Abstract
Serum concentrations of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides (TGs) and total cholesterol (TC) are important heritable risk factors for cardiovascular disease. Although genome-wide association studies (GWASs) of circulating lipid levels have identified numerous loci, a substantial portion of the heritability of these traits remains unexplained. Evidence of unexplained genetic variance can be detected by combining multiple independent markers into additive genetic risk scores. Such polygenic scores, constructed using results from the ENGAGE Consortium GWAS on serum lipids, were applied to predict lipid levels in an independent population-based study, the Rotterdam Study-II (RS-II). We additionally tested for evidence of a shared genetic basis for different lipid phenotypes. Finally, the polygenic score approach was used to identify an alternative genome-wide significance threshold before pathway analysis and those results were compared with those based on the classical genome-wide significance threshold. Our study provides evidence suggesting that many loci influencing circulating lipid levels remain undiscovered. Cross-prediction models suggested a small overlap between the polygenic backgrounds involved in determining LDL-C, HDL-C and TG levels. Pathway analysis utilizing the best polygenic score for TC uncovered extra information compared with using only genome-wide significant loci. These results suggest that the genetic architecture of circulating lipids involves a number of undiscovered variants with very small effects, and that increasing GWAS sample sizes will enable the identification of novel variants that regulate lipid levels.
Keywords: serum lipids, polygenic, genome-wide association, polygenic score, pathway analysis
Introduction
Serum concentrations of low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides (TGs) and total cholesterol (TC) are highly heritable phenotypes associated with the risk of cardiovascular morbidity and mortality.1, 2, 3, 4 A number of genome-wide association studies (GWASs) successfully identified multiple genes influencing circulating lipid levels.5, 6, 7, 8, 9, 10, 11, 12 There are currently over 100 established loci that include both common variants with relatively small effects as well as a considerable number of rare variants with large effects.13 Despite these successes, a substantial proportion of the heritability of each trait remains unexplained, suggesting that many determinants have yet to be identified.14
Several plausible explanations may underlie the unexplained heritability of lipid traits, including the presence of both unknown common variants with small effects and novel rare variants with larger effects. The ENGAGE GWAS5 was one of the first large population-based studies designed to find variants associated with circulating lipid levels. The study, based on 16 European cohorts including up to 22 562 individuals, identified 6 novel loci, in addition to replicating 16 previously known loci. However, as demonstrated by the recent GWAS from the Global Lipid Genetics Consortium (GLGC), numerous additional variants passed the genome-wide significance threshold as a result of increased sample size.15 The GLGC GWAS, which included over 1 00 000 individuals of European ancestry, reported 95 loci, with 59 reaching genome-wide significance for the first time. These results raise an interesting question: if common variants remain to be discovered, how many should we expect? Are there still a limited number of loci or can we expect a polygenic mechanism that involves a very large number of variants with very small effects? In the latter case, these variants would contribute to a continuous spectrum of alleles spanning the genome and single genes involved in this complex polygenic model might not be detectable by GWAS, regardless of sample size.16 Evidence for this type of genetic architecture can be shown using a genome-wide scoring approach, as was recently demonstrated for a number of psychiatric outcomes.17, 18, 19 Additionally, these polygenic scores may provide extra information useful in determining P-value thresholds for pathway analysis.
The current study aimed to explore the extent to which common variation accounts for the unexplained heritability of circulating lipid levels using the genome-wide scoring method. We also evaluated the evidence for a common polygenic effect underlying different lipid traits, using the same risk scoring approach. Finally, we examined the utility of genome-wide polygenic scores for identifying pathways beyond those identified using a classical GWAS approach.
