SUMMARY
It has been suspected that cell cycle progression might be functionally coupled with RNA processing. However, little is known about the role of the precise splicing control in cell cycle progression. Here, we report that SON, a large Ser/Arg (SR)-related protein, is a splicing co-factor contributing to efficient splicing of cell cycle regulators. Down-regulation of SON leads to severe impairment of spindle pole separation, microtubule dynamics, and genome integrity. These molecular defects result from inadequate RNA splicing of a specific set of cell cycle-related genes that possess weak splice sites. Furthermore, we show that SON facilitates the interaction of SR proteins with RNA polymerase II and other key spliceosome components, suggesting its function in efficient co-transcriptional RNA processing. These results reveal a mechanism for controlling cell cycle progression through SON-dependent constitutive splicing at suboptimal splice sites, with strong implications for its role in cancer and other human diseases.
INTRODUCTION
Efficient and proper RNA splicing is one of the critical steps in gene expression and mutations in cis-acting splicing elements or trans-acting splicing regulators are well known to cause diseases in humans (Cartegni et al., 2002; Cooper et al., 2009; Garcia-Blanco et al., 2004). Interestingly, multiple cell cycle regulators initially identified as cell division cycle (cdc) genes were later found to impair splicing in S. cerevisiae and S. pombe, suggesting a functional connection between RNA splicing and cell cycle progression (Lundgren et al., 1996; Shea et al., 1994; Vijayraghavan et al., 1989). However, it has been unclear whether the cell cycle defect is a consequence of defective RNA splicing or whether individual cdc genes have a direct role in splicing regulation. In at least one case, the impact of the cdc5 mutant on cell cycle progression could be suppressed by removing the intron in the tubulin-encoding TUB1 gene, suggesting that cell cycle defects may be manifested by specific splicing defects (Burns et al., 2002). Growing evidence has demonstrated that the control of RNA splicing, especially alternative splicing of apoptotic regulators, contributes to cell survival (Schwerk and Schulze-Osthoff, 2005; Shin and Manley, 2004). However, direct connection, if any, between the control of constitutive splicing and cell cycle progression or cell survival has been lacking.
The RNA splicing process requires an accurate recognition of exon-intron boundaries, which are aided by conserved cis-elements, such as the 5′ splice site, 3′ splice site, branch site and polypyrimidine tract, as well as the coordinated action of multiple trans-acting factors, including small nuclear RNPs (snRNPs) and SR proteins (Cartegni et al., 2002). Recent studies demonstrated that SR proteins are recruited to the nascent RNA via direct or indirect interactions with the C-terminal domain (CTD) of RNA polymerase II (RNAP II) to facilitate co-transcriptional RNA processing (Bentley, 2005; Das et al., 2007; Zhong et al., 2009) and defects in this process may cause prolonged association of nascent RNA with template DNA, a configuration known as R-loops responsible for triggering double-stranded DNA breaks (Li and Manley, 2005). As the functional connection between co-transcriptional RNA processing and genome instability may underlie key aspects of tumorigenesis in humans, it has been a major battleground to understand the mechanism of efficient and specific coupling between transcription and RNA splicing.
We have been pursuing a large, poorly characterized protein called SON because of its ability to bind the leukemogenic protein AML1-ETO and modulate its activity in cell growth (Ahn et al., 2008). SON was first cloned from a human embryonic cDNA library and classified as a new member of the v-mos Moloney murine sarcoma viral oncogene family (Berdichevskii et al., 1988). Although initially characterized as a DNA binding protein (Sun et al., 2001), SON contains multiple structural features related to RNA processing, including a large arginine/serine-rich (RS) domain, a glycine-rich motif (G-patch), and a double-stranded RNA binding motif (DSRM) (Aravind and Koonin, 1999; Saitoh et al., 2004; Saunders and Barber, 2003). Consistent with a potential role in RNA metabolism, SON has been shown to localize to nuclear speckles, which are enriched in the splicing machinery (Mattioni et al., 1992; Saitoh et al., 2004; Sun et al., 2001; Wynn et al., 2000). These observations raise the possibility that SON may function as an SR-related splicing factor in regulated splicing.
Here, we have uncovered a major function of SON in regulating a large number of genes dedicated to cell cycle progression, as SON siRNA caused massive disarray of microtubules and impaired spindle pole separation, thereby arresting the cell at mitotic phase. Mechanistic analysis revealed that SON acts as a co-activator for efficient RNA processing of multiple structural components of the cell cycle apparatus and its signaling molecules. SON-dependent splicing substrates contain weak splice sites, predicting inefficient or unstable spliceosome formation on them. Our data also suggest that SON facilitates splicing through the recruitment of SR proteins, such as SC35, to RNAP II complexes. These results reveal an insight into the regulation of cell cycle progression by cofactor-mediated splicing at suboptimal splice sites associated with a large number of constitutive introns. Our findings, coupled with the recent documentation of another large SR protein-related splicing co-activator (nSR100) required for development of the nervous system (Calarco et al., 2009), have a broad implication for a large number of SR-related proteins in coordinated regulation of RNA processing to govern cell proliferation and differentiation.
RESULTS
SON deficiency causes severe defects in mitotic spindle pole separation, chromosome alignment and microtubule dynamics
Our previous work revealed a functional interaction between SON and the leukemogenic protein AML1-ETO in the regulation of cell proliferation (Ahn et al., 2008). We identified SON as an AML1-ETO NHR4 domain-interacting protein and showed that disruption of the interaction between endogenous SON and AML1-ETO could rescue the cell growth defect induced by the full-length AML1-ETO protein (Ahn et al., 2008). These findings suggest a critical role of SON in the regulation of cell proliferation. To further pursue the function of SON, we performed SON siRNA transfection (Fig. S1A), and found a significant growth inhibition (Fig. S1B) and an increase in the 4n population (Fig. S1C). Western blot and flow cytometric analysis for histone H3-Ser10 phosphorylation, a marker of mitotic cells, coupled with Wright-Giemsa staining, revealed that cells were arrested in mitosis after SON siRNA transfection (Figs. S1D, S1E and S1F). Collectively, these data demonstrate a critical role of SON in facilitating cell cycle progression through the mitotic phase.
