted by its ability to bypass developmental blocks caused by loss of PU.1 or E2A (Medina et al., 2004). An EBF1 deficient B cell progenitor line has also been created that maintains multi-lineage potential (Pongubala et al., 2008).
Much B cell work focused on the critical transcription factor, Pax5 based on the surprising discovery that Pax5-deficient pro B cells, ordinarily B lineage committed, maintain alternative lineage potential (Nutt et al., 1999). This can be further extended to show that induced deletion of Pax5 in mature B cells re-confers multi-lineage potential(Cobaleda et al., 2007). Other transcription factors including GABPα (Xue et al., 2007), Ikaros (Reynaud et al., 2008), and Gfi1b (Ramirez et al., 2010; Spooner et al., 2009) have also been shown to have critical roles in B cell lineage. Similar to critical factors in T cells, important questions remain regarding how these factors are induced, where are the corresponding cis-regulatory elements and how they interact with each other to regulate the immune activity. Recent analyses taking advantage of ChIP-Seq have provided insights into these questions.
EBF1 binds to more than 9,000 genomic regions in pro-B cells (Treiber et al., 2010). Using cells with targeted deletion of the Ebf1 gene or overexpression of EBF1 cDNA together with the binding data, it was demonstrated that EBF1 mainly acts as an activator of transcription. EBF1 binding sites tend to co-occur with motifs of several transcription factors including Ets1, and E2A. In addition, EBF1 binds a large fraction of genes that are regulated by Pax5. This argues that the core B cell transcription factor activities are coupled by mutual positive regulation and by binding the same sets of targets. Indeed, another study has provided genome-wide evidence that E2A binding sites significantly overlap with the binding sites of EBF1 or Foxo1 (Lin et al., 2010). Interestingly, compound heterozygosity for EBF1 and E2A, the factors that bind the most enriched DNA sequence motifs for a set of E2A binding sites, causes a cooperative block in B cell development (O’Riordan and Grosschedl, 1999).
Based on this previous work and the discovery that Foxo1 and E2A characterized another set of E2A binding sites, Foxo1 and E2A compound heterozygous mice were created. These mice also generate very limited B cell numbers. Nearly 50% of the EBF1 binding sites in pro-B cells are flanked by an E2A binding site, suggesting that E2A may bind collaboratively by either interacting with EBF1 or preparing the chromatin for access by EBF1. Induction of the E2A isoform, E47, demonstrates that it is capable of inducing H3K4me1 and also displacing nucleosomes. This effect depends on the transactivation domain of E2A. Lastly, using a mix of data regarding bound or differentially expressed genes the authors find a gene network that illuminates the interactions of E2A, Foxo1, and EBF1 in the B cell network (Lin et al., 2010).
Another key transcription factor required for B cell development is Pu.1. Pu.1 is broadly expressed and important for nearly every blood lineage, including hematopoietic stem cells (Iwasaki et al., 2005). ChIP-Seq analysis of Pu.1 binding in B cells and macrophages reveal a large number of binding sites (32,000 to 45,000) (Heinz et al., 2010). Controlled expression of Pu.1 directly induces H3K4 methylation and nucleosome reorganization at its binding sites in hematopoietic cells and also in 3T3 cells (Ghisletti et al., 2010), directing conversion of fibroblasts to macrophages (Feng et al., 2008). These results suggest that Pu.1 functions as a global genomic organizer by priming chromatin at regulatory regions for further downstream events in cellular differentiation programs(Natoli, 2010). There is very little overlap of Pu.1 binding sites between B cells and macrophages. In each cell type the Pu.1 sites are co-localized with sequence motifs corresponding to lineage-determining transcription factors, suggesting an extensive collaboration between transcription factors. Specifically, expression of E2A recruits Pu.1 to E2A/PU.1 shared binding sites in B cells, whereas C/EBPβ and Pu.1 cooperatively bind at a different set of sites in macrophages. Cooperative binding of these specific factors shapes the cistromes of both, surely directing lineage commitment (Heinz et al., 2010).
