Abstract
Abnormal vascular phenotypes have been implicated in neuropathologies ranging from Alzheimer's disease to brain tumors. The development of transgenic mouse models of such diseases has created a crucial need for characterizing the murine neurovasculature. Although histologic techniques are excellent for imaging the microvasculature at submicron resolutions, they offer only limited coverage. It is also challenging to reconstruct the three-dimensional (3D) vasculature and other structures, such as white matter tracts, after tissue sectioning. Here, we describe a novel method for 3D whole-brain mapping of the murine vasculature using magnetic resonance microscopy (μMRI), and its application to a preclinical brain tumor model. The 3D vascular architecture was characterized by six morphologic parameters: vessel length, vessel radius, microvessel density, length per unit volume, fractional blood volume, and tortuosity. Region-of-interest analysis showed significant differences in the vascular phenotype between the tumor and the contralateral brain, as well as between postinoculation day 12 and day 17 tumors. These results unequivocally show the feasibility of using μMRI to characterize the vascular phenotype of brain tumors. Finally, we show that combining these vascular data with coregistered images acquired with diffusion-weighted MRI provides a new tool for investigating the relationship between angiogenesis and concomitant changes in the brain tumor microenvironment.
Keywords: angiogenesis, brain tumor microenvironment, diffusion tensor imaging, magnetic resonance microscopy, vasculature
Introduction
Knowledge of the three-dimensional (3D) architecture of blood vessels is crucial for several reasons: (1) Various neuropathologies ranging from Alzheimer's disease to brain tumors involve anomalous blood vessels (Iadecola, 2004; Jain et al, 2007). (2) The physiologic basis of image contrast in functional magnetic resonance imaging critically depends on the underlying microvasculature (Moonen et al, 1990; Pathak et al, 2003, 2008c). (3) The cerebral vasculature is central to understanding the hemodynamics and the pharmacokinetics of novel therapies in the brain (Reichold et al, 2009). (4) The widespread development of transgenic mouse models of neurologic disease has created a need for characterizing the vasculature of the murine brain (Raman et al, 2007).
In the case of brain tumors, the vascular architecture is a critical determinant of their pathophysiology. Vessel morphology of high-grade gliomas is highly heterogeneous and markedly different from the normal cerebral vasculature. It is also strongly dependent on tumor size, location within the tumor, and local angiogenic activity (Vajkoczy and Menger, 2000). Therefore, the vascular architecture provides insights into the ‘angiogenic' phenotype, which traditionally has been characterized using histologic techniques in terms of microvessel density (MVD) (Weidner, 1995). Although optical microscopy is a powerful tool for imaging the microvasculature at submicron resolutions, it offers only limited coverage, and recovering the 3D vascular and white matter fiber morphology after tissue sectioning requires complex reconstruction.
Multiphoton microscopy is a 3D optical imaging technique that has been used to image murine cortical capillaries at submicron resolution (Verant et al, 2007), but the imaging depth is limited to a few hundred microns. A recent study used optical coherence tomography to image vascular and lymphatic morphology, as well as tissue viability in various tumor models in vivo (Vakoc et al, 2009). Optical coherence tomography offers a wider field of view and greater depth of penetration at the expense of resolution relative to multiphoton microscopy, and does not require exogenous contrast agents. However, the maximum imaging depth is between 1 and 2 mm below the tissue surface; hence, coverage is still limited relative to modalities such as MRI and computed tomography (CT).
Studies have used micro-CT (μCT) to image the whole 3D mouse cerebral vasculature for various applications, including creating a vascular atlas integrated with an MRI brain atlas (Dorr et al, 2007), characterizing the vascular architecture of transgenic mice (Heinzer et al, 2008), and measuring cerebral blood volume (Chugh et al, 2009). Recently, a hierarchical imaging method was developed that involved using synchrotron radiation μCT (SRμCT) to image corrosion casts of 3D capillary networks at 1.4 μm resolution in select regions of interest (ROIs) 1 mm3 in size (Heinzer et al, 2006). However, constructing an intact corrosion cast without damaging microvessels remains technically challenging, and any complementary information is lost during tissue maceration (Hossler and Douglas, 2001). Another study used SRμCT imaging of small cylindrical tissue samples to compare normal and tumor vasculatures in a rat brain 9L gliosarcoma model (Risser et al, 2007). Although exquisite 3D images of the microvasculature can be acquired, this technique is not widely accessible because of the need for synchrotron-generated high-energy X-rays. Moreover, μCT does not offer soft tissue contrast or contrast based on aspects of the brain tumor microenvironment such as changes in cellularity caused by tumor growth and invasion. Such a capability is crucial for studying the interactions between tumor vasculature and its micromilleu.
Here, we describe a new method for phenotyping the vasculature of a brain tumor model using micro-MRI (μMRI), a rapidly evolving type of 3D MRI that offers very high ex vivo imaging resolutions (Johnson et al, 1993), and has previously been applied to phenotyping various transgenic mouse models (Benveniste and Blackband, 2002; Johnson et al, 2002). As μMRI is nondestructive and preserves 3D tissue and blood vessel architecture, it has been used to characterize the mouse embryonic vasculature (Berrios-Otero et al, 2009; Smith et al, 1994). When combined with different kinds of MR contrasts such as diffusion-weighted (DW) MRI, μMRI can provide unique information on the brain tumor microenvironment. For example, the apparent diffusion coefficient (ADC) and fractional anisotropy (FA) have been shown to reflect brain tumor cell density (Chenevert et al, 2000) and brain tumor invasion (Bennett et al, 2004), respectively. Furthermore, acquiring coregistered 3D vascular, anatomic, and diffusion data in one imaging session precludes the need for complex image-registration algorithms. In this study, we exploited the ability of μMRI to generate high-resolution 3D images of the vasculature of the entire mouse brain, and to characterize morphologic differences between the vascular architectures of a 9L brain tumor and the contralateral brain. We also characterized differences in angiogenesis between 9L brain tumors at two different growth stages (namely postinoculation day 12 (D12) and day 17 (D17)). Finally, we measured concomitant changes in ADC and FA at these time points, and assessed the ability of μMRI-derived parameters to phenotype each brain tumor microenvironment.
Materials and methods
Brain Tumor Model and Inoculations
In all, 10 female SCID (severe combined immunodeficiency) mice, weighing ∼25 g, were anesthetized with a ketamine/acepromazine cocktail. Their heads were immobilized in a stereotactic frame, and then using an aseptic technique, a 1-mm burr hole was drilled in the skull 1 mm anterior and 2 mm lateral to the bregma on the right side. A 10-μL gas-tight syringe (Hamilton Co., Reno, NV, USA) was used to inject 105 9L cells in 2 μL RPMI cell culture medium (Invitrogen, Carlsbad, CA, USA) into the right frontal lobe at a depth of 3 mm relative to the dural surface. The 9L cell line was grown in Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) with 10% fetal calf serum and expanded immediately before inoculation. The injection time was 5 minutes, after which the needle was retracted slowly for an additional 5 minutes. The skin was then closed with surgical staples that were removed before MRI. All animal studies were performed in accordance with the institutional Animal Care and Use Committee guidelines.
