Abstract
Congenital heart defects (CHDs) occur mostly sporadic, but familial CHD cases have been reported. Mutations in several genes, including NKX2.5, GATA4 and NOTCH1, were identified in families and patients with CHD, but the mechanisms underlying CHD are largely unknown. We performed genome-wide linkage analysis in a large four-generation family with autosomal dominant CHD (including atrial septal defect type I and II, tetralogy of Fallot and persistent left superior vena cava) and low atrial rhythm, a unique phenotype that has not been described before. We obtained phenotypic information including electrocardiography, echocardiography and DNA of 23 family members. Genome-wide linkage analysis on 12 affected, 5 unaffected individuals and 1 obligate carrier demonstrated significant linkage only to chromosome 9q21–33 with a multipoint maximum LOD score of 4.1 at marker D9S1690, between markers D9S167 and D9S1682. This 48-c critical interval corresponds to 39 Mb and contains 402 genes. Sequence analysis of nine candidate genes in this region (INVS, TMOD1, TGFBR1, KLF4, IPPK, BARX1, PTCH1, MEGF9 and S1PR3) revealed no mutations, nor were genomic imbalances detected using array comparative genomic hybridization. In conclusion, we describe a large family with CHD and low atrial rhythm with a significant LOD score to chromosome 9q. The phenotype is representative of a mild form of left atrial isomerism or a developmental defect of the sinus node and surrounding tissue. Because the mechanisms underlying CHD are largely unknown, this study represents an important step towards the discovery of genes implied in cardiogenesis.
Keywords: atrium, congenital heart defects, sinus node
Introduction
Congenital heart defects (CHDs) are among the most common congenital defects, occurring with an incidence of 8/1000 live-births.1 The aetiology of CHD is generally complex, environmental exposures, chromosomal abnormalities and gene defects can all contribute. Most CHD occur sporadic, but in recent years an increasing number of familial cases with various types of CHD have been reported.2, 3, 4 Although mutations in several genes have been identified in a small subset of patients and families with CHD, eg, NKX2.5,2 GATA43 and NOTCH1,5 the mechanisms underlying human cardiogenesis and CHDs remain largely unknown. Some CHD patients and families also display cardiac arrhythmias, which can occur due to the anatomical defect itself or sometimes due to surgical interventions.6 Moreover, in some patients arrhythmias are the direct consequence of the underlying genetic defect, in absence of any structural defect.2, 7
Ectopic atrial rhythms originate when a focus outside the sinus node takes over the pacemaker function. Consequently, the direction of atrial activation may be altered, which can be seen as an abnormal P-wave axis on the electrocardiogram (ECG). The appearance of the P-wave depends on the site of origin of the ectopic rhythm. The P-wave can be decreased in amplitude, or it can be biphasic or negative in leads II, III and AVF. The rhythm is regular and the P–R interval is usually normal or, at times, prolonged.8 Usually, ectopic atrial rhythms are haemodynamically insignificant. They can be isolated and idiopathic, especially in children, but can also co-occur with CHD.9 A P-wave frontal axis oriented in a superior direction, reflecting atrial pacemaker tissue located in the lower part of the atrium, is referred to as low atrial rhythm. This can be present due to sinus node dysfunction in patients with left atrial isomerism, a laterality disorder characterized by bilateral left sidedness.9, 10
Here, we describe a large four-generation family with an autosomal dominant condition, comprising various types of CHD and low atrial rhythm, a unique phenotype not described before within a family. A genome-wide linkage analysis yielded linkage to a region of 39 Mb on chromosome 9q with a maximum LOD score of 4.1.
Methods
Clinical details
This study was approved by the Medical Ethical Committee at the Academic Medical Centre in Amsterdam. Written informed consent was obtained from all participants. Subjects were clinically evaluated by analysis of medical records, physical examination with attention to syndromic features, cardiologic examination, 12-lead ECG and two-dimensional echocardiography. Previously performed ECGs were obtained from the medical records if available. The number of available ECGs ranged from one to 15 in the evaluated persons. One experienced paediatric cardiologist (JL) reviewed the ECGs and echocardiographic images. A total of 23 family members were examined. One patient (II-2) refused cardiologic examination for this study and therefore only previous medical records (including ECGs and echocardiography records) were obtained. We used magnetic resonance imaging (MRI) for evaluation of situs in the proband (IV-10). Abdominal ultrasound for evaluation of abdominal situs was performed in three individuals, and chest X-rays for evaluation of situs of the lungs were available for six individuals. Computed tomography (CT) of the thorax was performed in patient III-3.
