Abstract
Nitrogen (N) is the mineral nutrient required in the greatest amount and its availability is a major factor limiting growth and development of plants. As sessile organisms, plants have evolved different strategies to adapt to changes in the availability and distribution of N in soils. These strategies include mechanisms that act at different levels of biological organization from the molecular to the ecosystem level. At the molecular level, plants can adjust their capacity to acquire different forms of N in a range of concentrations by modulating the expression and function of genes in different N uptake systems. Modulation of plant growth and development, most notably changes in the root system architecture, can also greatly impact plant N acquisition in the soil. At the organism and ecosystem levels, plants establish associations with diverse microorganisms to ensure adequate nutrition and N supply. These different adaptive mechanisms have been traditionally discussed separately in the literature. To understand plant N nutrition in the environment, an integrated view of all pathways contributing to plant N acquisition is required. Towards this goal, in this review the different mechanisms that plants utilize to maintain an adequate N supply are summarized and integrated.
Keywords: Bacteria, nitrogen, nitrogen acquisition, plants
Introduction
Plants are sessile organisms and cannot escape adverse environmental conditions. In order to cope with constant and diverse challenges, plants must adjust their physiology, growth, and development. One of the most important challenges for plants is to maintain an adequate nutrient supply under fluctuating environmental conditions. Nitrogen (N) is the mineral nutrient required in the greatest amount and its availability is a major factor limiting plant growth in natural (Marschner, 1995; Epstein and Bloom, 2005) as well as agricultural (Galloway and Cowling, 2002) environments.
N is present in the biosphere in various chemical forms. Molecular nitrogen (N2) represents ∼80% of the atmosphere composition (Sanhueza, 1982). However, plants cannot directly use this form of N. N2 enters the biological N cycle in three main ways: through biological fixation (prokaryotic conversion of N2 to ammonia); by atmospheric fixation (lightning and photochemical conversion of N2 to nitrate); and by the Haber–Bosch industrial fixation of N2 to produce ammonia (Marschner, 1995). Once N is fixed as nitrate or ammonia, it can have two main fates: (i) nitrate and ammonia can undergo biochemical processes that transform them back to N2 (Marschner, 1995); or they can be reduced and/or assimilated for the biosynthesis of N-containing metabolites. Amino acids, urea, small polypeptides, and other N-containing biomolecules can be released back to the environment by secretion, excretion, or by the decay of organic matter. These organic forms of N can also be used as N sources by plants and other organisms (Jones et al., 2005).
Plants have evolved inorganic and organic N uptake systems to cope with the heterogeneous N availability in the soil. For nitrate (Crawford and Glass, 1998) and ammonium (Ludewig et al., 2007), two types of uptake system have been described: low-affinity transport systems (LATS), which operate at high nutrient concentrations (>1 mM); and high-affinity transport systems (HATS) that predominate in the micromolar range (Wang et al., 1993). Modulation of HATS and LATS function in coordination with changes in the pattern of growth and development allows plants to cope with heterogeneous N availability in the soil (Robinson, 1994; Zhang and Forde, 2000; López-Bucio et al., 2003; Zhang et al., 2007b; Vidal and Gutiérrez, 2008; Forde and Walch-Liu, 2009; Vidal et al., 2010b). In this review, these uptake systems will be referred to as the autonomous pathway of N acquisition. In natural environments, plants can also interact and associate with many and functionally diverse microorganisms that can also contribute to an adequate N supply (Gage, 2004; You et al., 2005). In this review, these mechanisms will be referred to as the association pathways. From this perspective, a single plant interacting with multiple microorganisms over time, e.g in the rhizosphere, may be considered as an ecosystem (Pickett and Cadenasso, 2002; Martin et al., 2004). These different pathways of N acquisition need to be integrated to provide causal relationships of plant N nutrition in ecosystem-level studies.The autonomous pathways for N acquisition have been extensively reviewed (Crawford and Glass, 1998; Forde and Walch-Liu, 2009; Krouk et al., 2010a; Vidal et al., 2010b). The latest advances regarding the autonomous pathways will be reviewed and the less frequently covered aspects of the association pathways (beyond nodulation) will be focused on. In this review perspectives are also provided on how these different mechanisms for N acquisition are integrated by the plant for optimal N nutrition.
Autonomous pathways for N acquisition
N nutrient uptake systems: molecular level
Two families of nitrate transporters, NRT1 and NRT2, have been identified in higher plants (Tsay et al., 2007). Both gene families code for symporters that transport nitrate concomitantly with protons (H+) in a mechanism that is driven by pH gradients across membranes (Miller et al., 2007). The NRT2 gene family codes for high-affinity nitrate transporters (Orsel et al., 2006) while NRT1 codes for low-affinity nitrate transporters, with the exception of NRT1.1 (also known as CHL1) which is a dual-affinity transporter involved in both low- and high-affinity nitrate uptake (Wang et al., 1998; Liu et al., 1999; Liu and Tsay, 2003). Two forms of nitrate HATS have been described, an inducible system that is stimulated by nitrate in the external medium (Crawford and Glass, 1998) and a constitutive system that works even when plants have not been previously supplied with nitrate (Crawford and Glass, 1998). The nitrate transporters studied in greatest detail are the Arabidopsis NRT2.1 and NRT1.1. NRT2.1 transcript is induced by low nitrate availability or N starvation and is repressed by high N provision (e.g. high ammonium or glutamine conditions) by a pathway involving the NRT1.1 transporter (Muños et al., 2004; Krouk et al., 2006). The transport activity of NRT1.1 is regulated by phosphorylation of its Thr101 (Liu and Tsay, 2003). Phosphorylated NRT1.1 functions as a high-affinity nitrate transporter and the dephosphorylated form of NRT1.1 functions as a low-affinity transporter (Liu and Tsay, 2003).
Uptake of ammonium/ammonia is mediated by the AMT/MEP/Rh family of membrane proteins, found not only in plants but also in microorganisms and animals (von Wirén and Merrik, 2004). In plants, members of the AMT1 family mediate ammonium transport. These proteins have been described as ammonium uniporters that transport ammonium along the electrochemical gradient (Ludewig et al., 2002, 2003) or as NH3/H+ co-transporters (Mayer et al., 2006). Ammonium uptake is known to be repressed by high external N and to be induced under N deficiency, by mechanisms that may act at both the transcriptional and post-transcriptional levels (Lee et al., 1992; Gazzarrini et al., 1999; Rawat et al., 1999; Yuan et al., 2007; Lanquar et al., 2009).
Soil organic compounds can also contribute to plant N nutrition (Näsholm et al., 1998; Lipson and Näsholm, 2001; Näsholm et al., 2009). Amino acids represent the largest fraction of low molecular weight dissolved organic N in the soil (Jones et al., 2005). The amino acid pool is dynamic because it is quickly taken up by plants and microorganisms (Jones and Hodge, 1999). Several known and putative amino acid transporters have been described in plants (Lipson and Näsholm, 2001). In Arabidopsis roots, three amino acid transporters have been identified with a role in the uptake of amino acids: lysine–histidine transporter 1 (LHT1), amino acid permease 1 (AAP1), and amino acid permease 5 (AAP5) (Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2008). LHT1 and AAP5 have different amino acid specificities, function at amino acid concentrations seen in field conditions, and are thought to be important components of the root amino acid uptake system in Arabidopsis (Svennerstam et al., 2008). Cationic amino acid transport is mediated by AAP5 while neutral and acidic amino acid transport is mediated by LHT1 (Svennerstam et al., 2008). AAP1 has been shown to be important for root amino acid uptake only at high amino acid concentrations (Lee et al., 2007).