Whole-Brain Sample Preparation
Five 9L brain tumor-bearing mice were sacrificed on postinoculation D12 and another five on postinoculation D17. The mice were perfusion fixed, followed by perfusion with a silicone rubber compound called Microfil (FlowTech Inc., Carver, MA, USA) according to a method we developed previously (Pathak et al, 2008b). In brief, mice were deeply anesthetized with isoflurane and then perfused through the left ventricle—first with heparinized phosphate-buffered saline, and then with 10% buffered formalin (Bestvater et al, 2002) for fixation, and finally with a 1:2 mixture of Microfil and diluent added with 5% curing agent by volume. The Microfil was allowed to cure at room temperature for 90 minutes, and then the heads were fixed in cold formalin for 2 days. Next, the brains were excised from their skulls and stored in cold formalin. Twenty-four hours before imaging, each brain was immersed in phosphate-buffered saline doped with 1 mmol/L Gd-DTPA (Omniscan, GE Healthcare, Oslo, Norway) to enhance soft tissue contrast. Each brain was imaged in an nuclear magnetic resonance (NMR) tube filled with Fomblin (Solvay Solexis, Milano, Italy), which provides a dark, uniform image background because it does not contribute any proton signal to the MR images.
Micro-Magnetic Resonance Imaging Protocol
After fixation, the five D12 and five D17 brains were imaged on a 400-MHz spectrometer in a 10-mm volume radio-frequency coil (Bruker BioSpin Corp., Billerica, MA, USA) using the following sequences and parameters: (1) T2*-weighted multiple gradient echo (MGE), echo time=4.9/9.7/14.5/19.3/24.0/28.8 milliseconds, repetition time=100 milliseconds, resolution=64 μm × 64 μm × 62 μm, flip angle (θ)=30°, averages=20, and total acquisition time ∼11 hours. (2) T2-weighted rapid acquisition with refocused echoes, echo time/repetition time=34.5/500 milliseconds, resolution=129 μm × 125 μm × 125 μm, θ=40°, averages=4, and total acquisition time ∼4 hours. (3) Diffusion tensor imaging (DTI), echo time/repetition time=34.38/2,000 milliseconds, resolution=129 μm × 125 μm × 1,000 μm, θ=90°, averages=20, and total acquisition time ∼11 hours; 1 non-DW image and 6 DW images were acquired with a b-value of 1,700 seconds/mm2 and diffusion-sensitizing gradient orientations: [1,1,0], [1,0,1], [0,1,1], [−1,1,0], [1,0,−1], [0,−1,1].
Image Processing
For each brain, a 3D tumor ROI was manually delineated on the T2-weighted images in the Amira visualization software (Visage Imaging, San Diego, CA, USA). A homologous contralateral ROI was defined by reflecting the tumor ROI across the brain midline. As some of the D17 tumors had grown past the midline, manual modifications to contralateral ROIs were necessary to exclude any tumor tissue.
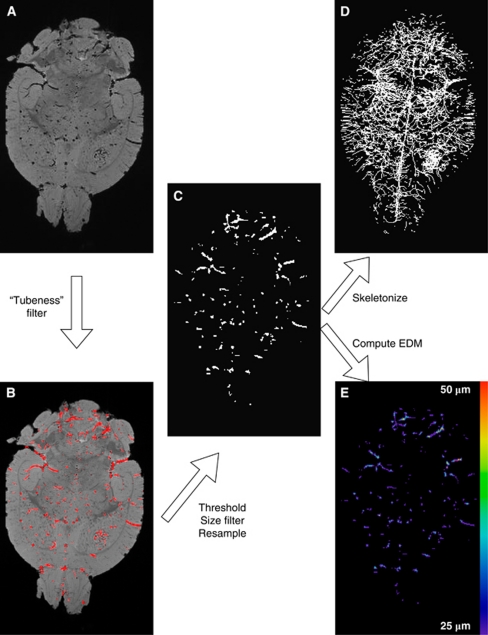
Three-dimensional MGE images corresponding to the first echo time were imported into ImageJ (Rasband WS, National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/) for blood vessel extraction and computation of vascular morphologic parameters. Initially, we intended to calculate voxelwise R2* (1/T2*) maps from the MGE data to segment the vasculature based on the R2* enhancement caused by the susceptibility difference between tissue and Microfil. However, the lack of mobile water protons within microfilled vessels precluded reliable curve fitting to calculate R2*. Blood vessels appear dark in MGE images because of the lack of free water in the microfilled lumina (Figure 1A). We segmented the vasculature using images from the first echo because they exhibited the highest contrast-to-noise ratio and the least susceptibility-related artifacts. The image processing procedure for vessel extraction is summarized in Figure 1. In brief, the vasculature was automatically segmented from the brain using a Hessian-based ‘tubeness' filter (Sato et al, 1998). This filter determines how ‘tube-like' a voxel is by convolving the image with a spherical Gaussian kernel with s.d. σ, computing the Hessian matrix at each voxel, and calculating a ‘tubeness' metric from the Hessian eigenvalues (Figure 1B). The sensitivity of the ‘tubeness' filter to tubular structures of varying radii can be tuned by varying σ. Therefore, we implemented a multiscale integration of filter responses using three σ values (σ=0.8, σ=1.0, and σ=1.2) to optimally extract vessels of all radii in the μMRI image (Sato et al, 1998). Next, a binary 3D vascular structure was obtained by applying an iterative threshold algorithm (Ridler and Calvard, 1978) on the result of the ‘tubeness' filter (Figure 1C). To eliminate small, disconnected objects and gaps in the vasculature, the binary image was subjected to a 3D size filter to remove isolated background and foreground regions smaller than three voxels.
Figure 1.
Image processing flowchart depicting an axial slice of a D12 brain. (A) First-echo MGE image of a D12 brain. (B) Output of the ‘tubeness' filter in red overlaid on the original image. (C) Three-dimensional binary vascular structure after thresholding, filtering, and resampling. (D) Maximum intensity projection (MIP) of the skeletonized vascular structure. (E) Three-dimensional EDM map showing the minimum distance in μm between each vessel voxel and the background. The color reproduction of this figure is available on the html full text version of the manuscript. D12, postinoculation day 12; MGE, multiple gradient echo.