Low atrial rhythm was defined as a P-wave frontal axis oriented in a superior direction. We defined the P-wave frontal axis to be horizontal when it was ∼0°, which is abnormal, although not a true low atrial rhythm. A wandering atrial pacemaker was defined as a change in P-wave axis of 60° or more on subsequent ECGs.
Linkage analysis, mutation screen and array comparative genomic hybridization
Genomic DNA of family members was extracted from peripheral blood according to standard procedures. Linkage analysis was performed using the ABI linkage set v2.5 MD10 set on an ABI 3700 Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA).11 Phenotype, genotype and pedigree information were combined for multipoint linkage analysis with the use of the easyLinkage software package12 running Simwalk v2.9113 with the assumption of an autosomal dominant pattern of inheritance, a disease-allele frequency of 0.0001 and a penetrance of 0.9. Gene frequency was assumed to be equal between males and females. Both parametric and nonparametric linkages were calculated.
PCR amplification of all the coding exons of candidate genes inversin (INVS), transforming growth factor-β receptor 1 (TGFBR1), tropomodulin 1 (TMOD1), Kruppel-like factor 4 (KLF4), inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IPPK), BARX homeobox 1 (BARX1), patched homologue 1 (PTCH1) and sphingosine 1-phosphate receptor 3 (S1PR3), multiple epidermal growth factor-like domains 9 (MEGF9) (only exon 12b, as this is specifically expressed in brain and heart14) was performed using primers located in flanking intronic sequences (available on request). Subsequently, PCR products were analysed by direct sequencing, using the BigdyeTerminator v3.1 Kit on an ABI 3700.
MLPA probes were designed for INVS and TGFBR1 using MAPD, a probe design suite for multiplex ligation-dependent probe amplification assays.15 The MLPA procedure and analysis were carried out as described.16
Array comparative genomic hybridization (array CGH) was performed using 4 × 180K slides (average 13 kbp spacing between probes), AMADID 023363 (Agilent, Santa Clara, CA, USA) according to the manufacturer's protocol (Oligonucleotide Array-Based CGH for Genomic DNA Analysis V5.0, June 2007) with adaptations. No amplification or restriction was used on the genomic patient and reference DNA. Fluorescent labelling of gDNA was performed using the CGH labelling kit for Oligo Arrays (Enzo Life Sciences Inc., Farmingdale, NY, USA) and purified using the MinElute PCR Purification Kit (Qiagen, Valencia, CA, USA). Hydridization was performed according to the manufacturer's protocol (Oligonucleotide Array-Based CGH for Genomic DNA Analysis V5.0, June 2007) and slides were scanned using an Agilent G2250C 2 μm scanner with Agilent Scan Control software (Version A.8.1.3.) using default settings. Spot intensities were measured with Agilent Feature Extraction software (V10.7), and further data analysis was performed using DNA Analytics software (V4.0.76) with algorithm ADM-2 using a filter of minimal three subsequent clones with a minimal absolute ratio of 0.3.
Results
Clinical details
In the presented family, several individuals had CHD with or without low atrial rhythm, whereas others had isolated low atrial rhythm. The pedigree of the family is shown in Figure 1. Table 1 summarizes the clinical features of the family members.
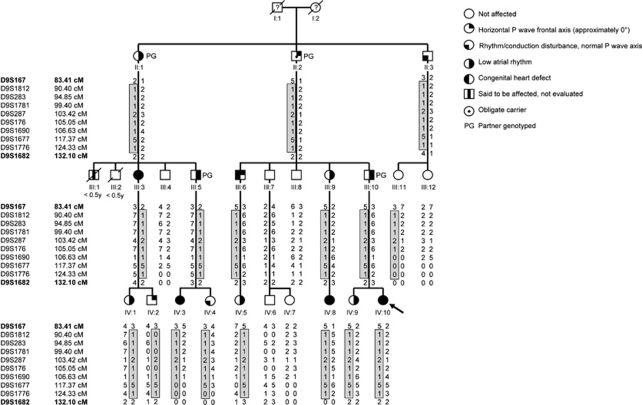
Figure 1.