Urea is excreted into the environment by a variety of organisms and represents a readily available nitrogen source in soils. In addition, urea is one of the major N forms applied as fertilizer in agriculture. Physiological experiments have shown that plant roots can directly uptake urea from the soil (Krogmeier et al., 1989; Gerendas et al., 1998). The main transporter associated with urea uptake in Arabidopsis is AtDUR3, which co-transports urea and protons (Liu et al., 2003). AtDUR3 is a high-affinity urea transporter and its expression levels increase in N-deficient roots and decrease after re-supplementation with nitrate or ammonium (Kojima et al., 2007).
Although the significance of proteins for plant nutrition remains to be determined, plants that are not mycorrhizal symbionts, including Arabidopsis, may use proteins as N source without obvious assistance from other organisms (Paungfoo-Lonhienne et al., 2008). Two possible mechanisms could explain access of plants to N in soil proteins. Proteases present in root exudates may degrade proteins in the soil to amino acids (Paungfoo-Lonhienne et al., 2008). Alternatively, intact proteins in the soil can be taken up by the root through unknown transporters or by endocytosis (Paungfoo-Lonhienne et al., 2008).
Developmental adaptations for optimal N nutrition: organism level
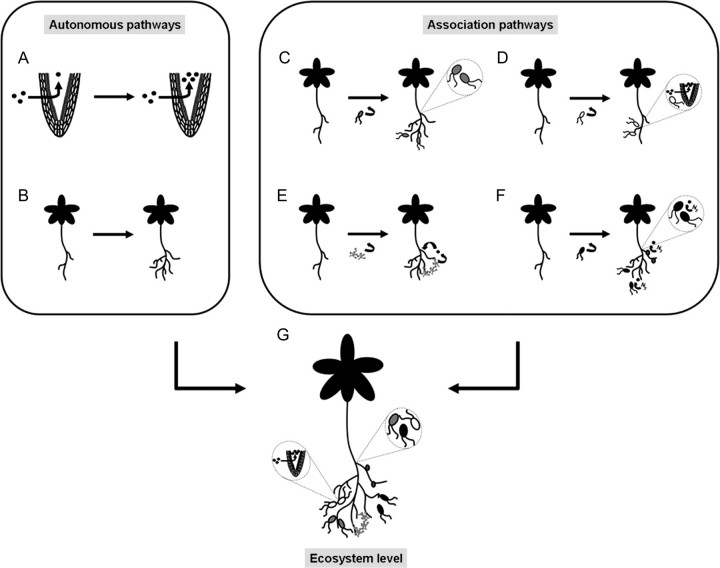
In addition to the regulation of the inorganic and organic N uptake systems (Fig. 1A), plants display considerable developmental plasticity in response to variations in the concentration and distribution of external nutrients. One of the most dramatic plant adaptations to ensure adequate N acquisition is the modulation of root system architecture (RSA) in response to N supply (Fig. 1B). Early studies by Drew et al. (1973) and Drew (1975) in barley (Hordeum sativum L.) demonstrated that seedlings subjected to a local high concentration of nitrate or ammonium had a dramatic proliferation of lateral roots (LRs) in the nutrient-rich zone. The proliferation of LRs within a localized nitrate-rich zone is a response that occurs in many plant species and represents a common adaptation phenomenon (Robinson, 1994; Hodge, 2004). Additional effects of N supply on root architecture and root developmental plasticity include changes in primary root growth (Walch-Liu et al., 2006b; Walch-Liu and Forde, 2008; Vidal et al., 2010a), LR initiation (Little et al., 2005; Remans et al., 2006b; Gifford et al., 2008), and LR elongation (Zhang and Forde, 1998; Zhang et al., 1999; Vidal et al., 2010a). Although different phenotypic impacts of N supply/source in plants have been identified, the N sensors and signalling pathways mediating these effects have yet to be fully characterized.
Fig. 1.
Simple model of pathways for N acquisition in plants: autonomous pathways regulate (A) N (black circle) uptake and/or (B) RSA. Association pathways allow plants to associate with (C) endophytic and/or (D) rhizospheric PGPB. These bacteria improve plant N nutrition by increasing root surface area (grey bacteria) and/or N uptake (white bacteria). (E) Plant association with mycorrhizal fungi improves plant N nutrition by modification of RSA. These fungi can also facilitate transfer of available N to the plant. (F) Plant association with NFB (black bacteria). Bacteria can be in the rhizosphere, inside the plant tissue forming, or not part of a nodule. In all cases, NFB bacteria can fix atmospheric N2 to NH3 available for plant use. Ecosystem level (G): autonomous and association pathways coexist in plants in the environment and act simultaneously and coordinately to ensure an adequate N supply.
In higher plants, the NITRATE REGULATED 1 (ANR1) gene was the first described regulatory factor involved in modulating root architecture in response to a localized nitrate supply (Zhang and Forde, 1998). ANR1 encodes a member of the MADS-box transcription factor gene family and was found in a reverse genetic screen designed to isolate genes whose expression is induced in nitrate-rich patches (Zhang and Forde, 1998). Transgenic plants in which ANR1 is repressed display a decreased root growth response to a localized nitrate supply (Zhang and Forde, 1998).
Reverse genetics approaches have suggested that NRT1.1 and NRT2.1 are components of the N signalling pathway. The role of these transporters has been supported mostly by work focusing on the effect of N availability in the modulation of RSA. Remans and colleagues (Remans et al., 2006a) found that NRT1.1 mutant plants exhibit a strongly decreased root colonization of localized high-nitrate patch and this effect is mediated by ANR1. In addition to the stimulatory effect on LR growth, nitrate also antagonizes the L-Glu effect on primary root elongation and this requires NRT1.1 (Walch-Liu and Forde, 2008). Interestingly, studies showed that Thr101-phosphorylated CHL1 is a high-affinity nitrate transporter, whereas Thr101-dephosphorylated CHL1 is a low-affinity transporter (Liu and Tsay, 2003). More recently, phosphorylation of this Thr101 by the calcineurin B-like protein-interacting kinase 23 (CIPK23) was shown to reduce nitrate primary response to low levels in low nitrate concentrations, whereas in high nitrate concentrations high expression of the primary response genes is correlated with a low phosphorylation status of the transporter (Ho et al., 2009). Thus, NRT1.1 phosphorylation generates different levels of expression of primary nitrate response genes according to nitrate availability. In addition, NRT2.1 has been implicated in LR initiation control in response to a low nitrate supply (Remans et al., 2006b) and in LR repression in response to high C/N ratio (Little et al., 2005).