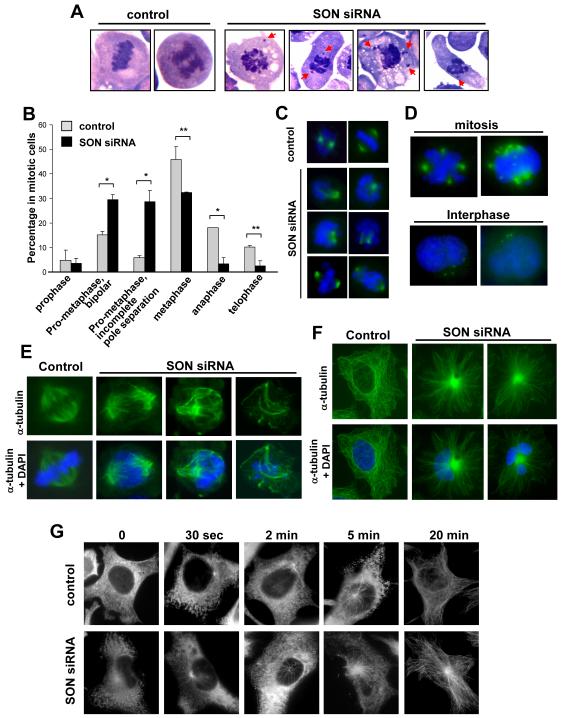
A close examination of mitotically arrested K562 cells revealed a strikingly irregular spread of chromosomes in SON siRNA-treated cells (Fig. 1A). A similar phenotype was also evident in HeLa cells where SON knockdown appears to arrest the cell at pro-metaphase as indicated by a significant increase in pro-metaphase cells with a concurrent decrease in metaphase, anaphase and telophase cells (Fig. 1B). This early mitotic defect prompted us to examine the status of mitotic spindle poles by staining the cell with an Aurora kinase A antibody, revealing failed mitotic pole separation in ~50% of pro-metaphase cells (Figs. 1B and 1C). We observed similar defects in spindle pole separation and chromosome alignment in K562 cells following SON knockdown (Figs. S2A and S2B). Strikingly, many SON-depleted cells exhibited multiple mitotic spindle poles and interphase centrosome amplification (Figs. 1D and S2C). These data revealed severe dysregulation of the spindle pole and centrosomes in SON-deficient cells.
Figure 1. SON knockdown causes multiple defects in mitotic chromosome arrangement, spindle pole separation and microtubule dynamics.
(A) Mitotic chromosome misalignment in SON-depleted cells. Wright-Giemsa staining revealed misaligned chromosomes in SON siRNA-transfected K562 cells. Chromosomes that have shifted to an abnormal location are marked with red arrows.
(B) Increase in pro-metaphase cells and incomplete separation of mitotic poles. HeLa cells were transfected with control siRNA or SON siRNA, and immunostained with an Aurora kinase A antibody and DAPI. Then, mitotic cells at different phases and with different mitotic pole status were counted under the fluorescence microscope. Bars represent mean ± SD from three independent experiments (n > 300 cells per experiment, *P < 0.005, **P < 0.02, t-test).
(C) Incomplete mitotic spindle pole separation and failure in chromosome alignment caused by SON knockdown. HeLa cells were prepared and immunostained as described above (green, Aurora kinase A for mitotic spindle poles; blue, DAPI for DNA).
(D) Abnormal spindle pole/centrosome amplification in SON siRNA-transfected K562 cells during mitosis and interphase. Cells were stained with an Aurora kinase A antibody (green) and DAPI (blue).
(E) Defects in mitotic spindle formation in SON siRNA-transfected cells. HeLa cells transfected with control siRNA or SON siRNA were fixed (day 3), and stained with anti-α-tubulin for microtubules (green) and DAPI for DNA (blue).
(F) Abnormal microtubule organization in interphase cells after SON depletion. Cells were prepared and stained as described in (E).
(G) Microtubule re-growth assay in HeLa cells transfected with SON siRNA or control siRNA (day 2). Cells were cold-treated to depolymerize microtubules and fixed at different time points after incubation in warm media (0, 30 sec, 2 min, 5 min, and 20 min). Microtubules were stained with α-tubulin antibody and analyzed by fluorescence microscopy.
Because chromosome alignment and segregation during mitosis require a tight control of microtubule assembly and disassembly, we next examined spindle microtubules in SON-depleted cells. By staining the cell with an α-tubulin antibody, we observed complete disruption of mitotic spindle organization. All mitotic spindles were irregular in shape, and microtubules associated with the mitotic spindles were totally disorganized and tangled without polarity with some exhibiting abnormally long fibrils while others were insufficiently elongated (Figs. 1E and S2B).
As microtubules also serve as critical cytoskeletal structures for organizing and maintaining the cell morphology required for various cellular functions, we further determined the functional requirement of SON for microtubule organization in interphase cells. Normally, microtubules are nucleated at the centrosome, and subsequently released from the initial nucleation site and reorganized to form a non-radial microtubule array during interphase. In contrast to this normal morphology observed in control siRNA-treated HeLa cells, microtubules in SON siRNA-transfected cells formed a dense aster originating from the centrosome, resulting in a completely radial array without forming any microtubule network/branching (Fig. 1F). This abnormal microtubule pattern failed to define the structural frame of the nucleus or contour of the cell. We performed a microtubule re-growth assay to further characterize the defect in microtubule assembly and disassembly after SON knockdown, observing that microtubules retained their ability to nucleate at the centrosome, but were abnormally elongated without forming networks as seen in normal cells (Fig. 1G). These results revealed severe defects in the regulation of microtubule biogenesis, organization, and dynamics throughout the cell cycle in response to SON knockdown.