Morphologic Characterization of the Vascular Phenotype
After segmenting the vasculature, we computed several morphometric parameters to characterize the 3D vascular architecture of tumor and contralateral ROIs. The fractional vascular volume (FV) of tumor and contralateral ROIs was determined by computing the fractional occupancy of the binary vascular structure within each ROI. All ROIs and binary vascular structures were resampled using bicubic interpolation to 50 μm isotropic resolution to allow consistent measurement of vascular morphologic parameters across all brains. The ImageJ ‘3D Skeletonization' plugin was used to automatically extract the vascular centerlines or 3D skeleton (Lee et al, 1994) (Figure 1D), to count the number of vessel branches, and to compute vessel branch lengths and Euclidean distances between branch vertices. From these parameters, we calculated the vascular tortuosity (vessel length/Euclidean distance), length per unit volume (LV=total summed vessel length/ROI volume) and MVD (number of branches/ROI volume). To measure vessel radius, a 3D Euclidean distance map (EDM) of the vasculature was computed using the ChamferMap module in Amira. This EDM represents the smallest distance between each vessel voxel and the background. Vessel radii were determined by multiplying the binary skeletonized image with the EDM (Figure 1E). These steps are summarized in Figure 1. All 3D visualizations were performed in Amira.
Apparent Diffusion Coefficient and Fractional Anisotropy Maps
As mentioned previously, a major advantage of μMRI over μCT is the availability of various MR contrast mechanisms. We resampled the high-resolution tumor and contralateral ROIs in ImageJ using bicubic interpolation to match the coarser resolution of the DTI data. Voxelwise ADC and FA maps were computed from the DTI data for each brain using DTI-Studio (H Jiang and S Mori, Radiology Department, Johns Hopkins University, Baltimore, MD, USA). For each voxel, we calculated the eigenvalues of the diffusion tensor (λ1, λ2, and λ3), from which ADC and FA were computed according to:
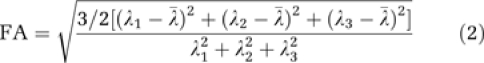 |
Finally, the median ADC and FA values of each ROI were computed using ImageJ.
Zonal Analysis of Tumor Regions of Interest
To further investigate the phenotypes of these tumors, we determined how the computed morphologic and diffusion parameters varied spatially within tumor ROIs. We divided each tumor ROI into three subregions of approximately equal volume by performing two successive 3D binary morphologic erosions in ImageJ. For the 50-μm isotropic ROIs, we used a spherical structuring element with a diameter of 9 voxels (450 μm). For tumor ROIs subsampled for DTI analysis, we used a two-dimensional structuring element with a diameter of five voxels (∼400 μm). In all cases, the result of the first erosion was subtracted from the original ROI to define the tumor rim; the result of the second erosion defined the tumor core; and the difference of the first and second erosion operations defined an intermediate zone. All of the vascular and diffusion parameters computed above were recomputed for the three zonal ROIs using the methods described above.
Cluster Analysis of Micro-Magnetic Resonance Imaging Parameters
To assess the feasibility of generating a ‘signature' of the angiogenic phenotype using the parameters obtained from our technique, we implemented an unsupervised hierarchical clustering algorithm on all 20 brain ROIs using vessel length, vessel radius, MVD, LV, FV, tortuosity, ADC, and FA as cluster variables. We used the ‘complete linkage' clustering algorithm, which defines the distance between two clusters as the distance between their two farthest-apart members based on the cluster variables and yields well-separated and compact clusters. We used the Manhattan distance metric, which is defined as:
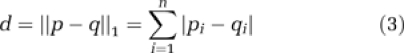 |
where p=(p1, p2,…, pn) and q=(q1, q2,…, qn). The cluster analysis was conducted using NCSS (Kaysville, UT, USA) for Windows.
Statistical Analysis
The median value of each μMRI parameter was determined for every tumor in each group of animals. A two-tailed, nonparametric Mann–Whitney U-test was used (α=0.05) to determine whether median μMRI parameters for tumor ROIs were significantly different from those for contralateral ROIs. The same test was used to determine whether there were any significant differences between μMRI parameters corresponding to D12 and D17.
Results
Morphologic Characterization of the Vascular Phenotype
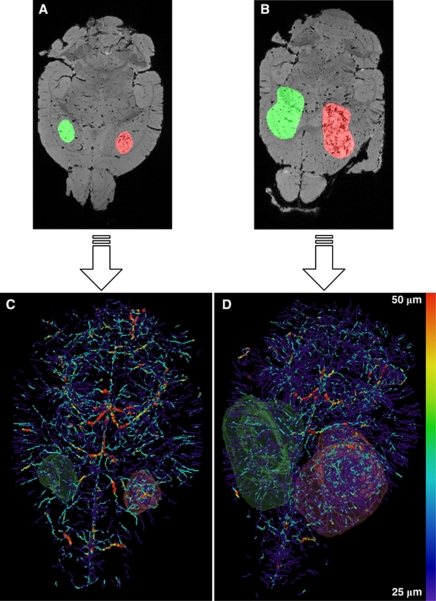
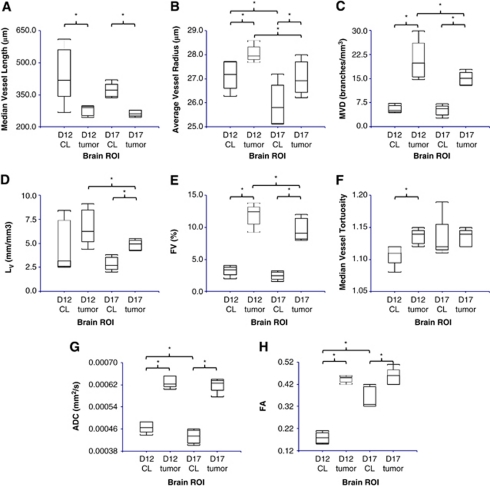
Figures 2A and 2B show the manually segmented tumor and contralateral ROIs overlaid on the MGE μMRI data, and Figures 2C and 2D show the resultant radius-encoded vascular maps for representative D12 and D17 9L tumor-bearing brains, respectively. For tumor and contralateral ROIs of each brain, six morphologic parameters were computed as summarized in Figures 3A to 3H: median vessel length, average vessel radius, MVD, LV, FV, and median vessel tortuosity. Figures 3A to 3H show that with the exception of D12 LV and D17 tortuosity, there was a significant (P<0.05) difference between these parameters for tumor and contralateral ROIs. For D17 brains, LV was significantly greater in tumors than in contralateral ROIs (Figure 3D); for D12 brains, tortuosity was significantly greater in tumors (Figure 3F). For both D12 and D17 groups, vessel radius, MVD, and FV were elevated in tumor with regard to contralateral ROIs (Figures 3B, 3C, and 3E), whereas vessel length exhibited the opposite relationship (Figure 3A). Finally, MVD, LV, and FV were found to be significantly higher in D12 tumors than in D17 tumors (Figures 3C to 3E).