Materials and methods
The polygenic risk score approach involves using results from a discovery set to explore the genetic architecture of an independent target sample. Our discovery set consisted of the meta-analysis of 16 European populations from the ENGAGE Lipid Consortium (N=17 798–22 562) (Table 1). A detailed description of this study, including populations, genotyping information and statistical analysis, was previously published.5
Table 1. Descriptive data of discovery and replication samples.
| ENGAGE | RS-II | |||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| Number of subjects | 8403 | 14 159 | 1061 | 1253 |
| HDL-C (mmol/l) | 1.3 (0.3) | 1.6 (0.4) | 1.2 (0.3) | 1.5 (0.4) |
| LDL-C (mmol/l) | 3.4 (0.9) | 2.3 (0.9) | 3.6 (0.9) | 1.5 (0.8) |
| TG (mmol/l) | 1.6 (1.1) | 1.1 (0.7) | 1.6 (0.9) | 1.5 (0.8) |
| TC (mmol/l) | 5.6 (0.9) | 5.9 (0.9) | 5.6 (1.1) | 5.7 (1.1) |
The target sample consisted of RS-II, an extension of the Rotterdam Study (RS), a prospective cohort study started in 1990 in the Ommoord district of the city of Rotterdam. RS-II, which was not a part of the ENGAGE discovery set, consists of 3011 participants (out of 4472 invitees) who were 55 years or older during the recruitment period (2000–2001).20 Of the 3011, 2540 persons were successfully genotyped with an Illumina 610K array. Fasting HDL-C, TG and TC were measured with enzymatic colorimetric tests on a Roche/Hitachi 911 analyzer (Roche Diagnostics, Meylan, France). LDL-C was estimated using the Friedewald formula.21
SNPs included in the construction of the polygenic scores were based on the results from the ENGAGE study. We selected different clusters of SNPs for the calculation of the scores using several P-value thresholds (Pdiscovery) ranging from 5 × 10−8 to 0.5. We calculated genetic scores for those various clusters of SNPs in the target sample by multiplying the number of risk alleles for each SNP (0, 1 or 2) by the effect sizes from the discovery set, and summing them up across all the SNPs in that cluster. For this analysis, we used the PLINK ‘profile scoring' option. SNPs that had a call rate <90% or HWE P-value <1 × 10−8 were excluded from these computations. A/T and G/C polymorphisms were also excluded to avoid potential strand inconsistencies. SNPs in linkage disequilibrium (LD) were pruned over 200 SNP sliding windows using a pair wise r2 threshold of 0.25 in PLINK.22 LD pruning was performed per SNP cluster. (See Supplementary Table 2 for the number of SNPs remaining in each cluster and used for analysis.)
The associations between these scores and serum lipid levels were tested in SPSS for Windows version 15 (SPSS, Chicago, IL, USA) using linear regression models, with sex, age and age2 as covariates (the same covariates as included in the discovery GWAS). The proportion of total variance explained by the genetic score, here referred to as the percentage of explained variance (PEV), was determined by comparing models with/without the risk score.
To evaluate whether the PEV results were driven by the GWAS hits, we also constructed a variable comprising only the significant GWAS variants and included it as a covariate in our original models. When calculating the polygenic scores for these analyses, we also removed SNPs within 2 Mb windows surrounding the GWAS hits. We employed exactly the same pruning approach for this analysis.
To search for evidence for a shared genetic background between various lipid traits, we tested additional models in which we used the polygenic score for a particular lipid and tried to predict the others, for instance, utilizing the HDL-C polygenic score to predict TG and vice versa.
The score which yielded the highest PEV for a given lipid trait ostensibly includes the most valuable genetic information; therefore, we selected these thresholds to utilize in pathway analysis (in contrast to using only genome-wide significant loci). For these analyses, we used the PANTHER tools (http://www.pantherdb.org).23 We first tested the genome-wide significant SNPs (Pdiscovery<5 × 10−8) from the ENGAGE GWAS in the pathway analysis. These results were then compared with those obtained using alternative Pdiscovery thresholds selected on the predictive ability of the polygenic scores. After SNP selection, SNPs within gene regions were converted to gene symbols using the ‘SCAN SNP and CNV annotation database' (http://www.scandb.org). Gene lists were tested for enrichment in three PANTHER categories: (1) pathways, (2) biological processes and (3) molecular functions. Testing for enrichment basically involves comparing one gene list to the reference list to statistically determine over- or under- representation of PANTHER classification categories. Based on the reference list, an expected value is computed (the number of genes one would expect in the list for a particular PANTHER category) and it is assumed that, under the null hypothesis, genes in the tested list are sampled from the same distribution as genes from the reference set. The Homo sapiens gene list from National Centre for Biotechnology Information was used as the reference gene list. To avoid bias caused by multiple testing, PANTHER's Bonferroni correction option was implemented. (See Supplementary Figure 1 for the overall flowchart of the study.)