“Average” behavior of transcription factors
Several properties characteristic of all transcription factors have been determined from genome-wide studies of transcription factor binding: (1) most transcription factors bind to one primary consensus motif that is conserved in all tissues; (2) binding is context dependent and the same factor recognizes distinct cistromes in different cells; (3) secondary motifs of tissue specific collaborating transcription factors distinguish cistromes of a factor in different cells; (4) although binding events are typically enriched near genes, most binding sites are located far away (>10kb) from known transcriptional start sites; (5) using loss or gain-of-function experiments, transcription of only a minor fraction of bound genes is influenced by the transcription factor (6) networks of transcription factors are established by positive and negative regulation of other critical regulators.
These properties reflect the typical analyses that are done with any ChIP-Seq data set. After calling binding sites, the top motifs and associated secondary motifs are scored to predict potential collaborating factors in the genome. It is important to test the functional importance of the binding events, typically by loss or gain of function of the transcription factor. Unfortunately for the ease in interpretation, the overlap of genes that are bound and genes that are differentially regulated is poor often 5–20%. This is significantly higher than expected by chance, but it means that the majority of binding events are not functional in that they do not immediately influence transcription.
There are several reasons why transcription factor binding is poorly coordinated with transcriptional output. First, the local binding of a transcription factor may regulate other genes through long-distance chromatin interaction. Visualization of direct interaction between enhancers and promoters by genome-wide 3C assays will help to solve this problem. Second, transcription factor binding may regulate the genes under various conditions the cells may encounter in vivo but which are not operative in vitro under the assay condition. Third, the primary role of a transcription factor may be to induce histone modification at an enhancer. Transcriptional activation of the target gene requires additional factors. Thus, deletion of the bound factor from the cells will not have apparent effect on the expression of the bound gene at that time. At different stages, when other relevant factors become present in the cell and the enhancer is active, loss of any of these factors may influence transcription of the target gene. Although demonstration of binding is not sufficient to say that a transcription factor regulates a gene, it provides evidence that the transcription factor may regulate the bound gene under appropriate conditions.
In addition to the classic model where transcription factors bind to promoters or enhancers and affect the levels of mRNA for proximal protein coding genes, the number of binding sites permits other functions. If a factor binds at greater than 30,000 genes, many of which are far from genes or in the middle of genes they may lead to transcription of microRNAs, other non-coding RNA species, or previously unannotated genes. Intragenic binding may control alternative promoter usage, splicing, or ensure proper direction of transcription. It is fairly likely that many sites have no function and result from the random occurrence of transcription factor binding motifs is expected in sequences as long as mammalian genomes. Whichever turns out to be the case, the binding profiles generated by ChIP-Seq are much more complex than could be predicted if transcription factors only controlled the on or off state of a gene.
Interplay of epigenetic modifications and transcription factors
ChIP-Seq analyses have revealed that the binding sites for most transcription factors are in enhancers far from promoter regions. Nucleosomes surrounding enhancers are marked by various histone modifications including H3K4 methylation and histone variant H2A.Z, suggesting that the chromatin structure at enhancers is prepared for binding of transcription factors when they become available in response to cell surface signaling. Indeed, previous studies have demonstrated that the inducible transcription factors such as NFATs and STATs migrate to the nucleus and become bound to chromatin a few minutes after TCR and cytokine stimulations of T cells. The ChIP-Seq studies also show that STATs and NF-κB binding to their target genes quickly following cytokine and LPS treatments, respectively(Barish et al., 2010; Liao et al., 2008). In particular, upon LPS stimulation, p300 and NF-κB become bound to thousands of potential enhancers that already display the H3K4me1 modification. This indicates that their target sites have been prepared for rapid binding of p300 and NF-κB upon signaling (Ghisletti et al., 2010).
The conclusion that cistromes of many factors are regulated by pre-existing histone modifications leads to the question of how these marks are deposited. Current data suggest that pioneer factors and cooperative binding of different factors direct binding in a sequence-dependent and chromatin-independent manner. During differentiation of hematopoietic stem cells to erythrocytes, GATA1 functions as a pioneering factor by binding to nucleosomal DNA and recruits the BAF chromatin remodeling complexes (Kim et al., 2009), which remodels chromatin to allow accessibility of chromatin. Another member of the GATA family transcription factors, GATA3, may have a similar role during Th2 cell differentiation, as it has been shown that the essential subunit BRG1 of the brahma associated factor (BAF) complexes is associated with several key Th2 cell cytokine enhancers that are known GATA3 target sites. STAT6 may also be involved in recruiting BRG1 to enhancers as deletion of STAT6 impaired its recruitment to distal enhancers of the Gata3 gene (De et al., 2011). Genetic targeting of Stat4 is correlated with globally decreased H3K4me3 at STAT4 target promoters (Wei et al., 2010), which suggests that STAT4 maintains the active H3K4me3 mark by recruiting histone modifying enzymes.