The phytohormone auxin plays an important role in the modulation of RSA in response to N. Studies with maize suggested that inhibition of root growth by high nitrate supply is correlated with reduced auxin concentration in the roots (Tian et al., 2008). It has been proposed that the auxin long-distance signal from shoot to root regulates the inhibition of early LR development by high rates of nitrate supply in Arabidopsis seedlings (Forde, 2002; Walch-Liu et al., 2006a). Recently, Krouk and colleagues have shown that NRT1.1 facilitates uptake of auxin and that nitrate inhibits NRT1.1-dependent auxin uptake, suggesting that transduction of nitrate signal by NRT1.1 is associated with a modification of auxin transport (Krouk et al., 2010b). Gifford and colleagues (Gifford et al., 2008) found a regulatory module including miR167 and its target the AUXIN RESPONSE FACTOR 8 (ARF8) involved in regulation of LR initiation and emergence in response to nitrate. More recently, a regulatory module that includes miR393 and the auxin receptor AFB3 was shown to mediate both LR and primary root growth in response to nitrate treatments in Arabidopsis roots (Vidal et al., 2010a). AFB3 expression was induced directly by nitrate and miR393 expression was induced by N metabolites generated after nitrate reduction. Because increased levels of miR393 lead to down-regulation of the AFB3 mRNA levels, this regulatory module provides a simple molecular mechanism to control RSA in response to internal and external N availability (Vidal et al., 2010a). There is also evidence that abscisic acid (ABA) plays a central role in mediating the regulatory effects of high nitrate concentrations on root branching in Arabidopsis. ABA signalling mutants abi4-1, abi4-2, and abi5 are insensitive to repression of LR growth by high nitrate, and the ABA biosynthesis mutants (aba1-1, aba2-3, aba2-4, and aba3-2) show reduced sensitivity to this high nitrate repression (Signora et al., 2001). The authors propose that there are two regulatory pathways mediating the inhibitory effects of nitrate in Arabidopsis roots. One pathway is ABA dependent and involves ABI4 and ABI5, whereas the second pathway is ABA independent (Signora et al., 2001).
Association pathways for N acquisition: ecosystem level
Plant-growth-promoting bacteria and N nutrition
Nutritionally beneficial plant–bacteria interactions (i.e. mutualistic symbiosis) can increase nutrient accessibility, uptake, or both (Bertrand et al., 2000; Park et al., 2009). Bacteria that contribute to plant nutrition have positive effects on plant growth and are generally referred to as plant-growth-promoting bacteria (PGPB). Some PGPB can produce phytohormones such as indole acetic acid, cytokinins, and gibberellins, increasing hormone levels inside the plant (Long et al., 2008; Islam et al., 2009). PGPB can also decrease ethylene levels enzymatically by 1-aminocyclopropane-1-carboxylate deaminase (Onofre-Lemus et al., 2009). By modulating hormone levels, PGPB can influence root morphology, increasing LR length, and hair number and length (Persello-Cartieaux et al., 2001). For example, Azospirillum spp. bacteria secrete high quantities of auxins, which could be an important factor contributing to the stimulation of root development in plants (Spaepen et al., 2007). Pseudomonas thivervalensis bacteria colonize and promote root development in Arabidopsis thaliana (Achouak et al., 2000; Persello-Cartieaux et al., 2001). Arabidopsis mutant plants in the auxin influx transporter gene AUX1 were insensitive to the effect of P. thivervalensis suggesting a role for bacterial auxin in inducing morphological modifications of roots (Persello-Cartieaux et al., 2001). Bacillus megaterium bacteria can promote growth of A. thaliana and Phaseolus vulgaris (common bean, Fabaceae) (Lopez-Bucio et al., 2007). B. megaterium increases root development independent of auxin or ethylene, because mutant plants defective in either auxin or ethylene signalling still show increased root growth when inoculated with the bacterium (Lopez-Bucio et al., 2007). Plants mutant in the cytokinin receptors revealed that the integrity of the cytokinin signalling pathway was essential for the bacterial effect in the plant and suggested that the increased root growth and plant growth promotion are due to cytokinin action (Ortiz-Castro et al., 2008).
The increased nutrient acquisition observed in response to PGPB inoculation can be explained not only by branching and enlargement of the root surface area (Fig. 1C), but also by increasing activity of nutrient uptake systems (Fig. 1D) (Bertrand et al., 2000). Studies with the PGPB genus Achromobacter in association with Brassica napus (rapeseed, Brassicaceae) revealed that the bacterium can increase plant growth by stimulating nitrate uptake by the plant (Bertrand et al., 2000). Electrophysiological measurements of nitrate net flux with ion-selective microelectrodes showed that inoculation resulted in a specific increase in net nitrate influx in the root zone that was morphologically similar in inoculated and uninoculated plants (Bertrand et al., 2000).
Phylobacterium strain STM196 affects both RSA and N nutrition in Arabidopsis (Mantelin et al., 2006). This bacterium elicits an increase in root branching and plant N status promoting plant growth under different N concentrations (Mantelin et al., 2006). The effect of the Phylobacterium inoculation leads to the abolition of the inhibition of LR elongation by high nitrate supply. This bacterium is able to optimize plant growth independently of the external nitrate concentration (Mantelin et al., 2006). However, the molecular mechanism by which this bacterium exerts this effect on the plant is still unknown.
Besides bacteria, other microorganisms such as mycorrhizal fungi can modify RSA and increase the area of interaction with the soil contributing to better nutrient acquisition (Fig. 1E). Many studies of arbuscular mycorrhizal (AM) fungi–plant associations have shown that AM fungi induce modification of RSA (Berta et al., 1995; Gamalero et al., 2004; Gutjahr et al., 2009). The importance of this association to plant nutrition has been mainly studied in the context of phosphorus uptake (Fitter and Hay, 2002; Plassard and Dell, 2010). However, a few studies have addressed the importance of mycorrhizal fungi for N nutrition. The AM fungi Glomus intraradices can increase inorganic N and total N content uptake capacity of carob trees (Ceratonia siliqua, Fabaceae) as compared with plants without this fungus (Cruz et al., 2004). Such an increase in plant N uptake was observed only in carob trees growing at low levels of N (Cruz et al., 2004). Stable isotope labelling experiments showed that inorganic nitrogen is taken up by the AM fungi and then transferred to the plant roots (Fig. 1E) (Govindarajulu et al., 2005).
Plant interactions with nitrogen fixing bacteria for N acquisition
The best known example of beneficial plant–bacteria association for N nutrition occurs in nodulating plants (Fig. 1F) (Sprent and James, 2007). Nodulating plants are able to obtain an important part of the N required to sustain their growth and development from nitrogen fixing bacteria (NFB) symbionts (Materona and Danso, 1991). NFB are able to reduce atmospheric N2 to ammonium by the action of an evolutionarily conserved enzyme complex called nitrogenase. This complex is composed of two enzymes: a dinitrogenase and a dinitrogenase reductase (Joerger et al., 1991; Zehr et al., 2003; Zhang et al., 2007a). Both bacteria and archea are able to carry out nitrogen fixation (Zehr et al., 2003). This symbiotic interaction occurs in plants of the Fabaceae family (legumes) and also in the plant genus Parasponia (Cannabaceae) (Sprent and James, 2007). Nodulating plants can interact with bacteria of the genera Rhizobium, Mesorhizobium, Sinorhizobium, Bradyrhizobium, and Azorhizobium of the Rhizobiaceae family (Gage, 2004). Legumes can also associate with some strains of the Methylobacterium, Cupriavidus, Shinella, Devocia, and Burkholderia genera (Chen et al., 2001; Sy et al., 2001; Rivas et al., 2003; Chen et al., 2005; Lin et al., 2008). Members of the Betulaceae, Casuarinaceae, Myricaceae, Elaegnaceae, Rhamnaceae, Rosaceae, Coriariaceae, and Datisticaceae nodulate with the actinomycetal genus Frankia (Gage, 2004; Sprent and James, 2007).