Pedigree of the family; circles represent females, squares males. Unaffected individuals are depicted with empty symbols and deceased individuals are indicated by slashes with age of death in years. Right-sided filled symbols denote individuals with low atrial rhythm, left-sided filled symbols represent those with congenital heart defects or persistent left superior vena cava, right upper quarter filled symbols represent individuals with horizontal P-wave frontal axis and left lower quarter filled symbols indicate those with rhythm/conduction disturbances with normal P-wave frontal axis. Dots denote obligate carriers. The proband is indicated by an arrow. Haplotypes are depicted under each individual, and the disease haplotype is demarcated by the box. For reasons of clarity not all parents are shown, markers according to the Marshfield map.
Table 1. Clinical details of family.
| Patient characteristics | Clinical features | Status in linkage analysis | |||||
|---|---|---|---|---|---|---|---|
| Sex/age | Age at diagnosis of CHD and/or low atrial rhythm | Age (years) at last available ECG | Low atrial rhythm | Rhythm/conduction disturbances | Congenital heart defects | Nuclear family scenario | |
| II-1 | F/83 | 74 years | 77 | + | AF | — | Affected |
| II-2 | M/89 | 84 years | 84 | ± | Bradycardia, atrial/ventricular arrhythmia, AFa | — | Obligate carrier |
| II-3 | M/87 | 78 years | 80 | − | Bradycardia, junctional escapes, AF, complete RBBB | — | Excluded |
| III-3 | F/57 | 50 years | 57 | + | Paroxysmal SVT | LSVC, aberrant right subclavian artery | Affected |
| III-4 | M/50 | 43 | − | — | — | Not affected | |
| III-5 | M/47 | 40 years | 40 | + | First-degree AV block, incomplete RBBB | — | Affected |
| III-6 | M/61 | 10 years | 36 | ± | Complete RBBB | Tetralogy of Fallot, LSVC | Affected |
| III-7 | M/59 | 58 | − | Sinus bradycardia | — | Not affected | |
| III-8 | M/54 | 53 | − | — | Not affected | ||
| III-9 | F/47 | 4 years | 38 | + | Bradycardia, atrial arrests with AV junctional escapes, intermittent complete AV dissociation | —b | Affected |
| III-10 | M/60 | 52 years | 52 | + | Bradycardia | — | Affected |
| III-11 | F/52 | 45 | − | — | — | Excluded | |
| III-12 | F/50 | 43 | − | — | — | Excluded | |
| IV-1 | F/28 | 21 years | 28 | + | — | — | Affected |
| IV-2 | M/25 | 24 years | 24 | ± | — | — | Excluded |
| IV-3 | F/23 | 2 months | 16 | + | Incomplete RBBB | ASD II, membrane in left atrium, LSVC, absent brachiocephalic vein | Affected |
| IV-4 | F/17 | Birth | 16 | − | Chronic SVT; atrial extrasystoles | — | Excluded |
| IV-5 | F/28 | 4 years | 28 | + | Incomplete RBBB | —c | Affected |
| IV-6 | M/34 | 33 | − | — | — | Not affected | |
| IV-7 | F/31 | 24 | − | — | — | Not affected | |
| IV-8 | F/18 | <0.5 years | 18 | + | Incomplete RBBB | Incomplete AVSD, ASD II, LSVC draining into left atrium, absent coronary sinus, absent brachiocephalic vein | Affected |
| IV-9 | F/36 | 32 years | 32 | + | — | — | Affected |
| IV-10 | F/33 | <0.5 years | 33 | + | Incomplete RBBB | Incomplete AVSD, common atrium, bilateral left atrial appendages | Affected |
Abbreviations: AF, atrial fibrillation; ASD II, secundum type atrial septal defect; AVSD, atrioventricular septal defect; F, female; LSVC, persistent left superior vena cava; M, male; RBBB, right bundle branch block; SVT, supraventricular tachycardia.
Low atrial rhythm+means that the P-wave frontal axis was oriented superiorly; low atrial rhythm±means that the P-wave frontal axis was horizontal (∼0°).
After myocardial infarction.
Suspicion of abnormal atrial appendage.