SON is required for cytokinesis and maintaining genome stability
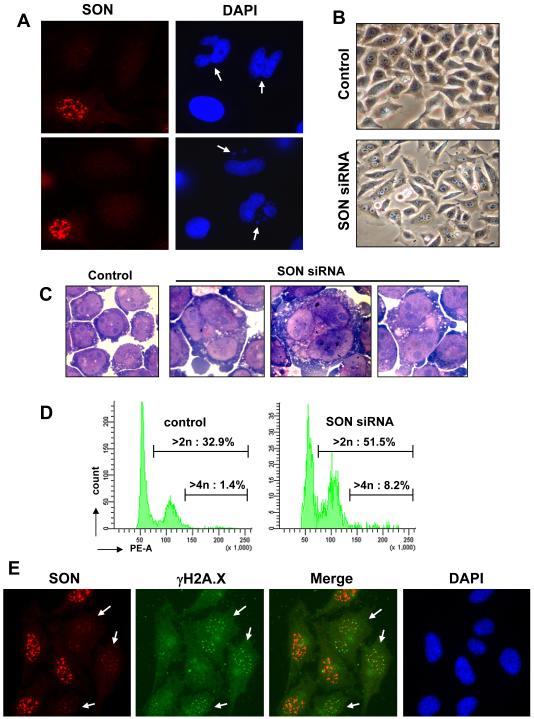
As expected from massive defects in microtubule organization and dynamics and multiple other cellular defects in SON knockdown cells, we found that about 10 to 15% cells were induced to commit apoptosis as detected 2 to 3 days after SON siRNA treatment (Fig. S1G). Among the cells that made it through mitosis, many exhibited abnormal nuclear structures, including the formation of protruding nuclear buds/lobes and micronuclei (Fig. 2A). We also observed micronuclei on BJ primary fibroblasts cells by DAPI staining (Fig. S2C), and flow cytometric analysis of DNA content revealed a dramatic increase in the population of cells containing 4n or higher DNA content (Fig. 2D). In addition, multinucleated cells were observed typically 6 - 7 days after siRNA transfection in HeLa (Fig. 2B) and U937 human leukemic monocyte lymphoma cells (Fig. 2C), suggesting defective cytokinesis among the cells that escaped the initial block at pro-metaphase.
Figure 2. SON knockdown causes abnormal nuclear structures, aneuploidy/polyploidy and DNA breaks.
(A) Abnormal nuclear structure caused by SON knockdown. HeLa cells were transfected with SON siRNA, and after 3 days, immunostained with SON antibody together with DAPI. SON knockdown causes nuclear buds/lobes and micronuclei (marked with arrows).
(B) Multinuclear HeLa cells were observed after transient SON knockdown. HeLa cells were transfected with SON siRNA and shown is a photo of representative multinuclear cells observed 7 days after SON siRNA transfection.
(C) Multinuclear cells were observed after SON siRNA transfection in U937 human leukemic monocyte lymphoma cell line (day 6).
(D) SON knockdown causes aneuploidy. BJ human primary fibroblasts were transfected with control siRNA or SON siRNA, and DNA content was measure by propidium iodide staining and flow cytometric analysis (day 7), showing that SON knockdown increases cells with >2n and >4n DNA.
(E) SON knockdown causes double-stranded DNA breaks. HeLa cells were transfected with SON siRNA, and stained with γH2A.X antibody. Cells with SON knockdown (indicated by arrows) showed γH2A.X foci.
Because protruding nuclear buds and micronuclei are hallmarks of DNA damage and abnormal chromosome rearrangement, we further examined whether SON deficiency might induce genome instability. We stained SON siRNA-treated HeLa cells with an antibody against phosphorylated histone H2A.X (γH2A.X), a marker of double-stranded DNA breaks, revealing a marked increase in γH2A.X foci (Fig. 2E). Together, these data indicate that SON is critical for maintaining genome stability by preventing aneuploidy/polyploidy and DNA breaks.
SON regulates the expression of a specific subset of genes involved in cell cycle progression and genome stability
Given the pleiotropic defects observed in SON knockdown cells and the fact that SON does not seem to be a structural component of the affected cellular structures, we hypothesized that SON might affect specific cellular functions through regulating gene expression. We tested this hypothesis by performing microarray analysis on HeLa cells transiently transfected with SON siRNA, revealing that expression levels of 659 genes were changed by more than 1.45-fold with statistical significance (p value <0.001) in response to SON knockdown.
By analyzing the biological functions of these 659 genes using the Ingenuity Pathway Analysis (IPA) software, we found that the most highly enriched functional categories are related to cancer, cell cycle and DNA replication/recombination/repair (Fig. 3A). Interestingly, down-regulated genes are most associated with DNA replication/recombination/repair and cell cycle (Fig. 3B), while up-regulated genes are linked to cell death/survival, cell signaling and molecular transport (Fig. S3A). Focusing on down-regulated genes, we noted many genes involved in repair of single-stranded and double-stranded DNA breaks, DNA damage responses, DNA rearrangement, DNA replication, G2/M checkpoint, cell cycle, mitosis and microtubule dynamics (Fig. 3C), suggesting that down-regulation of these genes may directly contribute to the observed phenotype. Quantitative real-time PCR, immunoblotting and immunostaining analyses validated the microarray results on a panel of down-regulated genes (Figs. 3D, 3E and S3B). These data thus suggest that SON is involved in regulated expression of many genes dedicated to maintaining genome stability and facilitating cell cycle progression.
Figure 3. SON knockdown causes down-regulation of genes involved in DNA maintenance/integrity and cell cycle progression.
(A) Analysis of top functions of 659 genes that showed significant changes after SON knockdown by Ingenuity Pathway Analysis (IPA) software. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that network is due to chance alone.
(B) Analysis of top function of 472 genes that are significantly down-regulated after SON knockdown using IPA, as described in (A).
(C) Representative genes that are down-regulated by SON knockdown. Down-regulated genes which belong to the functional groups for DNA replication/recombination/repair and cell cycle were determined by IPA, and representative genes were listed in the Venn diagram.
(D) Decrease in protein levels of TUBG1, TUBGCP2 and AKT1 after SON knockdown. Whole cell lysates from control or SON siRNA-transfected HeLa cells were prepared 3 days after transfection, and immunoblotted for SON, TUBG1 (γ-tubulin), TUBGCP2 (γ-tubulin complex protein 2) and AKT1. TUBA1B (α-tubulin) and TUBGCP4 (γ-tubulin complex protein 4) were also blotted as unaffected controls.
(E) Decrease in pericentrin (PCNT) level in the centrosome after SON knockdown detected by immunostaining (red for pericentrin, green for α-tubulin, blue for DNA).
SON is required for efficient RNA processing of affected genes
SON was previously reported to be present at nuclear speckles enriched with most splicing factors (Ahn et al., 2008; Saitoh et al., 2004; Wynn et al., 2000), suggesting that SON may play a role in RNA metabolism in mammalian cells. We confirmed that the pattern of SON localization exactly matches with that of typical pre-mRNA splicing factors throughout the cell cycle (Fig. S4A), and showed SON co-localizes with the SR splicing factor SC35 and small nuclear RNAs (snRNAs) (Figs. S4B and S4C). These findings strongly implicated SON as part of the pre-mRNA splicing machinery.