In addition to the importance of nodulation for plant nutrition, beneficial plant–bacteria interactions for N nutrition are also observed within plant species that do not nodulate (Fig. 1F) (Stone et al., 2001; Chi et al., 2005; Perin et al., 2006; Rosenblueth and Martinez-Romero, 2006). Interactions between non-nodulating plants and NFB are functional associations that have received considerably less attention than interactions leading to nodule formation (Egener et al., 1998; Iniguez et al., 2004; You et al., 2005). However, NFB can colonize the rhizosphere of the plant, as shown for the Burkholderia genus found associated with the rhizosphere of tomato plants (Caballero-Mellado et al., 2007). NFB have also been shown to colonize plant tissues and exhibit an endophytic lifestyle (Hurek et al., 1994b; Reinhold-Hurek and Hurek, 1998; You et al., 2005; Rosenblueth and Martinez-Romero, 2006). Endophytes capable of fixing N have been isolated from a wide diversity of non-nodulating plants in an order of up to 108 cells per gram of tissue (Reinhold-Hurek and Hurek, 1998; Chi et al., 2005; Perin et al., 2006). The endophytic population can vary depending on environmental factors such as the type of soil as well as plant characteristics such as genotype and developmental stage (Kuklinsky-Sobral et al., 2004; Rosenblueth and Martinez-Romero, 2006). Comparison of the rhizospheric and endophytic bacterial communities of cucumber plants (Cucurbitaceae) revealed higher diversity in the rhizospheric population than in the endophytic population (Mahaffee and Kloepper, 1997). In most cases, endophyte taxa can be found in the rhizosphere. However, there are examples of bacteria with strict endophytic lifestyle that can only be isolated from plants, such as Azoarcus sp. BH72, Herbaspirillum and Acetobacter species (Reinhold-Hurek and Hurek, 1998).
Non-nodulating plants and NFB can establish functional associations (Fig. 1F). Azoarcus sp. BH72 fixes N under microaerobic conditions. At nanomolar oxygen concentrations, these bacterial cells can shift into a state of higher activity of N fixation and respiratory efficiency in which intracytoplasmic membrane stacks (diazosomes) related to N fixation are formed, and the iron protein of the nitrogenase is highly enriched (Hurek et al., 1994a). Transcriptional fusion of the nitrogenase nifH gene promoter to green fluorescent protein reported high levels of nitrogenase gene expression in Azoarcus sp. BH72 within rice roots (Egener et al., 1998). In addition, molecular ecological methods were developed to assess nifH mRNA expression within Kallar grass (Leptochloa fusca, Poaceae) plants inoculated with this bacterium. Screening for the nifH gene by nifH-specific reverse transcription-PCR in root mRNA, showed that Azoarcus sp. BH72 expresses nitrogenase genes inside the plant root system (Hurek et al., 2002). Dry weight, total N content, and 15N/14N ratio were determined in plants inoculated with either wild-type bacteria or a nifK mutant strain BHNKD4 (unable to fix N) (Hurek et al., 2002). In N-deficient conditions, plants inoculated with strain BH72 grew better and accumulated more N with a lower 15N/14N ratio than non-N2-fixing control plants inoculated with the mutant strain (Hurek et al., 2002). Differences in N isotopic composition suggest that the plants in both treatments had access to different nitrogen sources (Hurek et al., 2002). It has been shown that nitrogenase discriminates against the heavier isotope (Hoering and Ford, 1960; Hurek et al., 2002). Therefore, the accumulation of more N with a decreased abundance of 15N suggests that the wild-type bacteria can provide N for plant use.
The significance of biological N fixation for wheat has been evaluated by the 15N dilution technique (Iniguez et al., 2004). In this technique, plants are grown with 15N isotopically labelled N sources and the increase in 14N relative to 15N content in the plant tissues under low N conditions is monitored over time. Wheat plants inoculated with NFB Klebsiella pneumoniae 342 assimilated up to 49% of the plant N from the atmosphere through biological N fixation (Iniguez et al., 2004). Indeed, plants grown under N-deficient conditions inoculated with a nifH mutant of K. pneumoniae (unable to fix N), showed signs of N deficiency, in contrast to plants inoculated with the wild-type bacterium (Iniguez et al., 2004). Similar experiments showed that some varieties of sugar cane (Sacharrum spp.) are also capable of obtaining a significant proportion of their N requirement from biological N fixation (Boddey et al., 1991). In fact, these plants can dispense with N fertilization under good conditions of water and the supply of other nutrients (Boddey et al., 1991).
N-mediated regulation of autonomous and association pathways
With the advent of genomic technologies, our understanding of plant transcriptional changes occurring upon exposure to different N conditions has grown considerably. Genome-wide gene expression analyses using nitrate and other forms of N, such as nitrite, or glutamic acid, revealed a large set of genes involved in a wide range of plant processes (Wang et al., 2003; Muños et al., 2004; Scheible et al., 2004; Gutiérrez et al., 2007; Vidal and Gutiérrez, 2008; Krouk et al., 2010a). Due to the importance of nitrate as primary N source for plants, the nitrate response has been the most thoroughly characterized. Roots are highly responsive to nitrate, with >1000 genes responding rapidly at very low concentrations of externally added nitrate (Wang et al., 2003). Some of the transcriptional changes caused by nitrate treatments have been shown to correlate with changes at the protein level, as observed by two-dimensional gel electrophoresis analysis (Prinsi et al., 2009). The expression of many genes involved in the autonomous pathway is regulated by these N treatments (e.g. ammonium and nitrate transporters, genes involved in the control of RSA). However, these genome-wide experiments also show N regulation of many plant genes that may impact the association pathways. It has been reported that low levels of nitrate and ammonium stimulate nodulation, whereas high concentrations of these nutrients inhibit nodule formation (Eaglesham, 1989; Zahran, 1999). The inhibitory effects of nitrate on different phases of nodulation, including the number of infection sites in the root, nodule development, N fixation in pre-existing nodules, and nitrogenase activity have been well documented (Bisseling et al., 1978; Caetano-Anolles and Gresshoff, 1991; Zahran, 1999). Moreover, nitrate can significantly decrease the number of rhizobial cells adhering to plant roots, which is an important step for root infection (Dazzo and Brill, 1978). Plant genes involved in the perception of nodulating factors, such as NFR1 and NFR5, as well as transcriptional regulators of nodulation, such as NSP1 and NSP2, are also regulated by N in plants exposed to nodulating factors (Barbulova et al., 2007). The transcription factor NIN was not induced by nodulating factors in the presence of nitrate or ammonium as compared with plants grown in the absence of N (Barbulova et al., 2007). The lack of NIN induction may represent an important event in nitrate-dependent inhibition of nodule development, since NIN factors are essential for nodule organogenesis (Schauser et al., 1998; Borisov et al., 2003; Oldroyd and Downie, 2008). The effect of nitrate on NIN gene expression was not observed in the hypernodulation aberrant root formation (har1) mutant plants treated with nodulating factors or with NFB, suggesting that NIN expression is controlled by HAR1 and that the nitrate effect is mediated by HAR1 (Nishimura et al., 2002; Barbulova et al., 2007). HAR1 is a key regulator involved in the systemic regulation that prevents nodule formation in the presence of nitrate. This process, termed autoregulation of nodulation (AON) is a universal inhibitory control mechanism conserved among legumes (Carroll et al., 1985; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003).