Systolic murmur, for which cardiologic follow-up until the age of 7 years, elevated pulmonary artery pressure at age 20 years.
The proband (IV-10) was diagnosed during her first year of life with an incomplete atrioventricular septal defect (AVSD), a common atrium and a small communication between the left and right ventricle. At 2.5 years of age, she underwent surgical correction including septation of the monoatrium, leaving the coronary sinus draining into the left atrium. During the surgical procedure, the presence of bilateral left atria with bilateral left atrial appendages was observed. At 33 years of age, she was asymptomatic. MRI at that age showed normal situs of the lungs and abdominal organs. ECG persistently showed left QRS axis deviation and low atrial rhythm.
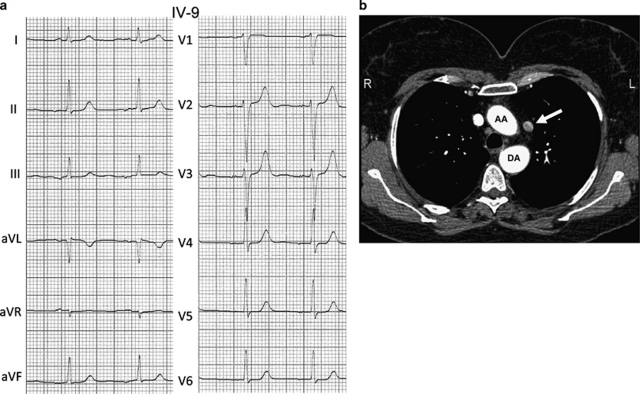
The sister of the proband (IV-9) was asymptomatic. Echocardiography did not reveal any abnormalities; however, ECG at age 32 showed low atrial rhythm (Figure 2). The asymptomatic father of IV-9 and IV-10 (III-10) also had normal echocardiography, and low atrial rhythm with bradycardia (45/min) on ECG.
Figure 2.
(a) ECG of patient IV-9 at age 32 years with isolated low atrial rhythm. The P-wave is negative in leads II, III and aVF. (b) Transverse computed tomography image of individual III-3. Persistent left superior vena cava (arrow). AA, ascending aorta; DA, descending aorta.
The paternal aunt of the proband (III-9) was diagnosed with congenital agenesis or malfunction of the sinus node at infant age because of bradycardia with AV-junctional escape rhythm and low atrial rhythm. At later age, she developed intermittent complete AV dissociation. Echocardiography repeatedly did not show any structural abnormalities. Because of chronic fatigue, a pacemaker was implanted at the age of 39 years. During implantation, the lead could not be placed in a stable position in the right atrial appendage, and therefore this appendage was assumed to be absent. Normal situs of the lungs and abdominal organs were confirmed by chest X-ray and abdominal ultrasound. The 18-year-old daughter of III-9 (IV-8) was born with a large incomplete AVSD and ASD II, which were surgically corrected at the age of 8 months. At surgery, absence of the coronary sinus and a persistent left superior vena cava (LSVC) connecting directly with the left upper corner of the left atrium were noted. The right superior vena cava was hypoplastic and the brachiocephalic vein was absent. ECGs persistently showed low atrial rhythm. Situs of the lungs and abdomen was normal.
One of the paternal uncles of the proband (III-6) had tetralogy of Fallot and LSVC draining into the coronary sinus, which was surgically corrected at the age of 27. P-wave frontal axis was horizontal. Normal situs of the lungs and bronchi was present. In his 28-year-old daughter (IV-5), a cardiac murmur was noticed shortly after birth and she was followed at a cardiology clinic until the age of 7 years. Echocardiography performed at age 20 showed mildly elevated pulmonary artery pressure, with unknown cause. Low atrial rhythm was also present.
The paternal grandfather of the proband (II-2) had a myocardial infarction at the age 76 years. He received a pacemaker at the age of 84 years because of atrial and ventricular arrhythmias with severe bradycardia. P-wave frontal axis was horizontal. He developed atrial fibrillation at the age of 85 years. Chest X-rays showed normal situs of the lungs and bronchi.