Figure 2.
Raw data and final results of image processing for representative D12 and D17 brains. Axial slices of the raw first-echo MGE μMRI data of a (A) D12 and (B) D17 brain. Tumor and contralateral ROIs are overlaid in red and green, respectively. (C, D) Three-dimensional vascular structures color coded by vessel radius and volume rendered ROIs corresponding to the μMRI data in panels A and B. D12, postinoculation day 12; D17, postinoculation day 17; MGE, multiple gradient echo;μMRI, micro-magnetic resonance imaging; ROI, region of interest.
Figure 3.
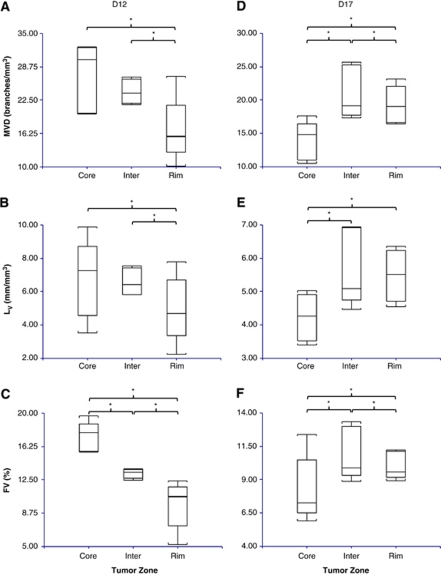
Box and whisker plots of six vascular and two diffusion parameters computed for D12 contralateral, D12 tumor, D17 contralateral, and D17 tumor ROIs (n=5 for all groups, *P<0.05). (A) The median vessel length, (B) average vessel radius, (C) MVD, (D) LV, (E) FV, (F) median vessel tortuosity (G) ADC, and (H) FA. ADC, apparent diffusion coefficient; D12, postinoculation day 12; D17, postinoculation day 17; FA, fractional anisotropy; FV, fractional vascular volume; LV, length per unit volume; MVD, microvessel density; ROI, region of interest.
Apparent Diffusion Coefficient and Fractional Anisotropy Maps
Figures 4A to 4D show coronal slices of representative ADC and FA maps for the same D12 and D17 tumor-bearing brains shown in Figures 2A and 2B. From these images, it is immediately apparent that ADC and FA were significantly higher in tumor than in contralateral ROIs for both D12 and D17 groups. This observation is summarized in Figures 3G and 3H. These data also show that ADC decreased and FA increased significantly in contralateral ROIs from D12 to D17. In contrast, there was no significant difference in these parameters between D12 and D17 tumor ROIs.
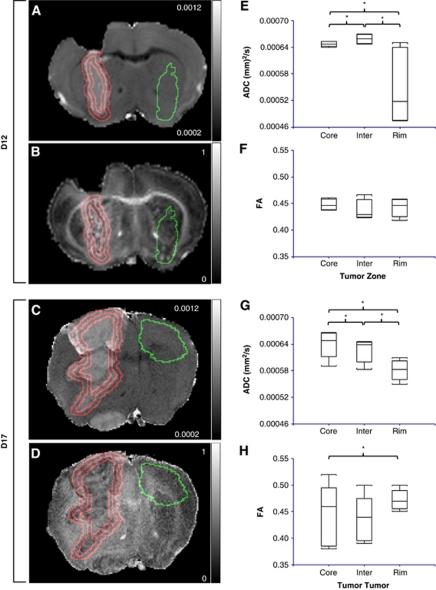
Figure 4.
ROI analysis of tumor ADC and FA. Coronal slices of the ADC and FA maps of representative D12 (A, B) and D17 (C, D) tumor-bearing brains. The contralateral ROI is outlined in green; the tumor core, intermediate (Inter), and rim zones are outlined in red. Box and whisker plots of D12 tumor (E) ADC, (F) FA, and D17 tumor (G) ADC, and (H) FA by zone (n=5 for all groups, *P<0.05): core, intermediate, and rim. Zones were defined by performing sequential 3D morphologic erosions. The color reproduction of this figure is available on the html full text version of the manuscript. ADC, apparent diffusion coefficient; D12, postinoculation day 12; D17, postinoculation day 17; FA, fractional anisotropy; ROI, region of interest; 3D, three dimensional.
Zonal Analysis of Tumor Regions of Interest
In both D12 and D17 tumors, ADC was significantly different in all three zones (Figures 4E and 4G). It was lowest in the rim for both groups, highest in the intermediate zone for D12 tumors, and highest in the core for D17 tumors. In contrast, there was no significant difference in FA across the three zones in D12 tumors (Figure 4F). In D17 tumors, FA was significantly greater in the rim than in the core (Figure 4H).
Vessel length and radius did not vary significantly between zones in D12 tumors. However, in the core of D17 tumors, vessel length was greater and vessel radius smaller than in the other two zones. In D12 tumors, MVD and LV were significantly lower in the rim than in the two inner zones, and FV decreased significantly from the core to the intermediate to the rim (Figures 5A to 5C). In contrast, in D17 tumors, MVD, LV, and FV were lower in the core than in the other zones (Figures 5D to 5F). Both MVD and FV were significantly greater in the intermediate zone than in the rim, as well. In D12 tumors, vessel tortuosity was significantly greater in the intermediate zone than in the rim, whereas no significant variation in tortuosity was seen between zones for D17 tumors.
Figure 5.
Box and whisker plots of select morphometric vascular parameters for D12 and D17 tumors by zone (n=5 for all groups, *P<0.05): core, intermediate (Inter), and rim. Zones were defined by performing sequential 3D morphologic erosions. D12 tumor (A) MVD, (B) LV and (C) FV. D17 (D) MVD, (E) LV and (F) FV. D12, postinoculation day 12; D17, postinoculation day 17; FV, fractional vascular volume; LV, length per unit volume; MVD, microvessel density.
Cluster Analysis of Micro-Magnetic Resonance Imaging Parameters
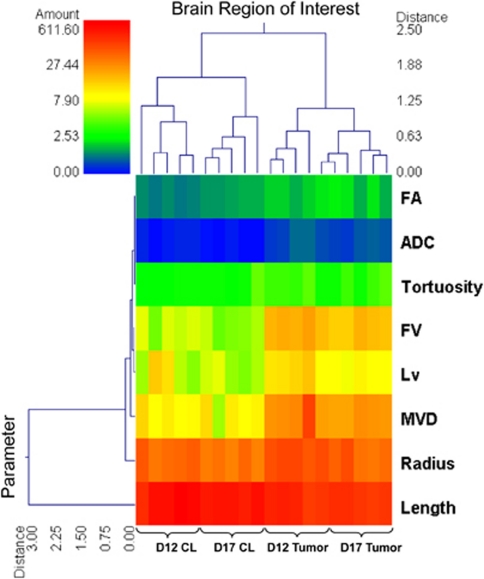
Figure 6 shows the feasibility of using μMRI-measured parameters to characterize and distinguish between the phenotypes of the 9L tumor model at D12 and D17. On the basis of six morphologic parameters and two diffusion parameters calculated in this study, unsupervised hierarchical clustering correctly classified every brain ROI as being D12 contralateral, D17 contralateral, D12 tumor, or D17 tumor, with the exception of one D12 tumor ROI being assigned to the D17 tumor cluster. Finally, Figures 7A to 7D illustrate the agreement between histology and the segmented vasculature. Figures 7E to 7H show the utility of μMRI in examining changes in the brain tumor microenvironment by combining vascular and DTI data. Such complementary contrast mechanisms permit assessments of the correlation between various microenvironmental variables.