N is also an important regulatory factor of plant and NFB associations in non-nodulating plants. However, little is known about the molecular mechanisms involved. Rice (Orysa spp.) plants treated with large doses of N fertilizers show a rapid decrease in NFB diversity in roots 15 d after treatment (Tan et al., 2003). Similarly, sorghum plants (Sorghum bicolor, Poaceae) grown under high-N fertilizer regimes showed decreased NFB associated with the rhizosphere (Coelho et al., 2009). In sugarcane (Saccharum spp., Poaceae), high N fertilization caused a decrease in the colonization of the plant by Acetobacter diazotrophicus as compared with plants grown under low N fertilization (Fuentes-Ramírez et al., 1999). In addition, nitrate or ammonium leads to the repression of nitrogenase genes and to inactivation of nitrogenase activity (Martin and Reinhold-Hurek, 2002). Therefore, N fertilization has an effect not only on the diversity and number of NFB associated with the plant, but also on the activity of the associated bacteria. Although the regulatory components of this interaction are unknown, A. thaliana (the best-studied non-nodulating plant) and other non-nodulating plants have genes homologous to those involved in nodulation of nodulating plants such as those for NFR1, NFR5, and SYMRK receptors and the transcription factors NIN, NSP1, NSP2, and EDF (Schauser et al., 1998; Stracke et al., 2002; Radutoiu et al., 2003; Kalo et al., 2005; Smit et al., 2005; Vernie et al., 2008; Hirsch et al., 2009). Whether the regulatory function of these genes has a role in non-nodulating plants and NFB interactions remains to be elucidated. According to this, Arabidopsis could be a good model to evaluate the role of these genes in non-nodulating plants, since it is the best plant system available so far for identifying and studying the role of gene functions.
Scaling up to the ecosystem level
To increase our understanding of how organisms function within ecosystems, it is necessary to resolve the underlying mechanisms of nutrient cycling at both the ecosystem and organism level. Such mechanisms may involve gene–environment interactions affecting community structure and ecosystem processes (Whitham et al., 2006). From an ecosystem-level perspective, much knowledge has been gained on how plant species can change ecosystem nitrogen cycling by controlling nitrogen input rates (Aerts and Chapin, 2000; Knops et al., 2002; Vitousek et al., 2002). However, little is known about symbiotic nitrogen fixation and even less regarding the degree of plant control over this phenomenon. This results in N cycling models of terrestrial ecosystems lacking mechanistic resolution between perspectives on plant–nutrient interaction at the level of ecosystem (e.g. forests) versus the level of individual plants (Hedin et al., 2009). Solving this problem requires the above discussed pathways operating at the molecular level to be explored at higher levels of organization (organisms) in order to demonstrate causal relationships across the gene-to-ecosytem continuum (Whitham et al., 2006). Recent advances in genomic techniques centred in Populus (Salicaceae) as a model system has allowed exploration of links at different levels of organization (Schweitze et al., 2004; Whitham et al., 2006). An additional step could be made by incorporating gene-to-ecosystem causal relationships into agent-based simulation models to provide mechanistic explanations of nutrient cycling in ecosystems.
Final remarks
Plants have evolved autonomous and association pathways that contribute to N acquisition (Fig. 1). In the environment, these pathways operate simultaneously and plants must integrate nutrient and other environmental signals impinging upon these pathways to effectively regulate the same processes: modulation of RSA and activity of N uptake systems (Fig. 1G). Albeit independent lines of evidence have shown that the autonomous and association pathways indeed interact and must be coordinately regulated to ensure efficient N uptake, these pathways have been mostly studied independently. Therefore, to truly understand plant N acquisition in the environment deeper understanding is required of (i) the molecular mechanisms controlling autonomous and association pathways as well as their interactions; and (ii) how these mechanisms impact nutrient cycling at the ecosystem level. Integrating mechanisms operating at molecular, organism, and ecosystem level would enhance our understanding of the terrestrial nitrogen cycle. This knowledge is also the first step in developing effective and sustainable biotechnological solutions to enhance N acquisition by plants in natural or agricultural environments. Proper plant N nutrition in the environment will not only improve production but would also contribute to sustainable agricultural practices by diminishing the use of N fertilizers and thus reducing greenhouse gases, stratospheric ozone, acid rain, and nitrate pollution of surface and ground water.
Acknowledgments
Research in the R.A.G. laboratory is funded by FONDECYT (1100698), ANR-CONICYT (ANR-007), ICM-MIDEPLAN (MN-PFG P06-009-F), FONDAP (15090007), NIH-FIRCA (F6414-01), and CORFO (07Genoma01). D.E.G. and T.K. are supported by FONDECYT postdoctoral grant (3100069) and CONICYT PhD scholarship (21080821), respectively.