His sister (II-1) showed low atrial rhythm on ECG at the age of 74 years. She developed atrial fibrillation when she was 77 years old. Echocardiography at this age showed dilated atria and mildly elevated pulmonary artery pressure, with normal left ventricular systolic function and diastolic dysfunction. Ultrasound showed normal abdominal situs. Two children of II-1 (III-1 and III-2) had died in the first months of life: III-1 was said to have had aortic hypoplasia, detected at autopsy, and III-2 was said to have died of pneumonia.
The daughter of II-1 (III-3) was asymptomatic at the age of 57 years. Echocardiography and CT showed an LSVC draining into the coronary sinus (Figure 2), aberrant right subclavian artery arising from the aorta distal from the left subclavian artery (arteria lusoria) and elevated pulmonary artery pressure, whereas on ECG low atrial rhythm and paroxysmal supraventricular tachycardia were present. Pulmonary situs was normal. In the asymptomatic 28-year-old daughter of III-3 (IV-1) echocardiography showed abnormal interventricular wall movements, but no ventricular septal defect or other structural abnormalities were seen. She did have low atrial rhythm. The son of III-3 (IV-2), who was also asymptomatic at the age of 25 years, showed a horizontal P-wave frontal axis on ECG with normal echocardiography.
In the youngest son of II-1 (III-5), ECG at the age of 40 years showed low atrial rhythm and first-degree atrioventricular (AV) block. On echocardiography at the same age, a mildly dilated left ventricle was present as well as moderate regurgitation of calcified aortic and tricuspid valves. The 23-year-old daughter of III-5 (IV-3) was diagnosed in her first year of life with ASD II, membrane in the left atrium, absent brachiocephalic vein, LSVC draining into the coronary sinus and a small right superior vena cava. ECGs persistently showed low atrial rhythm. Situs of the lungs and bronchi was normal. The other daughter of III-5 (IV-4) was diagnosed with frequent paroxysmal supraventricular tachycardia and atrial extrasystoles with the origin in the lower right atrium at birth, with normal sinus rhythm in between. She was on anti-arrhythmic medication until the current age of 17 years because of persistence of the tachycardia. Echocardiography showed no structural abnormalities and chest X-ray showed normal pulmonary situs.
At the age of 78 years, the asymptomatic brother of II-1 and II-2 (II-3) was found to have bradycardia with arrests up to 2.6 s with junctional escapes, atrial extrasystoles and non-sustained ventricular tachycardia. He also had paroxysmal atrial fibrillation and complete right bundle branch block. P-wave frontal axis was normal. Echocardiography at the age of 80 years demonstrated left ventricular hypertrophy, dilatation of the aortic root (42 mm) and mild prolapse of the mitral valve, but no congenital abnormalities.
Seven other family members were evaluated in this study (III-4, III-7, III-8, III-11, III-12, IV-6, IV-7), all with normal echocardiography and ECG. None of the individuals with an abnormal P-wave axis had a wandering pacemaker. Clear dysmorphic features were not present in any of the family members.
Linkage analysis and mutation screen
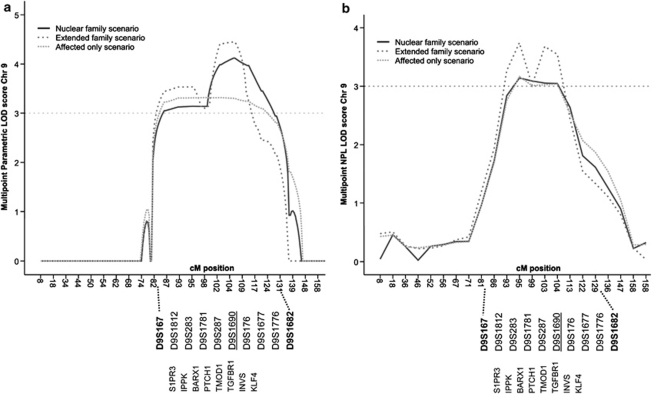
Before performing a genome-wide linkage, we excluded GATA4, TBX5 and NKX2-5 as the disease-causing gene in this family by linkage analysis. In first linkage scenario, termed nuclear family, subjects were considered affected if they had a structural heart defect and/or low atrial rhythm. Therefore, genome-wide linkage was performed on 12 affected (II-1, III-3, III-5, III-6, III-9, IIII-10, IV-1, IV-3, IV-5, IV-8, IV-9 and IV-10), 5 unaffected individuals (III-4, III-7, III-8, IV-6, IV-7) and 1 obligate carrier (II-2; Figure 1 and Table 1). The analysis demonstrated significant linkage to a single locus on chromosome 9q shared by all affected individuals. This locus was absent from unaffected individuals (Figure 1). The shared locus has a multipoint maximum LOD score of 4.1 at marker D9S1690, and is delineated by markers D9S167 and D9S1682 based on the haplotypes (Figures 1 and 3). No other loci with an LOD score higher than 1.0 was detected in this family genome wide.
