Abstract
Paramutation describes the transfer of an acquired epigenetic state to an unlinked homologous locus, resulting in a meiotically heritable alteration in gene expression. Early investigations of paramutation characterized a mode of change and inheritance distinct from mendelian genetics, catalyzing the concept of the epigenome. Numerous examples of paramutation and paramutation-like phenomena have now emerged, with evidence that implicates small RNAs in the transfer and maintenance of epigenetic states. In animals piRNA-mediated retrotransposon suppression seems to drive a vast system of epigenetic inheritance with paramutation-like characteristics. The classic examples of paramutation might be merely informative aberrations of pervasive and broadly conserved mechanisms that use RNA to sense homology and target epigenetic modification. When viewed in this context, paramutation is only one aspect of a common and broadly distributed form of inheritance based on epigenetic states.
The emergence of rogue traits
As mendelian genetics developed in the early 20th century, it was not uncommon for investigators to encounter traits that did not behave in the regular ways that were expected. Among the most commonly observed nonmendelian characteristics were patterns of pigmentation, including spotting in animals and striping in plants 1, 2. Most of these phenomena have never been explained, but a handful of cases 3, 4 seem to resemble what we now call paramutation. Our ability to ascribe any significance to those early reports stems from work begun separately by Brink 5 and by Coe 6 in the 1950s.
The phenomena studied by Brink and Coe were at odds with Mendelian orthodoxy and, perhaps for this reason, paramutation remained a mysterious and baffling phenomenon for decades, easy to dismiss as a curiosity with no broad significance. In light of recent developments, however, paramutation can be viewed as a byproduct of constitutive mechanisms that use small RNAs (sRNAs) to regulate epigenetic states. Its discoverers seem increasingly wise in their view that paramutation is a window into a mode of heredity operating alongside Mendelian genetics, but even Brink and Coe might find its extent surprising. Here we will discuss the history of the phenomenon, the evidence that it relies on deep-seated and pervasive mechanisms based on sRNAs, and how application of recent evidence provides insights into the mechanism of paramutation. The diversity of paramutation-like phenomena, and the very large numbers of sequences involved, suggests that epigenetic inheritance is much more common than previously supposed.
What is paramutation?
The term paramutation was coined by Brink, during his investigations of the R locus in maize, to describe “an interaction between alleles that leads to directed, heritable change at the locus with high frequency…” 3. Coe’s roughly contemporaneous investigations of the maize b1 locus produced evidence consistent with this definition 7. Although both loci encode proteins involved in anthocyanin pigment production, their structure, and their paramutation behaviors, are distinct. These idiosyncrasies are typical of paramutable loci 4, and thus paramutation has never had any equivalent to the rigor and predictive function of Mendel’s laws. Paramutation at the b1 locus is perhaps the best studied example of the phenomenon, and has a pattern that facilitates a general description (Figure 1). B1-Intense (B-I) and B’ are ‘epialleles’ of b1, with identical genetic structure but different expression states (lower in B’). B-I can spontaneously and heritably convert to B’, which is an epimutation, i.e. an epigenetically repressed allele. The B’ state is termed paramutagenic because when placed together with B-I, the B-I state invariably changes to B’; thus B-I is termed the paramutable allele. The newly acquired B’ state behaves like previously established B’ states: it is heritable and paramutagenic. Paramutation at R and other paramutable alleles is more difficult to describe; nevertheless they share the general features of epimutation, paramutagenicity, paramutability, and inheritance of the acquired state 3.
Figure 1. Paramutation made simple.