Results
Table 1 shows summary statistics for the discovery and target samples. The female/male ratio in the discovery set was significantly higher compared with the target set (1.6 vs 1.2, P<0.001). Genome-wide significant SNPs from the ENGAGE GWAS were checked for their associations in the target sample using linear regression. Generally, evidence of association between those SNPs and lipid levels were marginally significant or non-significant (Supplementary Table 1). The GWAS of circulating lipids in RS-II did not show any genome-wide significant findings except the CETP gene region SNPs, which were associated with HDL-C (rs7499892, P=3.4 × 10−13). Manhattan plots for the GWAS of the HLD-C, LDL-C, TG and TC can be found in Supplementary Figure 2.
Prediction
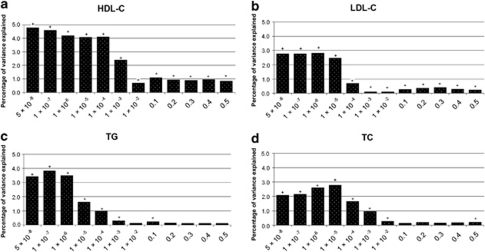
Figure 1 shows the PEV obtained for each lipid trait using the polygenic scores generated for a number of P-value thresholds in the target sample (RS-II). For HDL-C, the polygenic score computed using 19 genome-wide significant SNPs from 8 gene regions (Pdiscovery<5 × 10−8) resulted in the maximum PEV compared with the null model (4.75%, P=3.6 × 10−30; Figure 1a). For LDL-C (Figure 1b), the maximum PEV was observed with the polygenic score that included 21 SNPs with a Pdiscovery<1 × 10−6 (2.6%, P=5.1 × 10−16). Figure 1c shows PEVs for TG levels; the score that included 12 SNPs from 8 regions with Pdiscovery<1 × 10−7 (3.8%, P=2.8 × 10−21) was the best predictor. For these traits, the variance explained decreased with the inclusion of additional SNPs in the polygenic score selected using more liberal Pdiscovery thresholds (Figures 1a–c). Finally, for TC, the highest PEV was obtained using 46 SNPs from 24 regions with Pdiscovery<10−5 (2.7%, P=1.4 × 10−16). This was higher than the PEV obtained using only the genome-wide significant SNPs (PEV=2.1%, P=8.2 × 10−13, n=20 SNPs from 11 regions; Figure 1d). As with HDL-C, LDL-C and TG, the explained variance for TC dropped when more liberal Pdiscovery thresholds were used to construct the polygenic score. For LDL-C, HDL-C and TC, all scores were significant (up to a threshold of Pdiscovery<0.5). We observed similar patterns when we used unpruned data (Supplementary Figure 3).
Figure 1.
Graphs a–d show the PEV of circulating lipids with risk scores by different Pdiscovery thresholds. Adjusted for age, sex and age2. +P<5 × 10−8; *5 × 10−8<P<0.05.
Figure 2 shows the results from the second approach, in which models were adjusted for genome-wide significant variants. For HDL-C (Figure 2a), the PEV increased as SNPs were added, up to 0.5% with Pdiscovery<0.1 (P=1.0 × 10−4) and remained significant until Pdiscovery<0.5 (P=2.3 × 10−4). A similar pattern was observed with LDL-C (Figure 2b, explained variance was up to 0.4% (P=0.002)) with Pdiscovery threshold of 0.2. In contrast, the polygenic score for TG, when the effects of known variants were excluded, was not associated with TG levels in the target population (Figure 2c). For TC (Figure 2d), the maximum PEV was observed with Pdiscovery<1 × 10−5, (0.6%, P=1.8 × 10−4).