Figure 3.
Graphs of the multipoint parametric (a) and nonparametric, NPL (b) LOD scores of microsatellite markers on chromosome 9 for various analysis scenarios. The area of linkage is enlarged, depicting the position of markers and candidate genes screened by sequencing.
To reduce the risk of phenocopies, individuals IV-2, IV-4 and II-3 (and therefore his children III-11 and III-12) were not included in the nuclear family linkage analysis. IV-2 was excluded because he had a horizontal P-wave frontal axis, and although this is abnormal, it is not a clear low atrial rhythm. II-3 an IV-4 had significant rhythm and conduction abnormalities, but normal P-wave frontal axes. In a second linkage scenario, termed extended family, we presumed IV-2, IV-4 and II-3 to be affected and III-11 and III-12 to be unaffected. We found that all affected carried to locus, however III-11 did as well, who had normal echocardiography and ECG. The maximal LOD score for the extended family scenario is 4.45; the locus remained the same. A third linkage scenario, termed affected only, was run in which individuals II-1, III-3, III-5, III-6, III-9, III-10, IV-1, IV-3, IV-5, IV-8, IV-9 and IV-10 were included as affected. The maximal LOD score for this affected only scenario was 3.3, also delineated by markers D9S167 and D9S1682.
The shared locus comprises a 48-c critical interval, that corresponds to 39 Mb and contains 402 genes, of which 9 (INVS, TMOD1, TGFBR1, KLF4, IPPK, PTCH1, BARX1, MEGF9 and S1PR3) are known to be, directly or indirectly, involved in development and (left-right) patterning of the heart. Therefore, mutation analysis was performed in these genes in the proband (IV-10). Moreover, 10 conserved non-coding sequences within 5 kb up- or downstream of candidate genes were also screened. Nonetheless, no mutations were identified by direct sequencing. As large genomic rearrangements could also potentially underlie the phenotype, we also performed an array CGH on III-6 and IV-10, using a 180-K oligo array with genome-wide coverage. However, no genomic imbalances in or outside the co-segregating locus were detected, besides copy number variations are also present in controls (data not shown). Moreover, MLPA analysis of the coding exons of INVS and TGFBR1 did not reveal genomic imbalances for these genes.
Discussion
We describe a large family with an autosomal dominant condition with a wide spectrum of cardiovascular abnormalities. The phenotype comprises different types of CHD with an abnormal atrial rhythm. Remarkably, several family members have low atrial rhythm without detectable structural heart defects, suggesting that these individuals show a mild manifestation of the familial disorder. We identified a locus at chromosome 9q that co-segregates with this autosomal dominant disorder and excluded several candidate genes present within the shared 39-Mb disease locus.
The presence of bilateral left atrial appendages in the proband in combination with low atrial rhythm led us to hypothesize that the CHDs in this family might be the result of a defect in the establishment of left-right asymmetry, and might represent a mild or variant expression of left atrial isomerism. Left isomerism is a laterality disorder characterized by bilateral left sidedness, ie, two morphologic left cardiac atria and atrial appendages, often in association with bilateral bilobed (left) lungs, isomerism of the bronchi, abdominal situs abnormalities, polysplenia and abnormalities of the large systemic veins.17, 18 The internal architecture of the heart is often disrupted as well, and different forms of CHD are observed in left atrial isomerism.17, 18, 19 Besides CHD, the phenotype in this family is characterized by abnormal P-wave frontal axis on ECG, typically present in left atrial isomerism due to sinus node abnormalities. In normal hearts, the sinus node is located in the right atrium at the junction between the superior caval vein and the right atrial appendage, which results in a P-wave frontal axis orientated leftward and inferiorly.20 Histological studies in hearts with left atrial isomerism demonstrate absence, hypoplasia and/or ectopic location of nodal tissue in a large proportion of patients.21, 22 In correspondence with these histological abnormalities, abnormal atrial rhythms are commonly found in patients with left atrial isomerism,10, 23 and a low atrial rhythm with a superior P-wave frontal axis is frequently present.10, 23 In this family, four of the five individuals with CHD or isolated LSVC had low atrial rhythm, and one individual had a horizontal P-wave axis. In addition, seven family members without detectable structural heart defects also showed low atrial rhythm, and two had a horizontal P-wave axis. Indeed, rhythm and conduction disturbances have been described in patients with confirmed left isomerism without structural cardiac defects.24, 25 Taken together, the spectrum of CHD in this family is compatible with that seen in left isomerism. Importantly, we could not confirm signs of left isomerism outside the heart (eg, polysplenia) in the investigated family members, although imaging of the abdomen and lungs was not available in all subjects, so we cannot rule out laterality defects in every individual. In conclusion, the phenotype of this family resembles a developmental laterality defect, although affected individuals only show a mild expression, which appears to be restricted to the heart.