In Th17 cells, IRF4 prepares chromatin for STAT5 binding since it constitutively binds to sites overlapping with STAT5 sites and its deletion severely compromises the STAT5 binding following IL-21 treatment of CD4+ T cells (Kwon et al., 2009). In B cells, E2A, EBF1, and Pax5 are all likely factors that bind to their target sites during different stages of development and modulate chromatin structure to prepare their target genes for future action in response to environmental cues, and their binding induced H3K4 methylation at numerous sites (Heinz et al., 2010; Lin et al., 2010). In addition to establishing these marks, continuous binding is required as inhibition of Pu.1 in macrophage cells decreases H3K4me1 levels at its binding sites (Ghisletti et al., 2010). Even though these factors influence epigenomes, they still require a specific chromatin environment or presence of collaborating factors for binding to their target sites. For example, ectopically expressed B cell factor EBF1 bound to its target gene Egr3 in a CD4+CD8+ T cell progenitor line but not in NIH 3T3 fibroblasts (Treiber et al., 2010). Similarly, Pu.1 was associated with drastically distinct cistromes in B cells and macrophages, indicating the importance of cellular context for its targeting (Heinz et al., 2010).
Based on all of these findings it is obvious that histone modifications influence transcription factor binding which in turn influence further histone modification. All blood lineages are derived from hematopoietic stem cells and similarly, all T helper cells are derived from naïve CD4+ T cells. Each cell type is associated with a distinct epigenome that results from the interaction of the progenitor epigenome and the transcription factors in the cell. Differentiation could be initiated by environmental signals that trigger inducible transcription factors (Figure 2). The inducible transcription factors could collaborate with constitutive transcription factors, bind to target sites situated in open chromatin, and subsequently activate transcription of the poised target genes including other transcription factors and chromatin modifiers. The newly expressed transcription factors could bind to target sites with open chromatin and target sites without prior active modifications and over time modulate chromatin modification at novel sites for existing transcription factors. When the resulting transcription factor program and epigenome stabilize the cell will wait until further environmental cues cause further differentiation. This cycle will continue until the cell dies or reaches a terminal differentiation state. This is defined as a cell type that can give rise to no new cell types, but a definite feature based on epigenetics or transcription factor expression has not been defined.
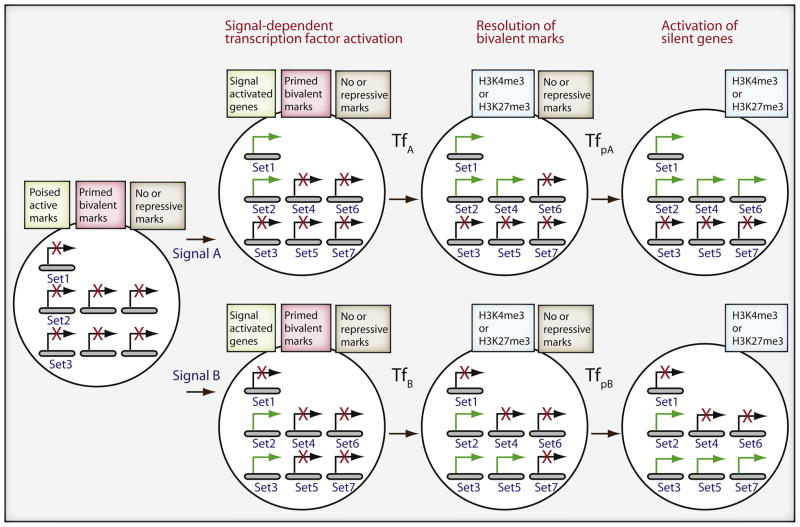
Figure 2. Signal dependent transcription factors lead to developmental decisions by sequentially activating epigenetically distinct sets of genes.