Figure 2.
Graphs a–d show the PEV of circulating lipids when the top regions are excluded. Adjusted for age, sex age2 and risk score computed from genome-wide significant findings. The lack of association in the first cluster of SNPs are due to the exclusion of SNPs within 2 Mb window region surrounding the top findings, as there were only a few SNPs to be included in the analysis after excluding the top regions. *P<0.05.
Cross-prediction
Table 2 shows the phenotypic correlations for the four outcomes studied, and additionally shows the correlations between the polygenic scores for different Pdiscovery thresholds. Correlations between the traits were modest, with the exceptions of TC and LDL-C, (r=0.9) and TG and HDL-C (r=−0.5). The correlations between the polygenic scores were weaker than the phenotypic correlations (0.8 for TC/LDL-C and −0.2 for TG/HDL for Pdiscovery<5 × 10−8)
Table 2. Correlation matrix of circulating lipids and genetic risk scores in RS-II.
| HDL-C | LDL-C | TG | TC | ||||
|---|---|---|---|---|---|---|---|
| Correlation between the phenotypes | HDL-C | 5 × 10−8 | 0.01 | −0.20** | 0.02 | Correlation between the genetic risk scores | |
| 1 × 10−7 | 0.01 | −0.17** | 0.03 | ||||
| 1 × 10−6 | −0.01 | −0.09** | 0.07** | ||||
| 1 × 10−5 | 0.02 | −0.04* | 0.05* | ||||
| LDL-C | 5 × 10−8 | −0.1** | 0.01 | 0.76** | |||
| 1 × 10−7 | 0.02 | 0.75** | |||||
| 1 × 10−6 | 0.05* | 0.81** | |||||
| 1 × 10−5 | 0.03 | 0.71** | |||||
| TG | 5 × 10−8 | −0.5** | 0.1** | 0.13** | |||
| 1 × 10−7 | 0.13** | ||||||
| 1 × 10−6 | 0.12** | ||||||
| 1 × 10−5 | 0.08** | ||||||
| TC | 5 × 10−8 | 0.1** | 0.9** | 0.3** | |||
| 1 × 10−7 | |||||||
| 1 × 10−6 | |||||||
| 1 × 10−5 |
Lower-left side of the matrix shows the phenotypic correlation between circulating lipid levels, adjusted by age, age2 and sex. Upper-right side of the matrix shows the correlation between the genetic risk scores of four circulating lipids, for the first four risk scores with Pdiscovery<5 × 10−8, Pdiscovery<1 × 10−7, Pdiscovery<1 × 10−6 and Pdiscovery<1 × 10−5. *Correlation significant at P<0.05. **Correlation significant at P<0.001.
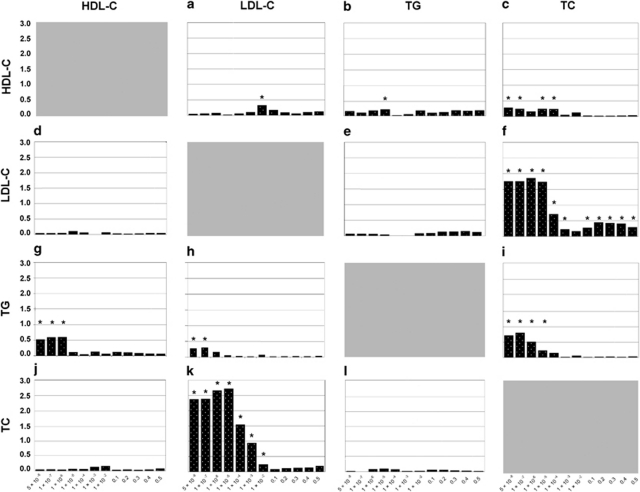
To evaluate the evidence for common polygenetic effects underlying lipid levels, we performed cross-prediction analyses (Figure 3). The highest PEV was based on the TC score at Pdiscovery (TC)<1 × 10−5, which explained up to 2.7% of the variance in circulating LDL-C (P=2.0 × 10−5; Figure 3k). Similarly, LDL-C risk profiles explained up to 1.8% of the variance in TC when we selected all SNPs with a Pdiscovery (LDL−C)<10−6 (P=1.4 × 10−11; Figure 3f). These findings are in line with the high phenotypic correlations between those variables. Figures 3g–i shows the predictions based on a TG score which explained up to 0.8% of the variance in other lipids. HDL-C scores explained up to 0.3% of the variance in other lipids (Figures 3a–c).