Alternatively, the familial phenotype might represent an underlying developmental defect of the sinus node, as most family members have an abnormal low atrial rhythm, caused by a dominance of a pacemaker located in the lower part of the right atrium over the normal sinus node. In its earliest stages, when the heart is no more than a simple tube, a dominant pacemaker activity is already found in the entire systemic inflow region of the heart (both left and right side). Subsequently, newly added myocardium at the inflow part, which differentiates from t-box transcription factor Tbx18 expressing precursor cells, will eventually give rise to the right-sided sinus node, among others.26 The developing sinus node also expresses the t-box transcription factor Tbx3, which acts to repress differentiation into working myocardium by imposing a conduction system phenotype.27 Thus, formation of the sinus nodes requires transcription factors, such as Tbx18 and Tbx3, and knockout of these genes leads not only to under- or maldevelopment of the sinus node, but also to various forms of CHD.26, 27 Consequently, it is possible that the phenotype seen in the current family is the direct result of developmental defects in the formation of the sinus node, possibly via pathways involving t-box transcription factors.
The co-segregating locus on chromosome 9q contains 402 genes (genome build 37), and some of these were selected as candidate genes based on current knowledge, literature and data mining analysis: INVS, S1PR3, TGFBR1, IPPK, BARX1, TMOD1, KLF4, MEGF9 and PTCH1. The coding regions and selected conserved non-coding regions of all these genes were sequenced, but no mutations were found. In addition, we looked for genome-wide genomic imbalances using array CGH; however, none were identified. No other genes known to have a role in left-right patterning or cardiac development map to the 9q region. The best option to identify the causative gene or region on the 9q locus would be to fully sequence it, and although advances in next generation sequencing have been tremendous, it is currently far from trivial to sequence our 39 Mb locus. Moreover, investigation of additional families with a similar phenotype might reduce the region of interest and could lead to the detection of the gene responsible for the phenotype in this family.
A concern of our study is the possibility of phenocopies. Ectopic atrial rhythms can be seen idiopathically in healthy individuals, especially in children. We cannot rule out that individuals in our family have P-wave abnormalities due to other reasons than the familial genetic defect. To minimize the risk of phenocopies, we performed linkage analysis on the most affected patients, ie, patients with low atrial rhythm, and excluded the patient with isolated horizontal P-wave frontal axis as well as the patients with significant rhythm disturbances and normal P-wave frontal axis. Though those patients all carried the disease locus, which implies that the familial disorder shows variable expression. Patient III-11, carrier of the risk allele, had normal echocardiography and ECG at the age of 45 years, which also indicates that the disorder is not 100% penetrant. Atrial fibrillation and other AV rhythm disturbances were mainly present in the older individuals in the family, suggesting age-dependant penetrance of these abnormalities. Although these abnormalities are common in the general population, it is unclear if they are a part of the disease phenotype.
In conclusion, we present a large four-generation family in which a condition, comprising of CHDs and low atrial rhythm, inherits as an autosomal dominant trait with variable expression, and which, to the best of our knowledge, has not been described before. A significant genome-wide linkage was demonstrated to a locus on chromosome 9q with an LOD score of 4.1. Although we did not uncover a causative mutation in this family, the mapping of this locus represents an important step towards the discovery of genes implied in cardiac development. Identification of the disease-causing gene will allow genetic screening and will ultimately provide fundamental insights in human cardiogenesis.