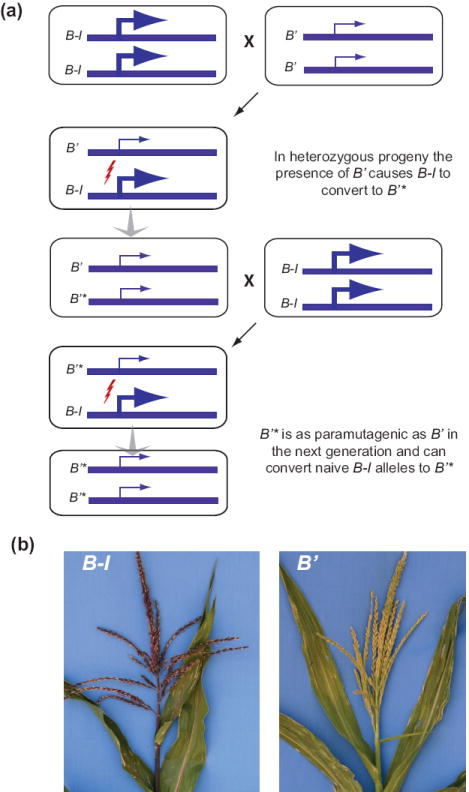
Paramutation at the maize b1 locus has simple characteristics that typify the phenomenon. B’ and B-I alleles are genetically identical but have distinct epigenetic states (i.e. they are epialleles); the paramutable allele B-I can spontaneously convert to the paramutagenic allele B’. (a) In the heterozygous progeny of homozygous parents, B’ invariably confers its epigenetic state on B-I (lightning bolt), which becomes B’*. The newly paramutant B’* epiallele rarely converts back to B-I, and is fully capable of paramutating a naïve B-I allele in subsequent generations. (b) Maize plants carrying the B-I (left) and B’ (right) epialleles; note purple pigmentation in B-I. B’* is indistinguishable from B’. Photographs kindly provided by Vicki Chandler and Lyudmila Sidorenko.
Early speculations on the mechanism
The mechanism(s) underlying paramutation remain incompletely understood, but Brink and Coe nevertheless were able to make some prophetic speculations. Brink’s investigations of R led to his astonishingly clear description 8 of what we now call the epigenome (Box 1), and to his view that paramutation is an aberration of normal processes. He saw specific alleles at r1 and paramutable alleles at other loci as peculiar variants whose behavior revealed the potential of the epigenome (which he termed ‘parachromatin’) to influence heredity. Coe’s insight, based on the B-I/B’ system, was more specific: he maintained that paramutation could be explained by transfer of a repressive substance from the paramutagenic to the paramutable allele, and he specifically proposed RNA as the repressor particle 9. The prescience of these ideas has been highlighted by modern developments.
Box 1. Brink’s conception of the epigenome.
In the 1950s R.A. Brink carried out investigations of paramutation at the R locus in maize 62, which together with his work on transposable elements led him to the earliest articulation of the relationship, and distinction, between the genome and the epigenome 8. Brink realized that the behavior of the R locus required a system of heredity distinct from the system (by then understood to be based on DNA) responsible for Mendelian inheritance. His views did not arise in isolation: by 1960, there had been decades of speculation concerning the means by which disparate phenotypes could arise from a single genome. In contrast to earlier discussions (including McClintock’s 63), Brink’s concept is an easily recognizable description of the nature and role of the epigenome 8. Brink discarded the terms euchromatin and heterochromatin because he felt that they carried confusing connotations, and wanted to avoid the sharp distinction often made between the two: the essence of Brink’s view is that many or most regions can exist as either heterochromatin or euchromatin. He instead proposed that a chromosome is composed of two general types of chromatin: orthochromatin and parachromatin. Orthochromatin is essentially synonymous with the genome, the substance that can be changed by mutation, is inherited in a regular pattern, and forms the basis of Mendelian genetics. By contrast, Brink proposed that parachromatin, which we now call the epigenome, is associated with orthochromatin (i.e. the genome), replicates itself in concert with the genome, and can act as a kind of genetic material. It is “…the meeting ground of the genetic and epigenetic functions of the chromosome” 8, responsible for functional changes in the genome, and reacting to the cellular environment to mediate differentiation. Brink recognized that the genome must remain constant, but that parachromatin must change to create a variety of reversible but heritable states of gene expression, and that cell differentiation involves the development of a succession of such states. Paramutation describes the normal process that occurs when parachromatin alters the function of a gene; the phenomena he and others had described were thus aberrant forms of a pervasive process.
Brink’s description of parachromatin makes it clear that he was, by 1960, in possession of a concept of the epigenome that is relevant even today. The striking clarity of his insight presaged the development of modern epigenetics, but has been little noted in recent times.