Figure 3.
Cross-prediction across different lipids. Evaluation of the evidence for a joint polygenic effect underlying various lipids: (a–c) Prediction based on HDL risk scores imposed on LDL, TGs and TC. (d–f) Prediction based on LDL risk scores imposed on HDL, TGs and TC. (g–i) Prediction based on TG risk scores. (j–l) Prediction based on TC risk scores. *P<0.05.
Pathway analysis
Pathways analyses using only genome-wide significant SNPs was compared with the analogous analyses using SNPs from the polygenic scores, which yielded the highest PEV for each trait (Figure 1). These scores used thresholds of P<1 × 10−6 for LDL-C, P<1 × 10−5 for TC, P<5 × 10−8 for HDL-C and P<1 × 10−7 for TG. Table 3 shows the findings from the pathway analysis, based on alternatives to a P-value threshold of 5.0 × 10−8. None of the pathways among categories defined by the PANTHER tool were significant after strict adjustment for multiple testing (Bonferroni correction). With respect to biological processes the lipid and fatty acid transport and lipid, fatty acid and steroid metabolism pathways were two biological processes enriched in the HDL-C and LDL-C GWAS findings. At the level of molecular function, genes with an apolipoprotein and transfer/carrier function were enriched in LDL-C, while genes with a lipase function were observed to be significantly enriched among the top GWAS results for HDL-C. For HDL-C and TG, we were not able to select alternative P-value thresholds as the highest PEVs were observed with P<5 × 10−8. With respect to LDL-C, the pathway analysis utilizing two different P-value thresholds (P<1 × 10−6 and P<5 × 10−8) resulted in the same findings. No additional pathways were identified by using extra information from the risk profiles for LDL-C, TG and HDL-C. For TC, on the other hand, the lipid, fatty acid and steroid metabolism, lipid and fatty acid transport and transport terms additionally emerged among biological processes tested using the alternative threshold (Table 3).
Table 3. Pathway analysis.
| NCBI | Observed | Expected | Over/under | P | P* | ||
|---|---|---|---|---|---|---|---|
| Pathways | n.s. | ||||||
| Biological process | |||||||
| HDL-C | Lipid, fatty acid and steroid metabolism | 770 | 5 | 0.42 | + | 4.05 × 10−5 | 1.26 × 10−3 |
| Lipid and fatty acid transport | 131 | 3 | 0.07 | + | 4.77 × 10−5 | 6.91 × 10−3 | |
| LDL-C | Lipid, fatty acid and steroid metabolism | 770 | 4 | 0.51 | + | 1.46 × 10−3 | 4.52 × 10−2 |
| Lipid and fatty acid transport | 131 | 3 | 0.09 | + | 8.81 × 10−5 | 1.28 × 10−2 | |
| TG | n.s. | ||||||
| TC | Lipid, fatty acid and steroid metabolism | 770 | 6 | 1.21 | + | 1.22 × 10−3 | 3.78 × 10−2 |
| Lipid and fatty acid transport | 131 | 4 | 0.21 | + | 5.55 × 10−5 | 8.05 × 10−3 | |
| Transport | 1306 | 8 | 2.05 | + | 8.47 × 10−4 | 2.63 × 10−2 | |
| Molecular function | |||||||
| HDL-C | Lipase | 75 | 3 | 0.04 | + | 9.11 × 10−6 | 1.47 × 10−3 |
| LDL-C | Apolipoprotein | 23 | 2 | 0.02 | + | 1.10 × 10−4 | 1.77 × 10−2 |
| Transfer/carrier protein | 327 | 3 | 0.22 | + | 1.26 × 10−3 | 3.66 × 10−2 | |
| TG | n.s. | ||||||
| TC | n.s. | ||||||
Enrichment of a particular ‘pathway', ‘biological process' or ‘molecular function' PANTHER categories were tested by pathway analysis. SNPs that are included in the pathway analysis are selected based on their Pdiscovery values, which were 10−6 for LDL-C, 10−5 for total cholesterol, 5 × 10−8 for HDL-C and 10−7 for triglycerides. NCBI, number of genes that belong to the particular category. Observed: number of genes that belong to the given particular category among GWAS results. Expected, expected value for number of genes that belong to the particular pathway among GWAS results. Over/under, stands for ‘over-represented/under-represented'. n.s., no significant findings. *P-value corrected for multiple testing.