Two different signals (A or B) activate, through pre-existing but signal-dependent transcription factors, shared gene set 2 and distinct gene set 1 or set 3, which are poised by active chromatin marks. Transcription factors, TfA or TfB, are induced and resolve the marks at bivalent genes, set 4 or set 5, allowing for expression of pioneer factors (TfpA or TfpB) that cause further epigenetic changes. The pioneer factors lead to epigenetic changes and induction of a new set, set 6 or set 7, of genes, resulting in a cell primed for a different set of responses.
Perspective
Despite certain limitations, ChIP-Seq is a powerful technique for characterization of status of chromatin and identification of target sites of epigenetic modification enzymes and transcription factors. The data derived from these analyses are of high predictive value for the potential of gene expression and comprehensive identification of cis- regulatory elements of transcription including enhancers and promoters. As the genome-wide data for transcription factors and other enzymes become available, combined with specific gene deletion, expression profiling, protein-protein interaction, and genome-wide chromatin interaction studies, these datasets can be integrated to obtain a comprehensive picture of how individual factors act and how they cooperate in the development, differentiation, and function of immune cells. In particular, this kind of analysis and data integration will aid us to address important questions of how basic mechanisms manifest to determine cell fate; how bivalent modifications at key transcription factors are established and maintained in non-expressing lineages; and how the developmental history of immune cells is remembered.
Most studies mentioned above seek to understand the regulation of message output of the nucleus, as this causally determines the effector function of the cell. Analysis of ChIP-Seq data has focused on creating an average gene profile and an average binding motif. This is most appropriate for those factors with enzymatic activity. p300 binding, PRC binding, and resulting histone modifications are excellent predictors of the functions for underlying stretches of DNA. Histone modification profiles and binding patterns of enzymes that write them can be presented as averages because they generally have one specific biological effect. This is in contrast with sequence specific transcription factors where the effect of binding is nuanced, and locus specific.
If the average binding profile of a transcription factor is not useful, it remains to be determined if there are intuitive and meaningful ways to integrate genome wide binding sets for sequence specific transcription factors. Genome-wide data sets have been used to determine genetic networks that control developmental decisions, but these efforts are still somewhat difficult to grasp and limited by the measurement of the input data sets. Because of the novelty of these techniques, the first studies have focused on known master regulators, while the features of less well characterized transcription factors aren’t known. Hopefully, after more genome-wide profiles are created, general features that distinguish master regulators or oncogenes from less important or harmful transcription factors will become clear.
Future mechanistic exploration will require the measurement of transcription factor binding in live cells. Static models imply large protein complexes nucleating around sequence specific factors. Current technologies are not sufficient, and ChIP is too inefficient to determine the true nature of transcription at single cell or single locus levels. Where it can be measured it is clear that transcription factors do not have long residence times on DNA (Stavreva et al., 2004). Histone modification may be a form of memory of transcription factor binding that overcomes the need to have all partner factors present at once. The time that a locus is bound by any factor may be nearly as important as which factors bind it. This mechanism of action could explain probabilistic, binary enhancer activity (Walters et al., 1995). The unlikely assembly of many components all required for transcription may give rise to transcriptional bursts (Suter et al., 2011).
In addition to the need for new methods to understand genome-wide data collectively, it is important that genome-wide data sets are supplied in ways that allow access to the whole community, in particular to biologists who do not have the technical ability to analyze ChIP-Seq data. It is a real possibility that each binding event must be considered independently since the sequence of every promoter, enhancer, etc. is unique. Because each sequence is bound independently, they also function independently. Although there is a possibility that a ChIP-Seq (or ChIP-PCR) signal for a transcription factor or chromatin modifier at one genomic locus is not due to direct binding of the factor at the locus but instead is generated by cross-linking with another genomic locus through long-distance interactions, the spatial proximity of the two genomic loci suggests a possible functional link. Thus, displaying ChIP-Seq data on genome browsers provides useful information and allows for spot -checking and permits in silico ChIP experiments . This is appropriate, as the correlation between ChIP-PCR and ChIP-seq is high.