Progress toward a molecular basis of paramutation
The detailed studies of paramutation carried out by Brink, Coe, and others did not succeed in discovering its mechanism; it remained a mysterious exception to Mendel’s laws through the early part of the technological revolution in genetics that began in the 1970s. After a fallow period, investigations of the mechanism of paramutation were renewed in the 1990s 10, and many more examples of paramutation-like phenomena have been found. These have been reported at both endogenous and transgenic loci, and commonly involve variants of pigmentation (e.g. a1, pl1, b2) and antibiotic resistance (e.g. nptII, hpt, spt) (see 4, 11 and references within). Two models, not necessarily incompatible, have been invoked to explain the mechanism of paramutation12. The ‘pairing’ model is based on models from other systems, such as transvection in Drosophila, that propose physical association of homologous loci 13, 14, and transfer of an epigenetic state from one to the other. There is however no experimental evidence supporting pairing as a mechanism for paramutation. It is extremely difficult to prove that pairing does not occur during paramutation, and this might in part explain the model’s persistence. Furthermore, by itself pairing does not explain transfer of an epigenetic state: something more is required. This ‘something’ might be a mechanism based on transfer of homologous sequence (i.e. RNA), which could also make physical pairing unnecessary. In contrast to pairing, experimental evidence for the role of RNA has emerged.
Work focused on the b1 locus (Figure 1) has established that paramutagenic action derives from a transcriptional enhancer consisting of seven tandem repeats 100 kb upstream of the b1 coding region, which might form a transcriptional enhancer element 15; other b1 alleles with only a single repeat are not paramutable. Paramutation is associated with methylation of the repeats and a reduction of their sensitivity to nuclease digestion, both indicative of change to a repressed chromatin structure 16. The tandem repeats are transcribed in both B-I and B’ epialleles 17, and 25nt sRNAs derived from the repeats have been detected in plants of both types 18. Paramutation relies on activity of an RNA-dependent RNA polymerase (RDRP), mediator of paramutation 1 MOP1 17, which is homologous to plant RDRPs that mediate RNA-directed DNA methylation (RdDM) 19-21. A specialized RNA polymerase (RNA polIV), which transcribes sequences destined to be processed into sRNAs 22, 23, is required for the establishment and maintenance of paramutant states 24. Although these observations strongly imply that paramutation is mediated by sRNAs, it remains unknown precisely how the change from B-I to B’ is effected. Establishment and/or maintenance of paramutant states at R and another paramutable allele (pl1) also requires factors associated with RdDM 19, 20, 25, 26.
In isolation, the evidence implicating RNA in paramutation might be difficult to interpret: the significance of this evidence is derived largely from work on other systems of trans-silencing. Recent evidence has demonstrated that eukaryotes use deeply conserved mechanisms that process RNA into short single-stranded segments of ~20-30 nucleotides in length and use the sRNAs as guides to direct proteins of the Argonaute family 27 and associated effector protein complexes to homologous sequences 26, 28, 29. The profusion of names for these sRNAs is related to their size, known protein partners, and presumed functions 28. sRNA-bound complexes can trigger degradation, translational repression, rearrangement, or epigenetic modification 28-30. The latter phenomenon might be considered most relevant to paramutation: sRNAs produced from one region induce epigenetic modification of homologous sequences, either at the locus from which the RNA originated or at an unlinked locus 29, 31-40. sRNAs can silence a broad variety of sequences, principally repeat elements, transgenes, and rearranged loci, and the rapid progress of discovery suggests that the scope of their activities might be much broader; nevertheless the precise molecular mechanisms that direct silencing remain to be elucidated. A seeming paradox is that epigenetic modification seems to promote transcriptional repression, yet transcription of the repressed sequence is required to produce the sRNAs. In fission yeast, sRNAs are produced from ‘silent’ heterochromatic repeats by transcription that occurs only in S phase of the cell cycle; the RNAi machinery uses the sRNAs to rapidly silence the repeats by the end of S phase 41. This cell-cycle dependent transcription of silent loci provides a mechanism for maintaining epigenetic silence through cell division, and might be more broadly distributed.
Although investigations of sRNA-mediated epigenetic modification have informed interpretations of the molecular basis of paramutation, it is equally true that paramutation provides some of the best evidence for involvement of sRNAs in heritable epigenetic states affecting genes (rather than repeat sequences or transgenes). It should be noted that there are other examples, termed epimutation, similar to paramutation in that a gene is unusually epigenetically silenced, but lacking the feature of transfer of the epigenetic state from one allele to another 34, 42-44; there is clear evidence for involvement of sRNA in some 34. For example, repression of silent SUPERMAN epialleles in Arabidopsis thaliana is alleviated in plants defective in ARGONAUTE 4, a central element of the RdDM pathway 45.