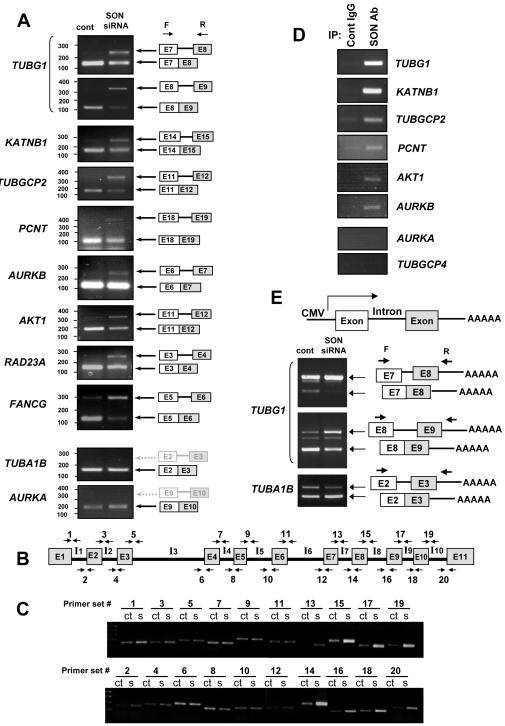
We therefore focused on testing the possibility that down-regulation of gene expression after SON knockdown might be due to inefficient pre-mRNA splicing. We designed a series of PCR primer pairs targeting two neighboring constitutive exons on a panel of affected genes, including TUBG1, KATNB1, TUBGCP2, AURKB, PCNT, AKT1, RAD23A, and FANCG, as well as on a few unaffected genes as controls. RT-PCR analyses revealed multiple intron retention events on genes that were down-regulated by SON siRNA, but no change on unaffected genes (Fig. 4A). To further confirm that SON is required for specific intron removal events, we designed PCR primers targeting each exon-intron (5′ splice site) or intron-exon (3′ splice site) junction within the entire TUBG1 gene (Fig. 4B), demonstrating that SON depletion caused inefficient intron removal at several, but not all, splice sites (Fig. 4C). In all affected cases, intron removal was impaired, but not abolished, suggesting strongly that SON acts as a co-activator for efficient intron removal at constitutive splice site on a selective group of genes and the resulting splicing defects are likely responsible for down-regulation of these genes in SON knockdown cells.
Figure 4. SON is required for efficient intron removal at constitutive splice site on a selective group of genes and binds to the RNA of those genes.
(A) Detection of the unspliced form of RNAs in SON-depleted cells by PCR analysis. RNAs were prepared from control or SON siRNA transfected HeLa cells, and RT-PCR analyses were done using primers targeting two neighboring constitutive exons for indicated genes (E, exon; F, forward primer; R, reverse primer). PCRs for TUBA1B and AURKA were done as controls that were not down-regulated by SON siRNA. Arrows with solid lines in black indicate detected bands and dotted lines in gray indicate the expected size of undetected bands.
(B) A schematic diagram of exons (E1 – E11) and introns (I1 – I10) of the TUBG1 gene and the primer sets designed for PCR shown in (C).
(C) PCR analysis using primer sets shown in (B) to compare splicing efficiencies of TUBG1 splicing junctions in control (ct) and SON siRNA-transfected (s) cells.
(D) Interaction of SON with RNA. UV-crosslinking and immunoprecipitation (CLIP) was performed with control IgG or SON antibody and associated RNAs were analyzed by RT-PCR.
(E) SON depletion causes impaired splicing of TUBG1 minigene. An intron and the flanking exons were cloned from the TUBG1 gene (exon 7 – 8 region and exon 8 – 9 region) and the TUBA1B gene (exon 2 – 3 region) to downstream of the CMV promoter to make minigene constructs. HeLa cells pre-treated with control or SON siRNA were transfected with the minigenes, and RT-PCR was performed to detect unspliced and spliced RNA.
Furthermore, we performed UV crosslinking and immunoprecipitation (CLIP) with SON antibody and analyzed associated RNAs by RT-PCR. The results showed that SON is associated with the RNAs from all examined down-regulated genes, including TUBG1, KATNB1, TUBGCP2, PCNT, AKT1 and AURKB. The RNAs from unaffected genes, such as TUBA1B, KATNA1, AURKA, TUBGCP4 and GAPDH, were not detected or marginally enriched in the parallel experiment (Fig. 4D and S4D). These results indicate that SON is directly involved in the processing of its target RNAs.
To further analyze SON-mediated splicing, we constructed several minigenes containing appropriate exonic and intronic sequences. Since the splicing efficiency between exons 7 and 8, and exons 8 and 9 in the endogenous TUBG1 pre-mRNA was inefficient and sensitive to SON down-regulation (Fig. 4A and 4C), we constructed two minigenes from these regions under the CMV promoter and tested their response to SON knockdown. RT-PCR analysis confirmed that splicing of these TUBG1 minigenes was indeed dependent on SON (Fig. 4E). In contrast, splicing of a minigene derived from the TUBA1B gene, which was not responsive to SON knockdown based on our microarray data, was independent of SON (Fig. 4E). These results demonstrated that SON-mediated splicing occurs in a gene-specific manner, which can be fully recaptured in the minigene assay.