As we discussed above, the binding sites identified by ChIP-Seq assays are subject to a number of complications including variations in cell source, ChIP conditions, control and parameter choices in peak identification algorithms. All of these can drastically affect the final definition of a cistrome associated with a specific factor. Conditions must be standardized in order to make meaningful comparisons between different cell types, conditions and results from different laboratories. It will be difficult to standardize everything, so it is also important to develop a set of library statistics. These statistics will allow an outside observer to quickly assess the quality of a data set. Useful parameters will likely include signal vs. noise and saturation estimates, but such standards have not been implemented.
The power and scope of the genome-wide data sets generated using ChIP-Seq and related techniques, in particular now that they are combined with other technologies, such as mass spectrometry, and manipulations, as in genetically-modified mice are changing the way of immunology research. Because so many observations can be made in parallel it is reasonable to claim that high quality data presents a complete picture of the genome as observed by each transcription factor. The main difficulty remaining is to establish novel, holistic paradigms to understand and manipulate the resulting immune phenomena.
Acknowledgments
The Division of Intramural Research, National Heart, Lung and Blood Institute supported the research in the authors laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Daniel L. Northrup, Email: northrupd@nhlbi.nih.gov.
Keji Zhao, Email: zhaok@nhlbi.nih.gov.
References
- Adamson AS, Collins K, Laurence A, O’Shea JJ. The Current STATus of lymphocyte signaling: new roles for old players. Curr Opin Immunol. 2009;21:161–166. doi: 10.1016/j.coi.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Akimzhanov A, Krenacs L, Schlegel T, Klein-Hessling S, Bagdi E, Stelkovics E, Kondo E, Chuvpilo S, Wilke P, Avots A, et al. Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling. Am J Pathol. 2008;172:215–224. doi: 10.2353/ajpath.2008.070294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Muir TW. Spreading Chromatin into Chemical Biology. Chembiochem. 2011 doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- Araki Y, Wang Z, Zang C, Wood WH, 3rd, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol Cell Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O’Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Wurster AL, Precht P, Wood WH, 3rd, Becker KG, Pazin MJ. Dynamic BRG1 Recruitment During T Helper Differentiation And Activation Reveals Distal Regulatory Elements. Mol Cell Biol. 2011 doi: 10.1128/MCB.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O’Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias KM, Laurence A, Davidson TS, Stephens G, Kanno Y, Shevach EM, O’Shea JJ. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–1020. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo LL, Jarvenpaa H, Tuomela S, Raghav S, Ahlfors H, Laurila K, Gupta B, Lund RJ, Tahvanainen J, Hawkins RD, et al. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 2010;32:852–862. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo G, Levy S, Bucci G, Romanenghi M, Fornasari L, Beeson KY, Goldberg SM, Cesaroni M, Ballarini M, Santoro F, et al. NA-Seq: a discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev Cell. 2009;16:466–481. doi: 10.1016/j.devcel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996a;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996b;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21:274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J, Knoechel B, Abbas AK. Regulatory T cells in the periphery. Immunol Rev. 2006;212:149–162. doi: 10.1111/j.0105-2896.2006.00414.x. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Miyara M, Sakaguchi S. Natural regulatory T cells: mechanisms of suppression. Trends Mol Med. 2007;13:108–116. doi: 10.1016/j.molmed.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22:177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–1903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian Genes Are Transcribed with Widely Different Bursting Kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Takaki H, Ichiyama K, Koga K, Chinen T, Takaesu G, Sugiyama Y, Kato S, Yoshimura A, Kobayashi T. STAT6 Inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem. 2008;283:14955–14962. doi: 10.1074/jbc.M801123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DI. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci U S A. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watford WT, Hissong BD, Durant LR, Yamane H, Muul LM, Kanno Y, Tato CM, Ramos HL, Berger AE, Mielke L, et al. Tpl2 kinase regulates T cell interferon-gamma production and host resistance to Toxoplasma gondii. J Exp Med. 2008;205:2803–2812. doi: 10.1084/jem.20081461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci U S A. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher-Reilley J, Wu Z, Spolski R, Jing X, Zhang YC, McCoy JP, Leonard WJ. The transcription factor GABP is a critical regulator of B lymphocyte development. Immunity. 2007;26:421–431. doi: 10.1016/j.immuni.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Anderson DE, Baecher-Allan C, Hastings WD, Bettelli E, Oukka M, Kuchroo VK, Hafler DA. IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature. 2008 doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]