All together, while it does not reveal how common these traits are, this evidence suggests that sRNA can mediate heritable epigenetic traits. The emphasis here on sRNA as a mediator of paramutation does not deprecate the possible involvement of other, perhaps separate, mechanisms in paramutation and similar phenomena, nor is it necessarily the case that every trans-sensing phenomenon is mediated by RNA. While the involvement of sRNAs in epigenetic phenomena is extensive, much remains to be understood; it is wise to doubt that they provide a complete explanation.
piRNA-mediated silencing: an example of adaptive paramutation in animals?
The evidence for paramutation in animals is scant in comparison to plants (as is typically the case for epigenetic phenomena), but sufficient to indicate that the phenomenon is possible (see Box 2). It seems likely, however, that more cases will emerge, because we now have evidence of a pathway of RNA-mediated homologous silencing in animals that carries out, on a large scale, functions strongly resembling paramutation.
BOX 2. Does paramutation occur in mammals?
There have been several reports suggestive of paramutation-like effects in mammals. While none of them are as fully investigated as the classic examples in plants, and there is no naturally occurring example that clearly involves paramutation, in at least one case the evidence is sufficient to conclude that a homology-mediated event has transferred an epigenetic state.
The first report involved polymorphic insulin alleles in patients with type I diabetes64. The predisposing effect of one allele is lost if it is paired with a protective allelic variant in the father. This implies that the untransmitted protective allele alters the state of the risk allele in the paternal germline, and that the altered state is transmitted to offspring. While suggestive of paramutation, this observation has not been confirmed, and the nature of the evidence leaves room for doubt as to its validity.
Mice heterozygous for a mutant c-kit locus created by lacZ insertion have white spotting and white bellies typical of c-kit mutations65. When bred to wild type mice, mutant mice produce wild type offspring with white tail spotting, which is then observed in variable proportions of wild type mice in succeeding generations. The authors proposed that transmission of tail spotting is a paramutation event mediated by aberrant c-kit RNAs observed in the testes of the mice. However this study did not recognize that white tail spotting occurs commonly in wild type C57Bl/6 mice, did not demonstrate that the progeny had a higher than normal incidence of tail spotting, and demonstrated no specific evidence for transmission of aberrant c-kit RNAs66-68.
The strongest example of a paramutation-like phenomenon in mammals comes from an investigation of sequences controlling imprinting in mice69. Soloway and colleagues replaced the region required for paternal methylation of the imprinted RAS protein-specific guanine nucleotide-releasing factor 1 (Rasgrf1) with the region controlling methylation of insulin-like growth factor 2 receptor (Igf2r). The mutant allele is paternally methylated and expressed like a wild-type allele. However paternal transmission of the mutant allele induces methylation and expression of a normally unmethylated and silent wild-type maternal allele in the next generation. As long as the allele is maternally transmitted it does not alter the wild-type allele. But once the wild-type allele has been altered by the mutant allele in the paternal germline, its acquired (methylated) state is maintained even when it is maternally transmitted and separated from the mutant allele.
The discovery of Piwi-interacting RNAs (piRNAs) points to a mechanism that could drive paramutation and similar phenomena in animals. piRNA functions resemble those of some plant sRNAs 23, 29, 31, 33-40, 46: sRNAs produced by transcription of repressed sequences feed back to induce epigenetic modification of homologous sequences. There is increasing evidence that piRNAs renew epigenetic silencing of retroelements in the animal germline 28, 31, 47-51; it currently is not known if they also regulate other, non-repetitive, sequences, but we speculate that they will be found to do so.
A specialized group of Argonaute 27 homologues, the Piwi proteins, partner with piRNAs in the germline. They are required for germ cell maintenance 27, 52, 53; mutations in Piwi family genes cause sterility, due at least in part to the activation of retrotransposons 31, 47, 49, 51, 52. Piwi homologues have been identified in multiple vertebrate species, where expression is prominent in germ cells but also occurs in other tissues 27, 52-54. Their ~25-30nt germline-specific piRNA partners are largely derived from retrotransposons 55-58. Investigations of I element control in Drosophila melanogaster (Figure I in Box 3) provide an example that resembles paramutation in some respects, as constitutively expressed piRNAs suppress I element activity and induce the formation of I element-derived piRNAs in germ cells 32, 49. In mammals, piRNAs are implicated in the methylation of retroelements they suppress, and they act upstream of cytosine methyltransferase 31, 47. Animals might also express other classes of sRNAs which mediate different homology-dependent silencing systems 28, 59.