The C-terminal domain containing the RS domain and the G-patch is necessary for SON’s activity in splicing
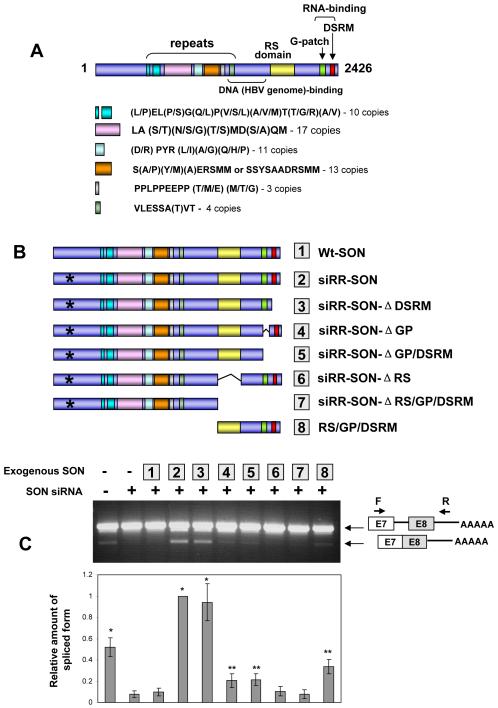
Taking advantage of this minigene system, we determined the domain requirement for SON-mediated splicing. SON is a large protein of 2,426 amino acids, containing various repeat sequences in the central part of the protein, and interestingly, its C-terminal domain carries a number of features related to the SR superfamily of splicing factors and regulators, including an RS domain and two putative RNA binding motifs, consisting of a G-patch and a double-stranded RNA binding domain (DSRM) (Fig. 5A). We constructed expression vectors for the full-length human SON cDNA (Wt-SON) and siRNA-resistant SON (siRR-SON) in which six nucleotides within the SON siRNA-target sequence were mutated without altering the coding capacity (Fig. S5A). When co-transfected with the TUBG1 minigene reporter, we found that siRR-SON, but not Wt-SON, could fully rescue the splicing defect of the minigene caused by SON siRNA (Fig. 5C), thus confirming the specific effect of exogenous SON on splicing of the minigene.
Figure 5. The C-terminal domain containing the RS domain and the G-patch is necessary for SON’s activity in splicing.
(A) Domains and unique amino acid repeats in the SON protein. Full length SON (known as isoform f) is composed of 2,426 amino acids. It contains an RS domain and two RNA-binding motifs at the C-terminus and a putative DNA-binding domain. In addition, SON contains unique amino acid repeats that span most of the N-terminal region. The features of each repeat are presented.
(B) Various SON fragments generated for the splicing rescue experiment. siRNA-resistant SON (siRR-SON) was generated by mutagenesis (marked by *), and various deletion mutants were generated from siRR-SON.
(C) Splicing rescue by different SON fragments. HeLa cells were transfected with control or SON siRNA, and then transfected with TUBG1 minigene (exon 7 - 8) together with various SON cDNA fragments listed in (B). RT-PCR was performed to detect unspliced and spliced RNA and the photo is the representative result. Relative amount of spliced form was calculated by measuring the density of spliced form (the amount of spliced form in the siRR-SON lane was set as 1). Bars represent mean ± SD from 5 independent experiments (*P < 0.005, **P < 0.03, t-test, when compared to SON siRNA only, the 2nd lane).
We next generated a series of deletion mutants (Fig. 5B and S5B) and tested their abilities to rescue splicing of the minigene. We found that DSRM deletion (construct 3) does not affect splicing activity of SON, indicating that this double-stranded RNA binding domain is dispensable for SON’s splicing activity (Fig. 5C). In contrast, the G-patch deletion mutants (constructs 4 and 5) reduced its activity. Deletion of the RS domain causes a complete loss of splicing activity (constructs 6 and 7). Strikingly, the C-terminal RS/GP/DSRM fragment (construct 8), which is only ~1/3 of the full-length protein, was sufficient to rescue SON-dependent splicing to a significant degree (Fig. 5C). We concluded from this analysis that the RS domain and the G-patch are the core motifs necessary for SON-mediated splicing, and the upstream sequences, which contain highly repetitive amino acid sequences, further support the function of SON in achieving efficient RNA splicing.
SON-dependent genes possess weak splice sites
To elucidate the specificity of SON regulation of RNA splicing, we examined the features of the splice sites associated with either SON-dependent or SON-independent genes, using the matrices for splice sites available in ESEfinder (http://rulai.cshl.edu/tools/ESE/, Cartegni et al., 2003). Interestingly, SON-dependent genes all possess a weak 5′ or 3′ splice site. Some of them showed low motif scores that are below the threshold score of typical constitutive splice sites (Fig. S6). Interestingly, some splice sites score as the standard constitutive splice sites, but correspond to so-called dual-specificity splice sites (Fig. S6), which may “confuse” the splicing machinery as they may be recognized as either 5′ or 3′ splice sites (Zhang et al., 2007). These observations suggest that the presence of weak or ambiguous splice sites may render splicing of those cell cycle genes particularly dependent on SON for their efficient recognition by the splicing machinery.
To experimentally test this hypothesis, we examined which sequence feature(s) contribute to SON dependency on the TUBG1 minigene model. We created mutant minigenes that possess stronger 5′ splice site with optimal sequences (5′ mt), stronger 3′ splice site with the improved polypyrimidine tract (3′ mt), or stronger splice sites at both ends (5′ mt + 3′ mt) (Figs. 6A and 6B). When a single site was converted to a strong one, the splicing efficiency of the mutant minigene was still SON-dependent. However, when both 5′ and 3′ splice sites were modified to stronger sites, splicing of the mutant minigene was no longer SON-dependent (Fig. 6C). To further confirm that consensus sequences within the intron are sufficient to render the minigene SON-independent, we replaced intron 7 of TUBG1 with intron 2 of the SON-independent TUBA1B gene (α2-Intron), which contains canonical splicing signals (Figs. 6A and 6B). Replacement of the intron indeed abolished SON dependency (Fig. 6C). We also performed a converse experiment by weakening the splice site on a SON-independent TUBA1B minigene, finding that weakening either the 5′ or the 3′ splice site impaired the splicing efficient and combination of these mutations abolished splicing; however, none of these mutants showed SON-dependency (Fig. S7). We conclude from these experiments that SON functions as a co-factor to institute correct splice site selection on a specific subset of genes containing weak or suboptimal constitutive splice sites. However, a weak splice site alone is clearly insufficient to confer SON-dependency, indicating that other sequence features in conjunction with weak splice sites contribute to SON regulated splicing.
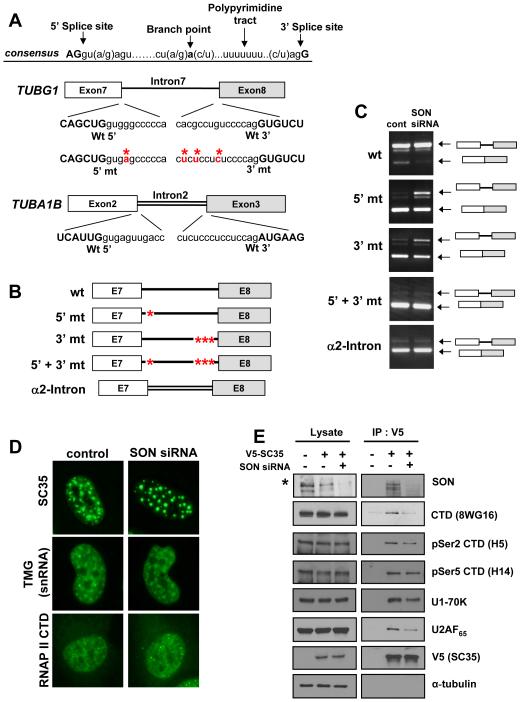
Figure 6. SON is required for processing of weak constitutive splice sites and facilitates the interaction of SR proteins with RNA polymerase II.