Glossary
Abbreviations
- AAP
amino acid permease
- ABA
abscisic acid
- AM
arbuscular mycorrhizal
- AON
autoregulation of nodulation
- ARF
auxin-response factor
- CIPK
calcineurin B-like protein-interacting kinase
- HATS
high-affinity transport system
- LATS
low-affinity transport systems
- LHT1
lysine-histidine transporter 1
- LR
lateral root
- NFB
nitrogen fixing bacteria
- PGPB
plant-growth-promoting bacteria
- RSA
root system architecture
References
- Achouak W, Sutra L, Heulin T, Meyer JM, Fromin N, Degraeve S, Christen R, Gardan L. Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov., two root-associated bacteria isolated from Brassica napus and Arabidopsis thaliana. International Journal of Systematic and Evolutionary Microbiology. 2000;50:9–18. doi: 10.1099/00207713-50-1-9. [DOI] [PubMed] [Google Scholar]
- Aerts R, Chapin FS. The mineral nutrition of wild plants revisited: a re-evaluation of processes and patterns. Advances in Ecological Research. 2000;30:1–67. [Google Scholar]
- Barbulova A, Rogato A, D'Apuzzo E, Omrane S, Chiurazzi M. Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Molecular Plant–Microbe Interactions. 2007;20:994–1003. doi: 10.1094/MPMI-20-8-0994. [DOI] [PubMed] [Google Scholar]
- Berta G, Trotta A, Fusconi A, et al. Arbuscular mycorrhizal induced changes to plant growth and root system morphology in Prunus cerasifera. Tree Physiology. 1995;15:281–293. doi: 10.1093/treephys/15.5.281. [DOI] [PubMed] [Google Scholar]
- Bertrand H, Plassard C, Pinochet X, Touraine B, Normand P, Cleyet-Marel JC. Stimulation of the ionic transport system in Brassica napus by a plant growth-promoting rhizobacterium (Achromobacter sp.) Canadian Journal of Microbiology. 2000;46:229–236. doi: 10.1139/w99-137. [DOI] [PubMed] [Google Scholar]
- Bisseling T, van den Bos RC, van Kammen A. The effect of ammonium nitrate on the synthesis of nitrogenase and the concentration of leghemoglobin in pea root nodules induced by Rhizobium leguminosarum. Biochimica et Biophysica Acta. 1978;539:1–11. doi: 10.1016/0304-4165(78)90115-0. [DOI] [PubMed] [Google Scholar]
- Boddey RM, Urquiaga S, Reis V, Döbereiner J. Biological nitrogen fixation associated with sugar cane. Plant and Soil. 1991;137:111–117. [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiology. 2003;131:1009–1017. doi: 10.1104/pp.102.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Mellado J, Onofre-Lemus J, Estrada-de Los Santos P, Martinez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Applied and Environmental Microbiology. 2007;73:5308–5319. doi: 10.1128/AEM.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Gresshoff PM. Alfalfa controls nodulation during the onset of Rhizobium-induced cortical cell division. Plant Physiology. 1991;95:366–373. doi: 10.1104/pp.95.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. Isolation and properties of soybean [ Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proceedings of the National Academy of Sciences, USA. 1985;82:4162–4166. doi: 10.1073/pnas.82.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, de Faria SM, Straliotto R, et al. Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Applied and Environmental Microbiology. 2005;71:7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Vandamme P. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. International Journal of Systematic and Evolutionary Microbiology. 2001;51:1729–1735. doi: 10.1099/00207713-51-5-1729. [DOI] [PubMed] [Google Scholar]
- Chi F, Shen SH, Cheng HP, Jing YX, Yanni YG, Dazzo FB. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Applied and Environmental Microbiology. 2005;71:7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MRR, Marriel IE, Jenkins SN, Lanyon CV, Seldin L, O'Donnell AG. Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Applied Soil Ecology. 2009;42:48–53. [Google Scholar]
- Crawford N, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends in Plant Science. 1998;3:389–395. [Google Scholar]
- Cruz C, Green JJ, Watson CA, Wilson F, Martins-Loucao MA. Functional aspects of root architecture and mycorrhizal inoculation with respect to nutrient uptake capacity. Mycorrhiza. 2004;14:177–184. doi: 10.1007/s00572-003-0254-5. [DOI] [PubMed] [Google Scholar]
- Dazzo FB, Brill WJ. Regulation by fixed nitrogen of host-symbiont recognition in the Rhizobium-clover symbiosis. Plant Physiology. 1978;62:18–21. doi: 10.1104/pp.62.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Saker LR. Nutrient supply and the growth of the seminal root system in barley. II. Localized compensatory increases in lateral root growth and rates of nitrate uptake when nitrate supply is restricted to only part of the root system. Journal of Experimental Botany. 1975;26:79–90. [Google Scholar]
- Drew MC, Saker LR, Ashley TW. Nutrient supply and the growth of the seminal root system in barley. I. The effect of nitrate concentration on the growth of axes and laterals. Journal of Experimental Botany. 1973;24:1189–1202. [Google Scholar]
- Eaglesham ARJ. Nitrate inhibition of root nodule symbiosis in doubly rooted soybean plants. Crop Science. 1989;29:115–119. [Google Scholar]
- Egener T, Hurek T, Reinhold-Hurek B. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. BH72, a grass-associated diazotroph, on rice roots. Molecular Plant–Microbe Interactions. 1998;11:71–75. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- Epstein E, Bloom A. Mineral nutrition of plants: principles and perspectives. 2nd edn. Sunderland, MA: Sinauer Associates, Inc; 2005. [Google Scholar]
- Fitter A, Hay R. Environmental physiology of plants. 2nd edn. London: Academic Press; 2002. [Google Scholar]
- Forde BG. Local and long-range signaling pathways regulating plant responses to nitrate. Annual Review of Plant Biology. 2002;53:203–224. doi: 10.1146/annurev.arplant.53.100301.135256. [DOI] [PubMed] [Google Scholar]
- Forde BG, Walch-Liu P. Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant, Cell and Environment. 2009;32:682–693. doi: 10.1111/j.1365-3040.2008.01927.x. [DOI] [PubMed] [Google Scholar]
- Fuentes-Ramírez LE, Caballero-Mellado J, Sepúlveda J, Martínez-Romero E. Colonization of sugarcane by Acetobacter diazotrophicus is inhibited by high N-fertilization. FEMS Microbiology Ecology. 1999;29:117–128. [Google Scholar]
- Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiology and Molecular Biology Reviews. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JN, Cowling EB. Reactive nitrogen and the world: 200 years of change. Ambio. 2002;31:64–71. doi: 10.1579/0044-7447-31.2.64. [DOI] [PubMed] [Google Scholar]
- Gamalero E, Trotta A, Massa N, Copetta A, Martinotti MG, Berta G. Impact of two fluorescent pseudomonads and an arbuscular mycorrhizal fungus on tomato plant growth, root architecture and P acquisition. Mycorrhiza. 2004;14:185–192. doi: 10.1007/s00572-003-0256-3. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. The Plant Cell. 1999;11:937–948. doi: 10.1105/tpc.11.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerendas J, Zhu Z, Sattelmacher B. Influence of N and Ni supply on nitrogen metabolism and urease activity in rice (Oryza sativa L.) Journal of Experimental Botany. 1998;49:1545–1554. [Google Scholar]
- Gifford ML, Dean A, Gutiérrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences, USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature. 2005;435:819–823. doi: 10.1038/nature03610. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in. Arabidopsis. Genome Biology. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Casieri L, Paszkowski U. Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytologist. 2009;182:829–837. doi: 10.1111/j.1469-8137.2009.02839.x. [DOI] [PubMed] [Google Scholar]
- Hedin LO, Brookshire J, Menge DNL, Barron A. The nitrogen paradox in tropical forest ecosystems. Annual Review of Ecology, Evolution, and Systematics. 2009;40:613–635. [Google Scholar]
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. The Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Munoz A, Heckmann AB, Downie JA, Oldroyd GE. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. The Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Hodge A. The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytologist. 2004;162:9–24. [Google Scholar]