Box 3. Primordial paramutation: piRNA-mediated retroelement suppression.
The Drosophila Piwi proteins, members of the argonaute family, and their mammalian homologues (miwi, hiwi, etc.) are expressed in germ cells27, 52, 53; they complex with piRNAs and presumably guide effector function to homologous sequences51, 55-57. The ability of piRNAs to mediate paramutation-like effects is illustrated by their role in I-R dysgenesis in Drosophila. I is a LINE element present in most Drosophila strains but absent from some70, 71. Reactive (R) strains lack functional copies of the I element, while inducer (I) strains carry about silenced 10 copies70, 72; both I and R strains have degenerate I element fragments in their pericentromeric heterochromatin32, 49, 70, 71. Crossing of an I male and R female results in I-R hybrid dysgenesis: most female progeny are sterile70, 73, the I element is active in their ovaries, retrotransposes in the oocyte, and causes mutations and sterility 32, 49. In flies that are able to reproduce, this activity ceases after a few generations, once the I element has multiplied to about 10 copies70, 72.
Both I and R strains have piRNAs derived from I elements in their female germ cells32, 49. In I strains most of these piRNAs are derived from I elements, but in R strains they are much less abundant, and they are derived from the degenerate pericentromeric I fragments32, 49. As repression of the I element increases, so does the abundance of I element piRNAs32, 49.
These findings are consistent with a model in which I elements are suppressed in I strains by piRNAs derived principally from the I elements themselves (Figure 2). In a dysgenic cross, suppression is lost, but piRNAs derived from the degenerate heterochromatic I elements begin to induce the production of piRNAs from I element transcripts; eventually these piRNAs become sufficiently abundant to suppress the I elements again32, 49.
I-R dysgenesis corresponds to paramutation at key points: the degenerate I fragments are functionally equivalent to paramutagenic alleles, and the intact I elements to paramutable alleles. The paramutable intact I elements participate in the maintenance of their own silencing, through piRNAs derived from their own transcripts. Paramutation, and perhaps epimutations seen in plants and animals42-44, 60, 61, might occur when the genetic architecture of a locus predisposes it to produce RNAs that are incorporated into the piRNA system and then feed back to the locus and its homologues.
Paramutation might occur in animals, as it does in plants, when the genetic structure of a locus permits the formation of RNAs that can be adopted into the piRNA system, or similar systems, and which then act as guides for Argonaute family members to find homologous loci and induce their silencing. This view does not answer the many questions about how paramutation occurs, but it provides a framework that helps to unify the disparate phenomena.
The realization that homology-dependent silencing mediated by sRNAs is affecting a very large proportion of the genome might also say something about the likely participation of pairing in paramutation. As noted previously, there remains no direct evidence that physical pairing of homologous loci is part of the paramutation process. Physical pairing is difficult to reconcile with the very large number of paramutation-like events that are probably occurring – it would require extraordinary and perhaps impossible contortions of the genome.
Concluding remarks
Paramutation, once a curiosity of plants, can be viewed as just one facet of a phenomenon that is broadly distributed in multicellular eukaryotes. What insights can paramutation provide, given that we now know so much about the underlying mechanisms? It might be useful to review Brink’s interpretation that paramutation is simply an aberration of a regulated process (Box 1) 8. The examples Brink, Coe, and others uncovered are useful today because of what they say about the ability of paramutation to generate heritable epigenetic information; our new understanding of the scope of the underlying mechanisms tells us that there could be a great deal of this information indeed.
In animals, it appears that all retrotransposons are subject to epigenetic inheritance. What we know about paramutation indicates that genes or their regulatory elements can become incorporated into this system of sRNA-mediated epigenetic inheritance, and from innocent bystanders turn into full-fledged denizens of the epigenetic underworld. This is something that clearly occurs in plants, and the existence of piRNA-mediated silencing in animals predicts its occurrence in that phylum also. All that is required is that a transcript, homologous to a specific locus, is made into piRNAs, and a self-reinforcing mechanism comes into play (Box 3). Such an event could occasionally occur simply as an accident; indeed, this might be the origin of the germline epimutations of the tumor suppressor Mut L homolog 1 (MLH1) 44, of the peloric variant of toadflax described by Linnaeus 42, and other epimutations of seemingly pristine loci43, 60, 61.