Discussion
Using prediction modelling, we could explain up to 4.8% of the variance in HDL-C, 2.6% in LDL-C, 3.8% in TG and 2.7% in TC. These PEVs are very similar to those from similar studies5, 9 and much higher than the single SNP analysis of genome-wide significant SNPs from the ENGAGE GWAS (Supplementary Table 1).
However, these proportions are much lower than those identified by GLGC, which were estimated to explain 12.4% (TC), 12.2% (LDL-C), 12.1% (HDL-C) and 9.6% (TG) of the variance in the Framingham Heart Study sample, as mentioned by Teslovich et al.24 This is expected as increases in sample size lead to better estimation of the effect sizes of the SNPs and GLGC had a sample size 5 times larger than the ENGAGE sample, which we used as a discovery set in our study.
For all of the traits, the PEV reached a maximum and then decreased with the use of more liberal Pdiscovery thresholds to calculate the polygenic scores (Figure 1). This is most likely explained by the inclusion of more and more biologically non-relevant SNPs, so that the effects of true positive findings are diluted and this is reflected by the decreases in PEV. For all of the studied traits, we found the highest PEV when the polygenic score was based on SNPs with a low Pdiscovery value (5 × 10−8 for HDL-C, 1 × 10−7 for TG, 1 × 10−6 for LDL-C and 1 × 10−5 for TC). Including the top regions from the ENGAGE GWAS data set as a separate predictor in the models (Figure 2) uncovered a residual polygenic component which does not explain >1% of HDL-C, LDL-C and TC levels. These findings suggest that there are unknown genes with much smaller effects involved in determining these outcomes. However, the PEVs for these additional variants were small when compared with those for the top findings. For TG, on the contrary, excluding the top regions from the polygenic score resulted in non-significant findings. For TC, which is highly heterogeneous compared with the other traits, it seems that some variants remain to be discovered (Pdiscovery<1 × 10−5). It is of note that among newly discovered loci for HDL-C by GLGC, leading SNPs from 10 loci had P-values >0.05 in the ENGAGE HDL-C analysis. Similar findings were observed for 10 loci for LDL-C, 3 loci for TG and for 9 loci in TC.24 It is already known that monogenic disorders25 and rare variants also account for variation in circulating lipid levels.26, 27, 28, 29, 30, 31, 32 This may help to explain why the explained variance is small compared with the high heritability of the traits, especially as many rarer variants are population specific, and might not have been well represented in our European data set, or not well tagged by the common SNPs under study. For instance, APOE gene variations are tagged by the CEACAM16-TOMM40 region among the ENGAGE GWAS top findings, and SNPs from this region were not associated to LDL-C levels in RS-II, however, APOE ɛ2 carrier status explains 2.6% of the phenotypic variation in LDL-C levels in RS-II. Additionally, the gender ratio difference between the discovery and target samples may have been a limitation to the current study, as some loci show different effect sizes for males and females.5 Our findings have implications for gene discovery and suggest that GWAS of much larger samples may be needed to discover additional variants with small effects for HDL-C and LDL-C. However, at the same time, this study suggests that many of the unknown SNPs have relatively large effects and that is confirmed by the GLGC data. Our findings suggest that GWAS on serum lipids in the future will still be successful as sample sizes increase.14 Our cross-prediction results are interesting from a biological perspective. These findings showed very little overlap between the polygenic scores for different circulating lipids. A strong inverse relationship exists between low HDL-C and elevated plasma TG (r=−0.5 in RS-II). Low HDL-C levels are strongly associated with hypertriglyceridemia as high levels of plasma TGs drive an exchange reaction for HDL-C cholesteryl esters mediated by CETP.33 In addition, the TG and phospholipids in HDL-C are hydrolysed by LIPC.13, 33 However, using our genetic evaluation it was not possible to predict a large proportion of the variance in TG