Figure 6.
Double dendrogram showing the results of the unsupervised hierarchical clustering analysis. The clustering variables are organized by rows, and brain ROIs by columns. Based on ADC, FA, and six vascular morphometric parameters, the clustering algorithm separated contralateral from tumor, and ordered all 20 ROIs correctly by group (D12 contralateral, D17 contralateral, D12 tumor, D17 tumor), with D12 contralateral and D17 tumor being farthest apart in parameter space. ADC, apparent diffusion coefficient; D12, postinoculation day 12; D17, postinoculation day 17; FA, fractional anisotropy; FV, fractional vascular volume; LV, length per unit volume; MVD, microvessel density; ROI, region of interest.
Figure 7.
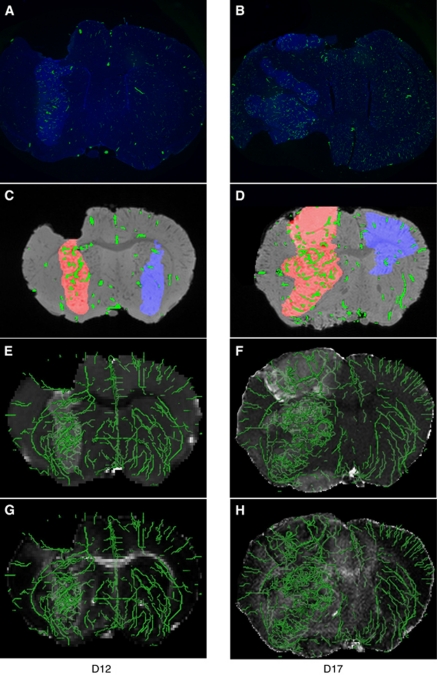
Comparison of histology and μMRI vascular and diffusion data. Representative histology of (A) D12 and (B) D17 9L tumor-bearing mouse brains. Nuclei are visible in blue, and microfilled vessels in green. (C, D) Corresponding slices of μMRI MGE images. Tumor and contralateral ROIs are shown in red and blue, respectively. Segmented vessels are shown in green. (E–H) Complementary diffusion data inherently coregistered to the vascular data. (Panels E and F) ADC and (panels G and H) FA maps of the slices shown in panels C and D), respectively, overlaid with 3D renderings of coregistered vasculature in green. Gray scale bars are omitted for clarity and are the same as shown in Figure 4. ADC, apparent diffusion coefficient; D12, postinoculation day 12; D17, postinoculation day 17; FA, fractional anisotropy; MGE, multiple gradient echo; μMRI, micro-magnetic resonance imaging; ROI, region of interest; 3D, three dimensional.
Discussion
Imaging the 3D neurovascular architecture with μMRI has many potential applications, a few of which will be discussed here. Traditionally, the neurovasculature has been characterized at two different spatial scales: the ‘cellular' or submicron scale using optical microscopy methods, and the ‘systemic' or submillimeter scale using methods such as in vivo MRI. However, coregistering histology with in vivo imaging data is challenging because of their vastly different spatial scales. Micro-MRI can potentially be used to bridge this resolution gap between optical imaging and in vivo MRI, facilitating coregistration of cellular factors (e.g., distribution of vascular endothelial growth factor) with in vivo biomarkers of angiogenesis, such as cerebral blood volume and vessel size index (Pathak et al, 2008a). Both cerebral blood volume and vessel size index measurements are derived from susceptibility-contrast MRI, but the relationship between brain tumor angiogenesis and susceptibility-induced contrast is not well understood. It has been shown that abnormal tumor vessel morphology can profoundly affect susceptibility-induced contrast (Pathak et al, 2003), and that computational models incorporating the actual vascular structure are required to elucidate this complex relationship (Kiselev, 2001; Pathak et al, 2003, 2008a). This is now possible with the μMRI data acquired in this work and the recent development of a computational model of MRI contrast known as the finite perturber method (Pathak et al, 2008c). Finally, recent evidence suggests that angiogenesis inhibition in brain tumors may promote a shift to a more invasive phenotype, providing brain tumor cells an avenue for evading antiangiogenic therapy (Chi et al, 2007; de Groot et al, 2010). Methods that are sensitive to both angiogenesis and brain tumor invasion, such as the μMRI method presented here, could prove indispensable for answering critical questions about this phenotypic shift in preclinical brain tumor models.
To the best of our knowledge, this is the first study to characterize the vascular phenotype of a mouse brain tumor model with μMRI. We showed that several commonly used measures of vascular geometry could be obtained from μMRI data and used to characterize vascular phenotypes in a mouse brain tumor model. The data presented here show the feasibility of this approach for differentiating the tumor vascular architecture from that of the contralateral brain, and for characterizing global and zonal changes in brain tumor vascular morphology with tumor progression.
The shorter vessel branch length and elevated vessel radius, MVD, LV, FV, and tortuosity in tumor compared with contralateral ROIs are hallmarks of brain tumor angiogenesis (Jain et al, 2007; Vajkoczy and Menger, 2000). Our results also show characteristic differences between the vasculatures of D12 and D17 tumors. The decreases in MVD, LV, and FV from D12 to D17 tumors are consistent with previous observations that larger tumors have lower vascular density in central and less angiogenic regions than do smaller tumors and highly angiogenic tumor peripheries (Vajkoczy and Menger, 2000). With 3D vascular data, we performed a zonal analysis, which suggested that D17 tumors consisted of well-vascularized rims and less vascularized cores, as expected of later-stage gliomas, which are characterized by higher angiogenic activity in their peripheries (Vajkoczy et al, 1998). In contrast, although MVD, LV, and FV were all higher in D12 versus D17 tumors, indicative of higher overall angiogenic activity, D12 tumors were less vascularized in the rim than in the core. This may be attributable to the build-up of interstitial fluid in the cores of D17 tumors with tumor growth, which in turn, creates an outward interstitial fluid pressure gradient that leads to redistribution of proangiogenic growth factors toward the tumor rim (Vajkoczy et al, 1998). It is also possible that larger D17 tumors contained vessels that were intermittently or poorly perfused because of abnormal hemodynamics or elevated interstitial fluid pressure, which could affect their degree of microfilling.