Adaptive mechanisms underlie paramutation, but is paramutation itself a mechanism of variation that can be operated on by selection? If stable and heritable traits are generated by sRNS-mediated mechanisms (as seems to be the case), then there is every reason to suppose that in some circumstances such traits are selected and become fixed in a population. Although such a scenario is inconsistent with Mendelian genetics, it is entirely compatible with Darwinian evolution. The big questions raised by this insight concern the amount of epigenetic information that has been generated by paramutation-like mechanisms and is maintained in the animal germline, and the extent to which this information shapes the stable characteristics of species.
Figure 2. piRNAs suppress hybrid dysgenesis.
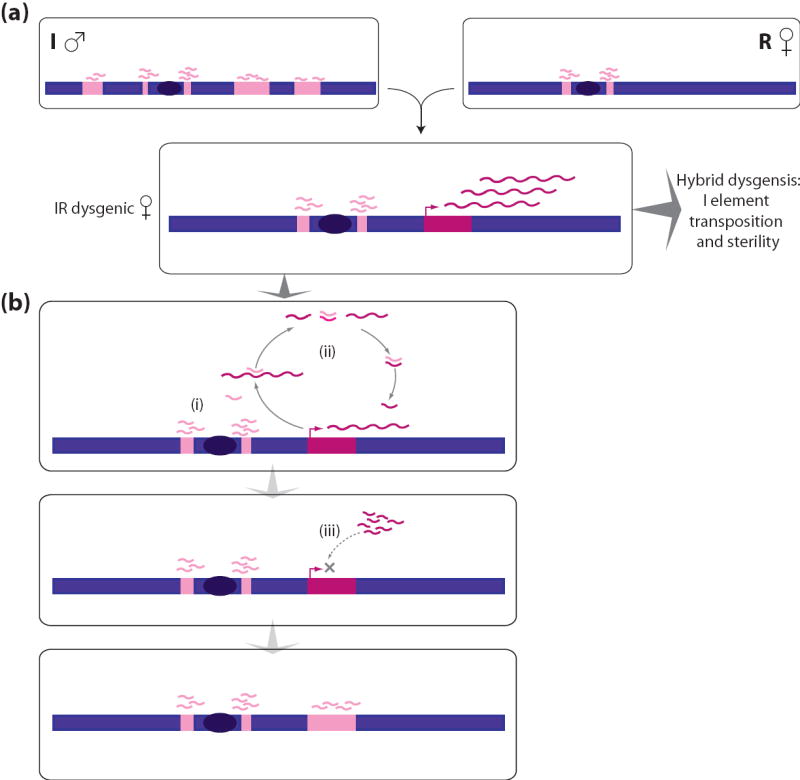
(a) In Drosophila, crossing of an inducer (I) male and reactive (R) female results in I-R hybrid dysgenesis, where paternally contributed I elements (dark pink) become active and retrotranspose in the oocyte, causing sterility. Both inducer (I) and reactive (R) Drosophila strains express piRNAs derived from I elements (pale pink fragments), but piRNAs are derived only from degenerate pericentromeric I fragments in R strains. (b) Most female progeny are sterile due to mutations caused by I element retrotransposition, but some are able to reproduce. In these females, piRNAs derived from pericentromeric I elements (i) direct new piRNA formation from the newly integrated paternal I elements by the ‘ping-pong’ mechanism (ii); eventually these new piRNAs become sufficiently abundant to direct stable I element silencing (iii).