(A) Splice site sequences in primary transcripts of wild type and mutant minigenes. Consensus sequences in splice sites are presented. The wild type and modified 5′ and 3′ splice site sequences between exon 7 and exon 8 of TUBG1, as well as 5′ and 3′ splice site sequences between exon 2 and exon 3 of TUBA1B, are presented. Upper cases indicate exon sequences, lower cases indicate intron sequences. Mutated nucleotides are marked in red with an aster.
(B) Various mutant minigene constructs generated for splicing assay. Asters indicate nucleotide changes shown in (A). Intron 7 of TUBG1 was replaced by intron 2 of TUBA1B to generate α2-Intron.
(C) SON is required for processing of weak splice sites. Minigenes shown in (B) were transfected into HeLa cells pre-treated with control or SON siRNA. RT-PCR was performed to detect unspliced and spliced RNA.
(D) SON knockdown altered SC35 localization, resulting in completely round shaped SC35 dots, and moderately affects snRNP localization, but does not affect localization of the CTD of RNAP II. Cells were immunostained for SC35, 2,2,7-tri-methylguanosine (TMG for snRNA) and the CTD of RNAP II (detected by 8WG16).
(E) SON facilitates SC35 interaction with the CTD of RNAP II, U1 snRNP and U2AF65. HEK293 cells expressing V5-tagged SC35 were transfected with control or SON siRNA, and V5-immunoprecipitation was performed to pull down SC35 complex. HEK293 cells without V5-SC35 expression were included as a control. Cell lysates and V5-immunoprecipites were immunoblotted with antibodies indicated. Due to the weak affinity of SON antibody in immunoblot, more concentrated lysates were used to detect SON from the lysates (panel marked with *).
SON facilitates the interaction of SR proteins with RNA polymerase II
A common mechanism for SR-like factors is to recruit other splicing factors and regulators to the splice sites, which may be an underlying mechanism to achieve regulated splicing on a subset of substrates containing a combination of specific cis-acting elements. Interestingly, in SON-depleted cells, we observed that nuclear speckles stained with anti-SC35 rounded up, but the cellular localization of RNAP II was not affected. Localization of the splicing machinery labeled by the tri-methylguanosine cap was also moderately affected (Fig. 6D). The fact that SC35 speckles become round upon SON knockdown suggests impaired shuttling of SR proteins between nuclear speckles and the site of active transcription for co-transcriptional RNA processing (Lamond et al., 2003).
To test this hypothesis, we generated a HEK293 cell line stably expressing V5-tagged SC35 as a model for SR protein recruitment and determined how the recruitment of SC35 to RNAP II is affected by SON siRNA. Immunoprecipitation with V5 antibody from cell lysate efficiently pulled down SON, demonstrating that SON is indeed associated with SC35 or SC35-containing complexes. We also detected the association of SC35 with RNAP II, U1-70K, and U2AF65 in control siRNA-treated cells (Fig. 6E). In contrast, SC35 interaction with RNAP II (detected by 8WG16; note that this antibody is known to cross-react with both hypo- and hyper-phosphorylated RNAP II, although the former seems to be the preferred antigen) was significantly attenuated in SON-depleted cells (Fig. 6E). Notably, the association of SC35 with Ser2-phosphorylated RNAP II (detected by the H5 antibody), which has been linked to transcriptional elongation, appears to be selectively affected, relative to Ser5-phosphorylated RNAP II (detected by the H14 antibody), which is predominantly associated with transcriptional initiation. The interaction of SC35 with U1-70K and U2AF65 was also attenuated in SON-depleted cells (Fig. 6E). Together, these results provide strong support to a model where SON facilitates co-transcriptional assembly of SR proteins and other key spliceosome components, such as U1 and U2AF to elongating RNAP II complexes, thereby ensuring efficient co-transcriptional RNA splicing (Fig. 7).
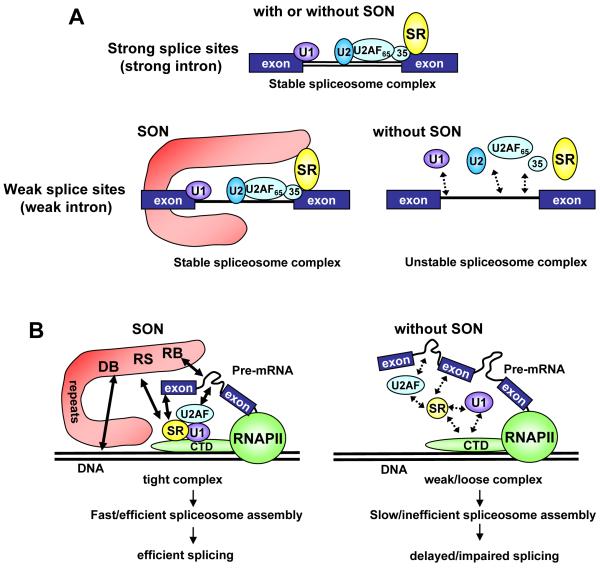
Figure 7. Proposed models for the role of SON as a splicing co-factor.
(A) If splice sites are strong (which can be due to optimal sequences in the intron), interactions between pre-mRNA and spliceosome components are strong and stable. Therefore, spliceosome complex is formed for splicing regardless presence or absence of SON. When splice sites are weak (due to suboptimal sequences), spliceosome components form weak and unstable interactions (represented by dotted lines) with pre-mRNA in the absence of SON, while stable interactions can be assured by presence of SON. The process may involve interaction(s) between SON and other critical splicing factors, including SR proteins.