Furthermore, combining vascular data with other MR-contrast mechanisms provides a powerful tool for examining the interactions between vascular and neuronal structures. For example, our findings of elevated ADC in tumor compared with contralateral ROIs, and the independence of tumor ADC from tumor volume and growth are in agreement with previous DTI studies of 9L tumors in rat brains (Kim et al, 2008; Zhang et al, 2007). The decrease in contralateral ADC from D12 to D17 is probably attributable to compression of the contralateral brain caused by extensive tumor growth. The lateral compression from the tumor may also preferentially restrict diffusion in this direction, thereby contributing to the increase in contralateral FA between D12 and D17. The increase in contralateral FA with tumor progression may also be attributed to the inclusion of white matter tracts in the larger contralateral D17 ROIs as seen in Figure 4D.
Zonal analysis showed lower ADC in the rim than in the rest of the tumor for both D12 and D17 groups; D17 tumors also exhibited higher FA in their rims than in their cores. This may be indicative of higher cell density (Chenevert et al, 2000) and, for D17 tumors, greater anisotropic cellular organization in the tumor periphery (Zhang et al, 2007). As discussed previously, the vascular morphometric parameters we measured show that D12 tumors have less vascularized rims relative to the other zones, whereas D17 tumors have relatively higher vascularized rims. Thus, for this tumor model and the spatial resolution used in this study, increased tumor cellularity did not always spatially correlate with elevated vascularity. In addition, vascularization decreased from D12 to D17 tumors as a whole, whereas ADC and FA did not change significantly with tumor progression.
It must be mentioned that, as this is an ex vivo study using fixed specimens, one cannot preclude the effects of aldehyde fixatives on ADC (Shepherd et al, 2009), and it is possible that blood vessels filled with polymerized Microfil affect ADC and FA differently than do perfused blood vessels in vivo. These ex vivo-specific factors may have different effects on tumor and normal tissue, causing the elevated tumor ADC seen in this and previous ex vivo studies.
The main technical challenge of using μMRI to image the vasculature of the whole mouse brain is achieving sufficient spatial resolution to resolve the vascular tree. The acquisition resolution of our μMRI scans ranged from 62 to 65 μm, whereas the diameter of cerebral capillaries is in the range of 3 to 5 μm. Thus, the capillaries were undetectable by μMRI, and the images acquired represent a subset of the total vessel population (radius ⩾25 μm). This is apparent when comparing the MVD visible in histology (Figures 7A and 7B, Supplementary Figures S1A and S1B) with μMRI (Figures 7C and 7D, Supplementary Figures S1C and S1D). When considering just the μMRI-visible tumor vessel population (Supplementary Figures S1E and S1F), one can see that the trends in μMRI-measured and histologically assessed radii between D12 and D17 tumors were similar. However, there is a slight rightward shift in the μMRI radius distribution relative to that assessed histologically, which may be attributed to spatial discretization caused by partial volume effects. With the increasing availability of high field magnets (⩾9.4 T) and more powerful gradient hardware, it is possible to achieve higher spatial resolution, e.g., 30 μm. However, there will be a concomitant increase in acquisition time, which decreases the throughput, and the increased size of the data also poses a challenge for analysis. The μMRI resolution used in this study enabled us to quantify relative changes in six morphologic vascular parameters with brain tumor progression, and between the tumor and the contralateral brain. Moreover, we were able to image the whole mouse brain vasculature in 3D. Acquiring the 3D structure of the intact neurovasculature is a significant advantage of this technique and, as we discuss below, can impact the calculation of vascular morphologic parameters.
Direct comparisons between parameters obtained from μMRI and those obtained using μCT or optical techniques are challenging because of differences in spatial resolution. For example, the median tumor vessel branch length is significantly shorter than that of the contralateral ROI for both D12 and D17 groups, which is consistent with observations reported in other studies (Heinzer et al, 2008; Zhang et al, 2009). However, the vessel lengths computed in this study are longer than those computed using other methods such as μCT and optical microscopy. This may be owing to the inability of μMRI to resolve vessel lengths below its resolution limit. Consequently, the loss of smaller branches may cause visible parent vessels to appear as longer branches instead of comprising shorter-length vessel segments. This effect is likely compounded by differences in ROI volumes used by different studies. For example, previous studies obtained their measurements from SRμCT images of 1 mm3 ROIs (Heinzer et al, 2008) or from laser scanning confocal microscopy images of 100-μm-thick sections (Zhang et al, 2009), whereas we performed our analysis on ROIs that encompass whole tumors with volumes ranging from ∼2.5 to 40 mm3. It is noteworthy that imaging the whole brain with μMRI permits us to measure the complete lengths of longer vessels without truncation as in other techniques.
The constraint on spatial resolution may also lead to underestimation of MVD and LV. Microvessel density is traditionally calculated from two-dimensional histologic sections and is reported in units of vessels/area instead of vessels/volume, making it difficult to directly compare our results with other studies. Boero et al (1999) reported LV values for normal mouse brains ranging from ∼430 to 1,300 mm/mm3, which is two orders of magnitude greater than the values measured here (1.99 to 9.15 mm/mm3).
Surprisingly, there is little consensus in the literature about the fractional blood volume of a normal mouse brain because each study uses different measuring techniques and mouse strains. Reported values range from ∼0.5% to 6% (Boero et al, 1999; Chugh et al, 2009; Heinzer et al, 2008; Tsai et al, 2009; Verant et al, 2007). The mean contralateral FV of 2.83%±0.78% calculated from our μMRI data falls within this range. Literature values of the fractional blood volume of orthotopic 9L tumors in the murine brain are scarce. Bremer et al (2003), using both in vivo MRI and nuclear imaging, reported a mean FV of 2% for D16 9L tumors implanted in the gluteal region of nude mice. Nomura et al (1994) also used nuclear imaging to measure blood volume and reported mean values of 12.4 μL/g for D8 9L tumors and 1.79 μL/g for the normal brain in Wistar rats. Assuming a brain tissue density of 1 g/mL, these values translate to 1.24% and 0.02% for tumor and normal brain, respectively. Pathak et al (2001) measured the FV of D10-D30 9L tumors implanted in Fisher rat brains using stereological techniques and obtained a mean value of 5.29% versus 1.89% for the normal brain. The mean tumor FV over all tumors in our study was 10.81%±3.04%. Although it is well established that 9L tumors exhibit increased blood volume relative to the normal brain, the results of these studies underscore the sensitivity of such measurements to the methodology used and the subsequent difficulty in comparing results derived from different studies.