- Hoering TC, Ford HT. The isotope effect in the fixation of Azotobacter. Journal of the American Chemical Society. 1960;82:376–378. [Google Scholar]
- Hurek T, Handley LL, Reinhold-Hurek B, Piche Y. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Molecular Plant–Microbe Interactions. 2002;15:233–242. doi: 10.1094/MPMI.2002.15.3.233. [DOI] [PubMed] [Google Scholar]
- Hurek T, Reinhold-Hurek B, Turner GL, Bergersen FJ. Augmented rates of respiration and efficient nitrogen fixation at nanomolar concentrations of dissolved O2 in hyperinduced Azoarcus sp. strain BH72. Journal of Bacteriology. 1994a;176:4726–4733. doi: 10.1128/jb.176.15.4726-4733.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. Journal of Bacteriology. 1994b;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez AL, Dong Y, Triplett EW. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Molecular Plant–Microbe Interactions. 2004;17:1078–1085. doi: 10.1094/MPMI.2004.17.10.1078. [DOI] [PubMed] [Google Scholar]
- Islam MR, Madhaiyan M, Deka Boruah HP, Yim W, Lee G, Saravanan VS, Fu Q, Hu H, Sa T. Characterization of plant growth-promoting traits of free-living diazotrophic bacteria and their inoculation effects on growth and nitrogen uptake of crop plants. Journal of Microbiology and Biotechnology. 2009;19:1213–1222. doi: 10.4014/jmb.0903.3028. [DOI] [PubMed] [Google Scholar]
- Joerger RD, Wolfinger ED, Bishop PE. The gene encoding dinitrogenase reductase 2 is required for expression of the second alternative nitrogenase from Azotobacter vinelandii. Journal of Bacteriology. 1991;173:4440–4446. doi: 10.1128/jb.173.14.4440-4446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Hodge A. Biodegradation kinetics and sorption reactions of three differently charged amino acids in soil and their effects on plant organic nitrogen availability. Soil Biology and Biochemistry. 1999;31:1331–1342. [Google Scholar]
- Jones DL, Healeya JR, Willetta VB, Farrarb JF, Hodge A. Dissolved organic nitrogen uptake by plants—an important N uptake pathway? Soil Biology and Biochemistry. 2005;37:413–423. [Google Scholar]
- Kalo P, Gleason C, Edwards A, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Knops JMH, Bradley KL, Wedin DA. Mechanisms of plant species impacts on ecosystem nitrogen cycling. Ecology Letters. 2002;5:454–466. [Google Scholar]
- Kojima S, Bohner A, Gassert B, Yuan L, von Wirén N. AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal. 2007;52:30–40. doi: 10.1111/j.1365-313X.2007.03223.x. [DOI] [PubMed] [Google Scholar]
- Krogmeier MJ, McCarty GW, Bremner JM. Phytotoxicity of foliar-applied urea. Proceedings of the National Academy of Sciences, USA. 1989;86:8189–8191. doi: 10.1073/pnas.86.21.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF. Nitrate signaling: adaptation to fluctuating environments. Current Opinion in Plant Biology. 2010a;13:266–273. doi: 10.1016/j.pbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell. 2010b;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A. Regulation of the high-affinity NO3– uptake system by NRT1.1-mediated NO3– demand signaling in. Arabidopsis. Plant Physiology. 2006;142:1075–1086. doi: 10.1104/pp.106.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Kuklinsky-Sobral J, Araujo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environmental Microbiology. 2004;6:1244–1251. doi: 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. The Plant Cell. 2009;21:3610–3622. doi: 10.1105/tpc.109.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB, Purves JV, Ratcliffe RG, Saker LR. Nitrogen assimilation and the control of ammonium and nitrate absorption by maize roots. Journal of Experimental Botany. 1992;43:1385–1396. [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M. AAP1 transports uncharged amino acids into roots of Arabidopsis. The Plant Journal. 2007;50:305–319. doi: 10.1111/j.1365-313X.2007.03045.x. [DOI] [PubMed] [Google Scholar]
- Lin DX, Wang ET, Tang H, Han TX, He YR, Guan SH, Chen WX. Shinella kummerowiae sp. nov., a symbiotic bacterium isolated from root nodules of the herbal legume Kummerowia stipulacea. International Journal of Systematic and Evolutionary Microbiology. 2008;58:1409–1413. doi: 10.1099/ijs.0.65723-0. [DOI] [PubMed] [Google Scholar]
- Lipson D, Näsholm T. The unexpected versatility of plants: organic nitrogen use and availability in terrestrial ecosystems. Oecologia. 2001;128:305–316. doi: 10.1007/s004420100693. [DOI] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proceedings of the National Academy of Sciences, USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO Journal. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. The Plant Cell. 1999;11:865–874. doi: 10.1105/tpc.11.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Ludewig U, Frommer WB, von Wirén N. AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. The Plant Cell. 2003;15:790–800. doi: 10.1105/tpc.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HH, Schmidt DD, Baldwin IT. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0002702. e2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Molecular Plant–Microbe Interactions. 2007;20:207–217. doi: 10.1094/MPMI-20-2-0207. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology. 2003;6:280–287. doi: 10.1016/s1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Ludewig U, Neuhäuser B, Dynowski M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Letters. 2007;581:2301–2308. doi: 10.1016/j.febslet.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Ludewig U, von Wirén N, Frommer WB. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. Journal of Biological Chemistry. 2002;277:13548–13555. doi: 10.1074/jbc.M200739200. [DOI] [PubMed] [Google Scholar]
- Ludewig U, Wilken S, Wu B, et al. Homo- and hetero-oligomerization of ammonium transporter-1 NH4+ uniporters. Journal of Biological Chemistry. 2003;278:45603–45610. doi: 10.1074/jbc.M307424200. [DOI] [PubMed] [Google Scholar]
- Mahaffee WF, Kloepper JW. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.) Microbial Ecology. 1997;34:210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- Mantelin S, Desbrosses G, Larcher M, Tranbarger TJ, Cleyet-Marel JC, Touraine B. Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta. 2006;223:591–603. doi: 10.1007/s00425-005-0106-y. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. , 889 pp. [Google Scholar]
- Martin DE, Reinhold-Hurek B. Distinct roles of P(II)-like signal transmitter proteins and amtB in regulation of nif gene expression, nitrogenase activity, and posttranslational modification of NifH in Azoarcus sp. strain BH72. Journal of Bacteriology. 2002;184:2251–2259. doi: 10.1128/JB.184.8.2251-2259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Tuskan GA, Difazio SP, Lammers P, Newcombe G, Podila GK. Symbiotic sequencing for the Populus mesocosm. New Phytologist. 2004;161:330–335. doi: 10.1111/j.1469-8137.2004.00982.x. [DOI] [PubMed] [Google Scholar]
- Materona LA, Danso SKA. Nitrogen fixation in two annual Medicago legumes, as affected by inoculation and seed density. Field Crops Research. 1991;26:253–262. [Google Scholar]
- Mayer M, Dynowski M, Ludewig U. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochemical Journal. 2006;396:431–437. doi: 10.1042/BJ20060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. Journal of Experimental Botany. 2007;58:2297–2306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. The Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsholm T, Ekblad A, Nordin A, Giesler R, Högberg M, Högberg P. Boreal forest plants take up organic nitrogen. Nature. 1998;392:914–916. [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytologist. 2009;182:31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual Review of Plant Biology. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Onofre-Lemus J, Hernández-Lucas I, Girard L, Caballero-Mellado J. ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Applied and Environmental Microbiology. 2009;75:6581–6590. doi: 10.1128/AEM.01240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiology. 2006;142:1304–1317. doi: 10.1104/pp.106.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz-Castro R, Valencia-Cantero E, López-Bucio J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signaling and Behavior. 2008;3:263–265. doi: 10.4161/psb.3.4.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KH, Lee CY, Son HJ. Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Letters in Applied Microbiology. 2009;49:222–228. doi: 10.1111/j.1472-765X.2009.02642.x. [DOI] [PubMed] [Google Scholar]