(B) A model for the role of SON as a co-factor in efficient transcription-splicing coupling. During transcription in wild type cells (left), SON interacts with DNA, RNA, SR proteins and other early spliceosome components through its DNA-binding domain (DB), RNA-binding motifs (RB) and RS domain (RS), thereby facilitating recruitment of early spliceosome components to the CTD of RNAP II. The long and unique amino acid repeats in SON may help this protein stretch and make contact with multiple components. Such organization and connection by SON may help efficient and immediate spliceosome assembly on the nascent pre-mRNA with a weak splice site, resulting in efficient splicing. In the absence of SON (right), SR proteins and other early spliceosome components are not efficiently recruited to the CTD of RNAP II, and DNA, RNA and proteins are not closely associated with each other. Therefore, co-transcriptional spliceosome assembly is not efficient, and splice site recognition/selection on a weak splice site is not accurate, resulting in delayed or impaired splicing.
DISCUSSION
A large number of SR-related proteins are encoded in mammalian genomes. Although a few large SR-related proteins have been shown to act as splicing co-activators, little is known about how various SR-related proteins are involved in specific cellular processes. In this study, we identified SON as an SR-related protein specifically devoted to RNA processing of many cell cycle/DNA repair-related genes. Although SON has recently been implicated as a regulator of alternative splicing (Moore et al., 2010), our findings uncover a link between SON-dependent fine-tuning of constitutive splicing and cell cycle progression.
SON as a mediator of specific cellular processes via regulated splicing
It has been documented that SR-related proteins, such as SRm160 and SRm300, function as co-activators of pre-mRNA splicing via interactions with SR family proteins and snRNPs (Blencowe et al., 1998; Eldridge et al., 1999). However, the biological significance of individual SR-related proteins in specific cellular processes or diseases is poorly understood. A recent study on an SR-related protein, nSR100, which is required for development of the nervous system (Calarco et al., 2009), suggests that SR-related proteins may play critical roles in specific gene expression programs. The requirement of SON for efficient intron removal from pre-mRNA of the genes associated with mitotic progression revealed SON as a critical mediator for the cell cycle machinery. Consistent with our finding on the importance of SON in cell cycle progression, SON was reported as one of the genes that showed elevated expression in proliferating areas of embryonic and postnatal brain as well as in postnatal developing teeth (McKee et al., 2005). Our data now unveil the underlying mechanism of cell cycle regulation via SON-mediated splicing. Our findings, together with the elucidated role of nSR100 in the nervous system (Calarco et al., 2009), reveal an emerging role of SR-related splicing co-activators in the regulation of specific cellular programs through coordinating the RNA splicing process.
Microarray analysis also revealed a set of genes up-regulated in response to SON knockdown (Fig. S3A). However, upon examining SON knockdown at an earlier point, we found that SON predominantly down-regulates gene expression without causing significant up-regulation of many genes (data not shown), indicating that most of those up-regulated genes may indirectly result from SON knockdown.
SON-mediated constitutive splicing of weak splice sites as a sensor for cell cycle progression
In eukaryotic cells, it is generally believed that weak or suboptimal splice sites are critical features for alternative splicing (Keren et al. 2010). However, little is known about the functional importance of processing suboptimal splicing signals present at constitutive splice sites. Interestingly, our results now reveal that such suboptimal splicing signals are built on a set of cell cycle-related genes, which renders their splicing particularly sensitive to SON levels, thus constituting a SON-mediated sensory system for the regulation of cell proliferation.
Our results demonstrate that SON is required for efficient processing of a specific group of genes with weak splice sites during constitutive splicing. The splicing of the TUBG1 minigene lost SON-dependency when both the 5′ and 3′ splice sites were modified to consensus sequences. This is entirely consistent with SON-independent splicing of the TUBG1 minigene when its less optimal intron was replaced with strong intron 2 of the TUBA1B gene, indicating that a weak splice site is necessary for modulation by SON. We attempted a converse experiment in which the TUBG1 intron 7 containing a weak splice site was inserted between the SON-independent TUBA1B exons 2 and 3, finding that this minigene could not undergo splicing at all even in wild type cells, indicating that the TUBG1 intron 7 in combination with the TUBA1B exons generates a pre-mRNA that is too weak to be recognized by the splicing machinery (data not shown). We thus took a different strategy to address whether a weak splice site is sufficient to confer SON-dependency by directly converting the strong splice sites in the SON-independent TUBA1B minigene to weak ones. The data clearly showed that a weak splice site alone is insufficient, indicating that a combination of other factors, such as the ratios of ESE/ESS/ISE/ISS, the length of polypyrimidine tract, and other protein modulators, may collectively contribute to SON-dependency. Although it is currently unclear which of these features are most critical, our data suggest that the association of SON on specific gene transcripts, instead of all pre-mRNAs with a weak splice site, may reflect mutually beneficial interactions between SON and other critical splicing factors, including SR proteins (Fig. 7A).
Structural features of SON for efficient coupling between transcription and splicing
SON is a large multi-domain protein, suggesting that it may serve as a landing pad for multiple protein-protein interactions to facilitate co-transcriptional splicing in the cell. Our data demonstrated that the RS domain and the G-patch in SON play important roles in splicing, but the DSRM is dispensable (Fig. 5). The G-patch is a conserved domain found in type D retroviral polyproteins and several eukaryotic RNA-binding proteins, but its precise role in RNA binding and/or protein-protein interactions remains a subject of future investigation (Aravind and Koonin, 1999). SON has also been implicated in DNA binding through a region upstream of the RS domain (Sun et al., 2001), raising an intriguing possibility that SON may use this function to provide a tight connection between transcription and RNA splicing.
It is also important to emphasize our finding that SR proteins become partially aggregated in rounded nuclear foci without impairing the general splicing machinery. It was previously reported that nuclear speckles become round up in response to inhibition of splicing and transcription (Mintz and Spector, 2000; O’Keefe et al., 1994). In our current study, we found that SR proteins became aggregated but the cellular distribution of snRNPs detected by a tri-methylguanosine cap antibody was not severely affected in SON knockdown cells, suggesting inefficient recruitment of SR proteins to nascent transcripts for co-transcriptional splicing. Recently, Sharma et al. reported that SON is required for correct localization of SR proteins and snRNPs in nuclear speckles, which appears to depend on the N-terminal region with repetitive amino acid sequences (Sharma et al. 2010). Our data indicate that this N-terminal fragment alone does not possess any RNA splicing activity; however, it is clearly important for the full activity of the protein in splicing (Fig. 5C). The multi-domain nature of SON, including its ability to bind to RNA and a potential role in inducing modification of SR proteins (i.e. phosphorylation), suggests that SON may nucleate diverse protein-protein and protein-RNA interactions to enhance the process of co-transcriptional splicing (Fig. 7B).