levels using HDL-C risk profiles despite the correlation between the two lipids. The polygenic score for TG was slightly better in predicting HDL-C than when we used the top SNPs, however, the PEV did not exceed 0.6% and was lower than the variance explained by HDL-C SNPs and also lower than the variance explained in circulating TG by TG SNPs. Thus, our data implies that common genetic variants involved in determining both TG and HDL-C levels do not explain the phenotypic correlation between these traits, suggesting that the correlation may be influenced strongly by environmental factors, and/or restricted to a few genes. An alternative explanation may be that we tested the polygenic effects of common variants weighted by their effect size from the initial GWAS. When there are strong causal variants among the top hits that are specific to HDL-C but not to TG, this may dilute the effect of genes with small effect sizes on both outcomes. Also, the current analyses do not account other forms of genetic variation, such as rare variants or copy number variations (CNVs). As expected, we also found evidence for a number of genes that regulate both HDL-C and LDL-C (Figure 3a) and a similar overlap between TG and LDL-C (Figure 3h). TC SNPs were able to explain up to 2.7% of the variation in LDL-C, suggesting that the genes determining LDL-C and TC are for a large part overlapping. This result is in line with the high phenotypic correlation between the two measures. Genome-wide significant findings from the ENGAGE GWAS harboured two loci (apolipoprotein B and LPL) influencing both HDL-C and TG, 2 loci influencing both TG and TC (DOCK7 and CEACAM16-TOMM40 regions) and 7 loci influencing both LDL-C and TC (CELSR2, APOB, ABCG5, HMGCR, FADS2/3, LDLR and CEACAM16-TOMM40). A limitation here is that LDL-C was not directly measured but calculated with the Friedewald formula in the RS-II sample and so, by definition, depends directly on TC, HDL-C and TG. This may cause a potential bias in findings for LDL-C and may inflate the association between lipids in cross-prediction findings with this phenotype. We investigated whether the polygenic score approach can be used as a tool for selecting SNPs of interest in order to further evaluate them in a pathway analysis. First, we evaluated the genome-wide significant SNPs from an existing GWAS and compared the results with those obtained using the SNPs from the polygenic model with the maximum PEV. Neither of the approaches yielded any novel pathways/biological processes (only those already known to be involved in lipid metabolism, such as cholesterol biosynthesis; lipid and fatty acid transport; and lipid, fatty acid and steroid metabolism). Also, we see that, although the use of the polygenic score approach did not provide extra information concerning LDL-C, HDL-C or TG, for TC, pathway analysis based on the best predicting polygenic score (with Pdiscovery<1 × 10−5) was more informative than analysis based solely on the genome-wide significant findings. Including TC SNPs up to a more liberal threshold of 1 × 10−5 suggested three processes, which are already biologically known but were not detectable with the 5 × 10−8 discovery threshold. This finding shows that for complex traits like TC, the risk scoring approach might be used to select the SNP cluster which harbours a large number of true positives that are not significant at the genome-wide level. Taken together with the polygenic component analysis results, it is likely that ENGAGE TC-GWAS results harbour undiscovered associated variants distributed between 1 × 10−6<Pdiscovery<1 × 10−5. Using a gene scoring approach, we tested the evidence of a polygenic component for the heritable circulating lipids. We concluded that a polygenic form of inheritance exists for HDL-C, LDL-C, TG and TC. These findings may be useful for future gene discovery efforts for lipids. We also tested for possible genetic overlap between biologically related lipid traits and compared two different approaches for pathway analysis. This study gives an example of utilizing the risk scoring approach to search for the common genetic background of different quantitative traits; thus, it may also be an example for more sophisticated future studies.