As expected, tumor vessels are significantly more tortuous than contralateral vessels in the D12 group, but there was no significant difference for the D17 group. Heinzer et al (2008) reported median tortuosity values between 1.2 and 1.25 for normal vessels >7.5 μm in diameter, whereas the median contralateral tortuosity calculated in this study was 1.12. Again, it is likely that partial volume effects led to lower tortuosity values because directional variations of blood vessels on a scale comparable with or smaller than the image resolution were undetectable.
On the basis of all μMRI-measured parameters discussed above, we characterized the phenotypic changes of the brain microenvironment that accompany tumor progression. This is evident from the double dendrogram in Figure 6, which shows that unsupervised hierarchical clustering sorted tumor and contralateral ROIs into two well-separated clusters. It then sorted D12 and D17 contralateral ROIs into two separate, smaller clusters. Within the tumor cluster, four D12 tumor ROIs were further distinguished as one subcluster, and the fifth D12 tumor ROI was assigned to a second subcluster with D17 tumor ROIs. This lone D12 tumor ROI was the closest ROI within that subcluster to the other D12 tumor ROIs in parameter space. We also found that removing ADC and FA from the cluster analysis did not appreciably affect the clustering of tumor ROIs, but did negatively impact the clustering of contralateral ROIs (data not shown). This indicates that the ‘vascular phenotypes' of the D12 and D17 tumors were unique, and that tumor growth from D12 to D17 caused a substantial mass effect in the contralateral brain.
In conclusion, μMRI has the potential to characterize the vascular phenotype of preclinical brain tumor models. Our method could differentiate between tumor and contralateral vasculatures, as well as between the vascular phenotype of D12 and D17 tumors. Although μMRI does not have submicron imaging capability, it is capable of ‘whole-brain' mapping, offering superior coverage to optical techniques. In addition, although μMRI cannot outperform μCT in terms of spatial resolution, it has the advantage of complementary contrast mechanisms such as DW imaging, which allows us to simultaneously measure changes in the brain tumor microenvironment. The work presented here shows the feasibility of using μMRI to study the relationship between angiogenesis and other components of the brain microenvironment in a range of pathologies involving the neurovasculature.
Acknowledgments
The authors gratefully acknowledge Kristin Donato for assistance with histology.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
This research was supported by the Toshiba Medical Systems/RSNA Research Seed Grant RSD0718, NIH P50CA103175 (Career Development Award), NIH R21CA128793-01, Komen Foundation Grant KG 090640, and JHU Institute of NanoBiotechnology (INBT) Junior Faculty Pilot Award.
Supplementary Material
References
- Bennett KM, Hyde JS, Rand SD, Bennett R, Krouwer HG, Rebro KJ, Schmainda KM. Intravoxel distribution of DWI decay rates reveals C6 glioma invasion in rat brain. Magn Reson Med. 2004;52:994–1004. doi: 10.1002/mrm.20286. [DOI] [PubMed] [Google Scholar]
- Benveniste H, Blackband S. MR microscopy and high resolution small animal MRI: applications in neuroscience research. Prog Neurobiol. 2002;67:393–420. doi: 10.1016/s0301-0082(02)00020-5. [DOI] [PubMed] [Google Scholar]
- Berrios-Otero CA, Wadghiri YZ, Nieman BJ, Joyner AL, Turnbull DH. Three-dimensional micro-MRI analysis of cerebral artery development in mouse embryos. Magn Reson Med. 2009;62:1431–1439. doi: 10.1002/mrm.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestvater F, Spiess E, Stobrawa G, Hacker M, Feurer T, Porwol T, Berchner-Pfannschmidt U, Wotzlaw C, Acker H. Two-photon fluorescence absorption and emission spectra of dyes relevant for cell imaging. J Microsc. 2002;208:108–115. doi: 10.1046/j.1365-2818.2002.01074.x. [DOI] [PubMed] [Google Scholar]
- Boero JA, Ascher J, Arregui A, Rovainen C, Woolsey TA. Increased brain capillaries in chronic hypoxia. J Appl Physiol. 1999;86:1211–1219. doi: 10.1152/jappl.1999.86.4.1211. [DOI] [PubMed] [Google Scholar]
- Bremer C, Mustafa M, Bogdanov A, Jr, Ntziachristos V, Petrovsky A, Weissleder R. Steady-state blood volume measurements in experimental tumors with different angiogenic burdens a study in mice. Radiology. 2003;226:214–220. doi: 10.1148/radiol.2261012140. [DOI] [PubMed] [Google Scholar]
- Chenevert TL, Stegman LD, Taylor JM, Robertson PL, Greenberg HS, Rehemtulla A, Ross BD. Diffusion magnetic resonance imaging: an early surrogate marker of therapeutic efficacy in brain tumors. J Natl Cancer Inst. 2000;92:2029–2036. doi: 10.1093/jnci/92.24.2029. [DOI] [PubMed] [Google Scholar]
- Chi A, Norden AD, Wen PY. Inhibition of angiogenesis and invasion in malignant gliomas. Expert Rev Anticancer Ther. 2007;7:1537–1560. doi: 10.1586/14737140.7.11.1537. [DOI] [PubMed] [Google Scholar]
- Chugh BP, Lerch JP, Yu LX, Pienkowski M, Harrison RV, Henkelman RM, Sled JG. Measurement of cerebral blood volume in mouse brain regions using micro-computed tomography. Neuroimage. 2009;47:1312–1318. doi: 10.1016/j.neuroimage.2009.03.083. [DOI] [PubMed] [Google Scholar]
- de Groot JF, Fuller G, Kumar AJ, Piao Y, Eterovic K, Ji Y, Conrad CA. Tumor invasion after treatment of glioblastoma with bevacizumab: radiographic and pathologic correlation in humans and mice. Neuro Oncol. 2010;12:233–242. doi: 10.1093/neuonc/nop027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr A, Sled JG, Kabani N. Three-dimensional cerebral vasculature of the CBA mouse brain: a magnetic resonance imaging and micro computed tomography study. Neuroimage. 2007;35:1409–1423. doi: 10.1016/j.neuroimage.2006.12.040. [DOI] [PubMed] [Google Scholar]
- Heinzer S, Krucker T, Stampanoni M, Abela R, Meyer EP, Schuler A, Schneider P, Muller R. Hierarchical microimaging for multiscale analysis of large vascular networks. Neuroimage. 2006;32:626–636. doi: 10.1016/j.neuroimage.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Heinzer S, Kuhn G, Krucker T, Meyer E, Ulmann-Schuler A, Stampanoni M, Gassmann M, Marti HH, Muller R, Vogel J. Novel three-dimensional analysis tool for vascular trees indicates complete micro-networks, not single capillaries, as the angiogenic endpoint in mice overexpressing human VEGF(165) in the brain. Neuroimage. 2008;39:1549–1558. doi: 10.1016/j.neuroimage.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Hossler FE, Douglas JE. Vascular corrosion casting: review of advantages and limitations in the application of some simple quantitative methods. Microsc Microanal. 2001;7:253–264. doi: 10.1017.S1431927601010261. [DOI] [PubMed] [Google Scholar]