- Paungfoo-Lonhienne C, Lonhienne TG, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S. Plants can use protein as a nitrogen source without assistance from other organisms. Proceedings of the National Academy of Sciences, USA. 2008;105:4524–4529. doi: 10.1073/pnas.0712078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin L, Martínez-Aguilar L, Paredes-Valdez G, Baldani JI, Estrada-de Los Santos P, Reis VM, Caballero-Mellado J. Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. International Journal of Systematic and Evolutionary Microbiology. 2006;56:1931–1937. doi: 10.1099/ijs.0.64362-0. [DOI] [PubMed] [Google Scholar]
- Persello-Cartieaux F, David P, Sarrobert C, Thibaud MC, Achouak W, Robaglia C, Nussaume L. Utilization of mutants to analyze the interaction between Arabidopsis thaliana and its naturally root-associated Pseudomonas. Planta. 2001;212:190–198. doi: 10.1007/s004250000384. [DOI] [PubMed] [Google Scholar]
- Pickett STA, Cadenasso ML. The ecosystem as a multidimensional concept: meaning, model, and metaphor. Ecosystems. 2002;5:1–10. [Google Scholar]
- Plassard C, Dell B. Phosphorus nutrition of mycorrhizal trees. Tree Physiology. 2010;30:1129–1139. doi: 10.1093/treephys/tpq063. [DOI] [PubMed] [Google Scholar]
- Prinsi B, Negri AS, Pesaresi P, Cocucci M, Espen L. Evaluation of protein pattern changes in roots and leaves of Zea mays plants in response to nitrate availability by two-dimensional gel electrophoresis analysis. BMC Plant Biology. 2009;23:113. doi: 10.1186/1471-2229-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Rawat SR, Silim SN, Kronzucker HJ, Siddiqi MY, Glass AD. AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. The Plant Journal. 1999;19:143–152. doi: 10.1046/j.1365-313x.1999.00505.x. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends in Microbiology. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences, USA. 2006a;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiology. 2006b;140:909–921. doi: 10.1104/pp.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martínez-Molina E, Gillis M, Velázquez Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Systematic and Aplied Mcrobiology. 2003;26:47–53. doi: 10.1078/072320203322337308. [DOI] [PubMed] [Google Scholar]
- Robinson D. The responses of plants to non-uniform supplies of nutrients. New Phytologist. 1994;127:635–674. doi: 10.1111/j.1469-8137.1994.tb02969.x. [DOI] [PubMed] [Google Scholar]
- Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Molecular Plant– Microbe Interactions. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- Sanhueza E. The role of the atmosphere in nitrogen cycling. Plant and Soil. 1982;67:61–71. [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J. Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Molecular and General Genetics. 1998;259:414–423. doi: 10.1007/s004380050831. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant physiology. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitze JA, Bailey JK, Rehill BJ, Martinsen GD, Hart SC, Lindroth RL, Keim P, Whitham T. Genetically based trait in a dominant tree affects ecosystem processes. Ecology Letters. 2004;7:127–134. [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. Long-distance signaling in nodulation directed by a CLAVATA1–like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. The Plant Journal. 2001;28:655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiology Reviews. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Sprent JI, James EK. Legume evolution: where do nodules and mycorrhizas fit in? Plant Physiology. 2007;144:575–581. doi: 10.1104/pp.107.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone PJ, O'Callaghan KJ, Davey MR, Cocking EC. Azorhizobium caulinodans ORS571 colonizes the xylem of Arabidopsis thaliana. Molecular Plant–Microbe Interactions. 2001;14:93–97. doi: 10.1094/MPMI.2001.14.1.93. [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Näsholm T. Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytologist. 2008;180:620–630. doi: 10.1111/j.1469-8137.2008.02589.x. [DOI] [PubMed] [Google Scholar]
- Sy A, Giraud E, Jourand P, et al. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. Journal of Bacteriology. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Hurek T, Reinhold-Hurek B. Effect of N-fertilization, plant genotype and environmental conditions on nifH gene pools in roots of rice. Environmental Microbiology. 2003;5:1009–1015. doi: 10.1046/j.1462-2920.2003.00491.x. [DOI] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. Journal of Plant Physiology. 2008;165:942–951. doi: 10.1016/j.jplph.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Letters. 2007;581:2290–2300. doi: 10.1016/j.febslet.2007.04.047. [DOI] [PubMed] [Google Scholar]
- Vernie T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P. EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. The Plant Cell. 2008;20:2696–2713. doi: 10.1105/tpc.108.059857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Current Opinion in Plant Biology. 2008;11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2010a;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutiérez RA. Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. WIREs Systems Biology and Medicin. 2010b doi: 10.1002/wsbm.87. Wiley Interdisciplinary reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek PM, Cassman K, Cleveland C, et al. Towards an ecological understanding of biological nitrogen fixation. Biogeochemistry. 2002;57:1–45. [Google Scholar]
- von Wirén N, Merrik M. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Topics in Current Genetics. 2004;9:95–120. [Google Scholar]
- Walch-Liu P, Forde BG. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. The Plant Journal. 2008;54:820–828. doi: 10.1111/j.1365-313X.2008.03443.x. [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov, Filleur S, Gan Y, Remans T, Forde BG. Nitrogen regulation of root branching. Annals of Botany. 2006a;97:875–881. doi: 10.1093/aob/mcj601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG. Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant and Cell Physiology. 2006b;47:1045–1057. doi: 10.1093/pcp/pcj075. [DOI] [PubMed] [Google Scholar]
- Wang MY, Siddiqi MY, Ruth TJ, Glass A. Ammonium uptake by rice roots (II. Kinetics of 13NH4+ influx across the plasmalemma) Plant Physiology. 1993;103:1259–1267. doi: 10.1104/pp.103.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proceedings of the National Academy of Sciences, USA. 1998;95:15134–15139. doi: 10.1073/pnas.95.25.15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham TG, Bailey JK, Schweitzer JA, et al. A framework for community and ecosystem genetics: from genes to ecosystems. Nature Reviews. 2006;7:510–523. doi: 10.1038/nrg1877. [DOI] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. The Plant Cell. 2007;19:2636–2652. doi: 10.1105/tpc.107.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Nishiguchi T, Saito A, Isawa T, Mitsui H, Minamisawa K. Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light-dark cycle. Applied and Environmental Microbiology. 2005;71:8183–8190. doi: 10.1128/AEM.71.12.8183-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran HH. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiology and Molecular Biology Reviews. 1999;63:968–989. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environmental Microbiology. 2003;5:539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. Regulation of Arabidopsis root development by nitrate availability. Journal of Experimental Botany. 2000;51:51–59. [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences, USA. 1999;96:6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hurek T, Reinhold-Hurek B. A nifH-based oligonucleotide microarray for functional diagnostics of nitrogen-fixing microorganisms. Microbial Ecology. 2007a;53:456–470. doi: 10.1007/s00248-006-9126-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D. Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. Journal of Experimental Botany. 2007b;58:2329–2338. doi: 10.1093/jxb/erm114. [DOI] [PubMed] [Google Scholar]