Acknowledgments
ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney. For the MICROS study, we thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the research project. In South Tyrol, the study was supported by the Ministry of Health and Department of Educational Assistance, University and Research of the Autonomous Province of Bolzano and the South Tyrolean Sparkasse Foundation. Genome-wide genotyping of the Rotterdam Study was supported by NWO (175.010.2005.011). The ERF study was supported by grants from The Netherlands Organisation for Scientific Research, Erasmus MC and the Centre for Medical Systems Biology (CMSB). We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P Veraart for her help in genealogy, J Vergeer for the supervision of the laboratory work and P Snijders for his help in data collection. The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project no. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V Struchalin for their support in creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. We are grateful to the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Isaacs A, Sayed-Tabatabaei FA, Aulchenko YS, et al. Heritabilities, apolipoprotein E, and effects of inbreeding on plasma lipids in a genetically isolated population: the Erasmus Rucphen Family Study. Eur J Epidemiol. 2007;22:99–105. doi: 10.1007/s10654-006-9103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J., III Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- Kuulasmaa K, Tunstall-Pedoe H, Dobson A, et al. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 2000;355:675–687. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- Namboodiri KK, Green PP, Kaplan EB, et al. The Collaborative Lipid Research Clinics Program Family Study. IV. Familial associations of plasma lipids and lipoproteins. Am J Epidemiol. 1984;119:975–996. doi: 10.1093/oxfordjournals.aje.a113818. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner JS, Chambers JC, Aguilar-Salinas CA, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu MS, Waterworth DM, Debenham SL, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- Demirkan A, Penninx BWJH, Hek K, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts Mol Psychiatrye-pub ahead of print 22 June 2010. [DOI] [PMC free article] [PubMed]
- International Schizophrenia Consortium Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N, van Duijn CM, Janssens AC. Genetic scoring analysis: a way forward in genome wide association studies. Eur J Epidemiol. 2009;24:585–587. doi: 10.1007/s10654-009-9387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman A, Breteler MM, van Duijn CM, et al. The Rotterdam Study: 2010 objectives and design update. Eur J Epidemiol. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert B, Rouis M, Griglio S, de Lumley L, Laplaud P. Lipoprotein lipase (LPL) deficiency: a new patient homozygote for the preponderant mutation Gly188Glu in the human LPL gene and review of reported mutations: 75% are clustered in exons 5 and 6. Ann Genet. 2001;44:25–32. doi: 10.1016/s0003-3995(01)01037-1. [DOI] [PubMed] [Google Scholar]
- Kotowski IK, Pertsemlidis A, Luke A, et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simha V, Garg A. Inherited lipodystrophies and hypertriglyceridemia. Curr Opin Lipidol. 2009;20:300–308. doi: 10.1097/MOL.0b013e32832d4a33. [DOI] [PubMed] [Google Scholar]
- Slatter TL, Jones GT, Williams MJ, van Rij AM, McCormick SP. Novel rare mutations and promoter haplotypes in ABCA1 contribute to low-HDL-C levels. Clin Genet. 2008;73:179–184. doi: 10.1111/j.1399-0004.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- Talmud PJ. Rare APOA5 mutations--clinical consequences, metabolic and functional effects: an ENID review. Atherosclerosis. 2007;194:287–292. doi: 10.1016/j.atherosclerosis.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Genest JJ, Jr, Martin-Munley SS, McNamara JR, et al. Familial lipoprotein disorders in patients with premature coronary artery disease. Circulation. 1992;85:2025–2033. doi: 10.1161/01.cir.85.6.2025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.