- Iadecola C. Neurovascular regulation in the normal brain and in Alzheimer's disease. Nat Rev Neurosci. 2004;5:347–360. doi: 10.1038/nrn1387. [DOI] [PubMed] [Google Scholar]
- Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Benveniste H, Black RD, Hedlund LW, Maronpot RR, Smith BR. Histology by magnetic resonance microscopy. Magn Reson Q. 1993;9:1–30. [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL, Hedlund LW. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 2002;222:789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Kim S, Pickup S, Hsu O, Poptani H. Diffusion tensor MRI in rat models of invasive and well-demarcated brain tumors. NMR Biomed. 2008;21:208–216. doi: 10.1002/nbm.1183. [DOI] [PubMed] [Google Scholar]
- Kiselev VG. On the theoretical basis of perfusion measurements by dynamic susceptibility contrast MRI. Magn Reson Med. 2001;46:1113–1122. doi: 10.1002/mrm.1307. [DOI] [PubMed] [Google Scholar]
- Lee TC, Kashyap RL, Chu CN. Building skeleton models via 3-D medial surface/axis thinning algorithms. Graph Models Image Processing. 1994;56:462–478. [Google Scholar]
- Moonen CT, van Zijl PC, Frank JA, Le Bihan D, Becker ED. Functional magnetic resonance imaging in medicine and physiology. Science. 1990;250:53–61. doi: 10.1126/science.2218514. [DOI] [PubMed] [Google Scholar]
- Nomura T, Inamura T, Black KL. Intracarotid infusion of bradykinin selectively increases blood-tumor permeability in 9L and C6 brain tumors. Brain Res. 1994;659:62–66. doi: 10.1016/0006-8993(94)90863-x. [DOI] [PubMed] [Google Scholar]
- Pathak AP, Hochfeld WE, Goodman SL, Pepper MS. Circulating and imaging markers for angiogenesis. Angiogenesis. 2008a;11:321–335. doi: 10.1007/s10456-008-9119-z. [DOI] [PubMed] [Google Scholar]
- Pathak AP, Jones M, Zhang J.2008bNew techniques for 3D, high-resolution, whole brain mapping of murine vasculatureProc Intl Soc Mag Reson Med16284 [Google Scholar]
- Pathak AP, Rand SD, Schmainda KM. The effect of brain tumor angiogenesis on the in vivo relationship between the gradient-echo relaxation rate change (DeltaR2*) and contrast agent (MION) dose. J Magn Reson Imaging. 2003;18:397–403. doi: 10.1002/jmri.10371. [DOI] [PubMed] [Google Scholar]
- Pathak AP, Schmainda KM, Ward BD, Linderman JR, Rebro KJ, Greene AS. MR-derived cerebral blood volume maps: issues regarding histological validation and assessment of tumor angiogenesis. Magn Reson Med. 2001;46:735–747. doi: 10.1002/mrm.1252. [DOI] [PubMed] [Google Scholar]
- Pathak AP, Ward BD, Schmainda KM. A novel technique for modeling susceptibility-based contrast mechanisms for arbitrary microvascular geometries: the finite perturber method. Neuroimage. 2008c;40:1130–1143. doi: 10.1016/j.neuroimage.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman V, Pathak AP, Glunde K, Artemov D, Bhujwalla ZM. Magnetic resonance imaging and spectroscopy of transgenic models of cancer. NMR Biomed. 2007;20:186–199. doi: 10.1002/nbm.1136. [DOI] [PubMed] [Google Scholar]
- Reichold J, Stampanoni M, Lena Keller A, Buck A, Jenny P, Weber B. Vascular graph model to simulate the cerebral blood flow in realistic vascular networks. J Cereb Blood Flow Metab. 2009;29:1429–1443. doi: 10.1038/jcbfm.2009.58. [DOI] [PubMed] [Google Scholar]
- Ridler TW, Calvard S. Picture thresholding using and iterative selection method. IEEE Trans Syst Man Cybern. 1978;8:630–632. [Google Scholar]
- Risser L, Plouraboue F, Steyer A, Cloetens P, Le Duc G, Fonta C. From homogeneous to fractal normal and tumorous microvascular networks in the brain. J Cereb Blood Flow Metab. 2007;27:293–303. doi: 10.1038/sj.jcbfm.9600332. [DOI] [PubMed] [Google Scholar]
- Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, Koller T, Gerig G, Kikinis R. Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal. 1998;2:143–168. doi: 10.1016/s1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med. 2009;62:26–34. doi: 10.1002/mrm.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BR, Johnson GA, Groman EV, Linney E. Magnetic resonance microscopy of mouse embryos. Proc Natl Acad Sci USA. 1994;91:3530–3533. doi: 10.1073/pnas.91.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, Lyden PD, Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J Neurosci. 2009;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajkoczy P, Menger MD. Vascular microenvironment in gliomas. J Neurooncol. 2000;50:99–108. doi: 10.1023/a:1006474832189. [DOI] [PubMed] [Google Scholar]
- Vajkoczy P, Schilling L, Ullrich A, Schmiedek P, Menger MD. Characterization of angiogenesis and microcirculation of high-grade glioma: an intravital multifluorescence microscopic approach in the athymic nude mouse. J Cereb Blood Flow Metab. 1998;18:510–520. doi: 10.1097/00004647-199805000-00006. [DOI] [PubMed] [Google Scholar]
- Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, Munn LL, Tearney GJ, Fukumura D, Jain RK, Bouma BE. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15:1219–1223. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verant P, Serduc R, Van Der Sanden B, Remy C, Vial JC. A direct method for measuring mouse capillary cortical blood volume using multiphoton laser scanning microscopy. J Cereb Blood Flow Metab. 2007;27:1072–1081. doi: 10.1038/sj.jcbfm.9600415. [DOI] [PubMed] [Google Scholar]
- Weidner N. Intratumor microvessel density as a prognostic factor in cancer. Am J Pathol. 1995;147:9–19. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, van Zijl PC, Laterra J, Salhotra A, Lal B, Mori S, Zhou J. Unique patterns of diffusion directionality in rat brain tumors revealed by high-resolution diffusion tensor MRI. Magn Reson Med. 2007;58:454–462. doi: 10.1002/mrm.21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zheng X, Jiang F, Zhang ZG, Katakowski M, Chopp M. Dual-color fluorescence imaging in a nude mouse orthotopic glioma model. J Neurosci Methods. 2009;181:178–185. doi: 10.1016/j.jneumeth.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.