Involvement of SON in tumorigenesis and other diseases
Genome instability and deregulation of the cell cycle are hallmarks of cancer and fundamental to many types of human diseases. Our discovery of SON’s function in regulating the mitotic machinery, such as centrosome components and genes critical for microtubule dynamics, as well as the DNA repair machinery, provides mechanistic insights into the role of SON in tumorigenesis and other human diseases. For example, SON has been shown to be important for trafficking of H1N1 influenza virions to late endosomes during the early infection stage (Karlas et al. 2010). Because virus trafficking is a microtubule-dependent process (Marsh and Helenius, 2006), disruption of normal microtubule organization and dynamics may underlie the observed defects in viral infection in SON knockdown cells. Our findings also predict SON to be a master regulator of multiple cellular processes that depend on microtubules. In addition to the significant role of SON in RNA processing of cell cycle regulators, it is clear from our microarray experiments that SON has a broad role in multiple cellular processes. Because of its function as a splicing sensor for cell cycle-related genes, it may serve as a target for developing therapeutic strategies against cancer.
EXPERIMENTAL PROCEDURES
Microarray and Functional Analysis
HeLa cells were grown in 100 mm dishes and transfected with negative control siRNA or SON siRNA (400 pmol). Then, cells were harvested after 66 hours and RNAs were isolated, and microarrays were performed using GeneChip Human Genome U133A 2.0 (Affymetrix). Data were obtained from three independent transfections and sample preparations, and analyzed by The Scripps Research Institute DNA Array Core Facility by two-sample t-test with random variance model. The data have been deposited in Gene Expression Omnibus (GEO) as the series accession number GSE26888. Functional analysis of the genes significantly changed by SON siRNA was performed using Ingenuity Pathway Analysis (IPA) software.
Construction of minigenes and analysis of minigene splicing
For construction of minigenes, the following regions were amplified by PCR from HeLa genomic DNA; TUBG1 exon 7- intron 7- exon 8, TUBG1 exon 8 – intron 8 – exon 9, and TUBA1B exon 2 – intron 2 – exon 3. These fragments were inserted into the pcDNA3.1(+) vector. The minigene fragment α2-Intron was synthesized (Integrated DNA Technologies), and other mutant minigenes were created by QuikChange site-directed mutagenesis (Stratagene). To analyze splicing of minigenes, HeLa cells were transfected with control siRNA or SON siRNA, and then minigene alone or minigene plus various SON constructs were transfected. RT-PCR was performed to detect spliced or unspliced forms.
Detection of SON-RNA interaction by UV corsslinking and immunoprecipitation (CLIP) and PCR
HeLa cells irradiated with UV and then SON antibody or control IgG were used to precipitate associated RNA. Details are described in Supplemental Experimental Procedures.
Immunoprecipitation of SC35 complexes
HEK293 cells stably expressing V5-tagged SC35 were generated by Flp-In System (Invitrogen). These cells were transfected with control siRNA or SON siRNA. After 48 hours, cells were harvested, lysed in RSB lysis buffer (10 mM Tris-HCl pH 7.6, 100 mM NaCl, 2.5 mM MgCl2) with 0.4% Triton X-100, protease inhibitors (Roche) and Phosphatase inhibitors (PhosSTOP, Roche), incubated on ice for 20 min and sonicated (5 sec × 3 times). After centrifugation, lysates were incubated with V5-antibody-conjugated agarose beads (Sigma) for 3 hours at 4 °C. Beads were washed 4 times with RSB with 0.4% Triton X-100 and twice with RSB without detergent.
Further detailed experimental procedures are described in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the Zhang lab members for discussion and help, and Dr. Andreas Merdes (Centre National de la Recherche Scientifique, France) for providing antibodies for TUBGCP2 and TUBGCP4. This work was supported by National Institute of Health grant CA096735, CA104509 (to D.E.Z.) and GM049369 (to X.D.F.) and the Lady TATA Memorial Trust Award Postdoctoral Fellowship (to E.Y.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn EY, Yan M, Malakhova OA, Lo MC, Boyapati A, Ommen HB, Hines R, Hokland P, Zhang DE. Disruption of the NHR4 domain structure in AML1-ETO abrogates SON binding and promotes leukemogenesis. Proc. Natl .Acad. Sci. USA. 2008;105:17103–17108. doi: 10.1073/pnas.0802696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Berdichevskii FB, Chumakov IM, Kiselev LL. Decoding of the primary structure of the son3 region in human genome: identification of a new protein with unusual structure and homology with DNA-binding proteins. Mol. Biol. (Mosk) 1988;22:794–801. [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CG, Ohi R, Mehta S, O’Toole ET, Winey M, Clark TA, Sugnet CW, Ares M, Jr., Gould KL. Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell Biol. 2002;22:801–815. doi: 10.1128/MCB.22.3.801-815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, Li Y, Sharp PA, Blencowe BJ. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA. 1999;96:6125–6130. doi: 10.1073/pnas.96.11.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–12. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioni T, Hume CR, Konigorski S, Hayes P, Osterweil Z, Lee JS. A cDNA clone for a novel nuclear protein with DNA binding activity. Chromosoma. 1992;101:618–624. doi: 10.1007/BF00360539. [DOI] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz PJ, Spector DL. Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 2000;129:241–251. doi: 10.1006/jsbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Toyn JH, Johnston LH. The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res. 1994;22:5555–5564. doi: 10.1093/nar/22.25.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Sun CT, Lo WY, Wang IH, Lo YH, Shiou SR, Lai CK, Ting LP. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Biol. Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- Wynn SL, Fisher RA, Pagel C, Price M, Liu QY, Khan IM, Zammit P, Dadrah K, Mazrani W, Kessling A, et al. Organization and conservation of the GART/SON/DONSON locus in mouse and human genomes. Genomics. 2000;68:57–62. doi: 10.1006/geno.2000.6254. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hastings ML, Krainer AR, Zhang MQ. Dual-specificity splice sites function alternatively as 5′ and 3′ splice sites. Proc. Natl. Acad. Sci. USA. 2007;104:15028–15033. doi: 10.1073/pnas.0703773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.