Acknowledgments
The authors wish to thank Jeremy Brown, Jennifer Cropley, Dario Boffelli and Michael Buckland for helpful comments and discussions, and JC for help with the figures. We apologize to those authors of primary papers we have not been able to cite due to space restrictions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan TH. Chromosomes and heredity. The American Naturalist. 1910;44:449–496. [Google Scholar]
- 2.Cuénot L. L’Hérédité de la pigmentation chez les souris (3me note) Arch Zool Exp Gén. 1904;4:45–56. [Google Scholar]
- 3.Brink RA. Paramutation. Annu Rev Genet. 1973;7:129–152. doi: 10.1146/annurev.ge.07.120173.001021. [DOI] [PubMed] [Google Scholar]
- 4.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 5.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coe EH. A Regular and Continuing Conversion-Type Phenomenon at the B Locus in Maize. Proc Natl Acad Sci U S A. 1959;45:828–832. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coe EH. The Properties, Origin, and Mechanism of Conversion-Type Inheritance at the B Locus in Maize. Genetics. 1966;53:1035–1063. doi: 10.1093/genetics/53.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brink RA. Paramutation and chromosome organization. Q Rev Biol. 1960;35:120–137. doi: 10.1086/403016. [DOI] [PubMed] [Google Scholar]
- 9.Coe EH., Jr Heritable repression due to paramutation in maize. Science. 1968;162:925. doi: 10.1126/science.162.3856.925. [DOI] [PubMed] [Google Scholar]
- 10.Chandler V, Alleman M. Paramutation: epigenetic instructions passed across generations. Genetics. 2008;178:1839–1844. doi: 10.1093/genetics/178.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittelsten Scheid O, et al. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. [DOI] [PubMed] [Google Scholar]
- 12.Stam M. Paramutation: a heritable change in gene expression by allelic interactions in trans. Mol Plant. 2009;2:578–588. doi: 10.1093/mp/ssp020. [DOI] [PubMed] [Google Scholar]
- 13.Grant-Downton RT, Dickinson HG. Plants, pairing and phenotypes--two’s company? Trends Genet. 2004;20:188–195. doi: 10.1016/j.tig.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 15.Stam M, et al. The regulatory regions required for B’ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stam M, et al. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 18.Chandler VL. Paramutation: from maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Dorweiler JE, et al. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erhard KF, Jr, et al. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 21.Matzke M, et al. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Wierzbicki AT, et al. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollick JB, et al. Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics. 2005;171:725–740. doi: 10.1534/genetics.105.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale CJ, et al. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollick JB. Sensing the epigenome. Trends Plant Sci. 2008;13:398–404. doi: 10.1016/j.tplants.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaratiegui M, et al. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambeyron S, et al. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 35.Janowski BA, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 36.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 37.Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- 38.Preuss SB, et al. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell. 2008;32:673–684. doi: 10.1016/j.molcel.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]
- 41.Kloc A, et al. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cubas P, et al. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 43.Jacobsen SE, Meyerowitz EM. Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 44.Suter CM, et al. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 45.Zilberman D, et al. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 46.Llave C, et al. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravin AA, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 48.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 49.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 52.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 53.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki T, et al. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 55.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 56.Girard A, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 57.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 58.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 59.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 60.Das OP, Messing J. Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics. 1994;136:1121–1141. doi: 10.1093/genetics/136.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 62.Brink RA, et al. Paramutation: directed genetic change. Paramutation occurs in somatic cells and heritably alters the functional state of a locus. Science. 1968;159:161–170. doi: 10.1126/science.159.3811.161. [DOI] [PubMed] [Google Scholar]
- 63.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Bennett ST, et al. Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. The IMDIAB Group. Nat Genet. 1997;17:350–352. doi: 10.1038/ng1197-350. [DOI] [PubMed] [Google Scholar]
- 65.Rassoulzadegan M, et al. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 66.Arnheiter H. Mammalian paramutation: a tail’s tale? Pigment Cell Res. 2007;20:36–40. doi: 10.1111/j.1600-0749.2006.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnheiter H. ‘A response to ‘Mammalian paramutation: a tail’s tale?’- a commentary by H. Arnheiter on our paramutation paper’- a reply. Pigment Cell Melanoma Res. 2009;22:142–143. doi: 10.1111/j.1755-148X.2008.00521.x. [DOI] [PubMed] [Google Scholar]
- 68.Rassoulzadegan M. A response to ‘Mammalian paramutation: a tail’s tale?’- a commentary by H. Arnheiter on our paramutation paper. Pigment Cell Melanoma Res. 2009;22:140–141. doi: 10.1111/j.1755-148X.2008.00521.x. [DOI] [PubMed] [Google Scholar]
- 69.Herman H, et al. Trans allele methylation and paramutation-like effects in mice. Nat Genet. 2003;34:199–202. doi: 10.1038/ng1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chambeyron S, Bucheton A. I elements in Drosophila: in vivo retrotransposition and regulation. Cytogenet Genome Res. 2005;110:215–222. doi: 10.1159/000084955. [DOI] [PubMed] [Google Scholar]
- 71.Jensen S, et al. Regulation of I-transposon activity in Drosophila: evidence for cosuppression of nonhomologous transgenes and possible role of ancestral I-related pericentromeric elements. Genetics. 2002;162:1197–1209. doi: 10.1093/genetics/162.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jensen S, et al. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 73.Bucheton A, et al. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]