Acknowledgments
We thank all family members for their kind participation. We also thank A Ilgun, A Mul, D van Gent, S de Jong, F Salehi and F Asidah for their excellent technical assistance, S Romeih, and M Groenink for the assessment of the MRI and CT images, and HACM de Bruin-Bon for performing echocardiographic examinations. We thank Dr RJ Oostra for helpful discussions. AFMM is supported by grants from the Netherlands Heart Foundation (96.002) and the European Community's Sixth Framework Program contract (‘HeartRepair' LSHM-CT-2005-018630).
The authors declare no conflict of interest.
References
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Benson DW, Sharkey A, Fatkin D, et al. Reduced penetrance, variable expressivity, and genetic heterogeneity of familial atrial septal defects. Circulation. 1998;97:2043–2048. doi: 10.1161/01.cir.97.20.2043. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Balaji S, Harris L.Atrial arrhythmias in congenital heart disease Cardiol Clin 200220459–468.vii. [DOI] [PubMed] [Google Scholar]
- Postma AV, van de Meerakker JB, Mathijssen IB, et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- Fisch CK, Knoebel SB.Atrial arrhythmiasin Fisch CK, Knoebel SB (eds): Electrocardiography of Clinical Arrhythmias Armonk, NY: Futura publishing company; 2000. Ch 2. [Google Scholar]
- Walsh EP, Cecchin F. Arrhythmias in adult patients with congenital heart disease. Circulation. 2007;115:534–545. doi: 10.1161/CIRCULATIONAHA.105.592410. [DOI] [PubMed] [Google Scholar]
- Momma K, Takao A, Shibata T. Characteristics and natural history of abnormal atrial rhythms in left isomerism. Am J Cardiol. 1990;65:231–236. doi: 10.1016/0002-9149(90)90090-n. [DOI] [PubMed] [Google Scholar]
- Reed PW, Davies JL, Copeman JB, et al. Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nat Genet. 1994;7:390–395. doi: 10.1038/ng0794-390. [DOI] [PubMed] [Google Scholar]
- Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- Uchikawa H, Toyoda M, Nagao K, et al. Brain- and heart-specific Patched-1 containing exon 12b is a dominant negative isoform and is expressed in medulloblastomas. Biochem Biophys Res Commun. 2006;349:277–283. doi: 10.1016/j.bbrc.2006.08.046. [DOI] [PubMed] [Google Scholar]
- Zhi J, Hatchwell E. Human MLPA Probe Design (H-MAPD): a probe design tool for both electrophoresis-based and bead-coupled human multiplex ligation-dependent probe amplification assays. BMC Genomics. 2008;9:407. doi: 10.1186/1471-2164-9-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmann TT, Alders M, Jongbloed RJ, et al. Long QT syndrome caused by a large duplication in the KCNH2 (HERG) gene undetectable by current polymerase chain reaction-based exon-scanning methodologies. Heart Rhythm. 2006;3:52–55. doi: 10.1016/j.hrthm.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Peeters H, Devriendt K. Human laterality disorders. Eur J Med Genet. 2006;49:349–362. doi: 10.1016/j.ejmg.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Norgard G, Berg A.Isomerism (Heterotaxia)in: Gatzoulis MA, Webb GD, Daubeney PEF (eds): Diagnosis and Management of Adult Congenital Heart Disease Edinburgh: Churchill Livingstone; 2003413–421. [Google Scholar]
- Webber SA, Uemura H, Anderson RH.Isomerism of the atrial appendagesin Anderson RH, Baker EJ, Macartney FJ, Rigby ML, Shinebourne EA, Tynan M (eds): Paediatric Cardiology2nd edn.New York: Churchill Livingstone; 2002813–850. [Google Scholar]
- Johnson WH, Moller JH.Diagnostic Methodsin Johnson WH, Moller JH (eds): Pediatric Cardiology1st ednLippincot: Williams and Wilkins; 2001 [Google Scholar]
- Ho SY, Seo JW, Brown NA, Cook AC, Fagg NL, Anderson RH. Morphology of the sinus node in human and mouse hearts with isomerism of the atrial appendages. Br Heart J. 1995;74:437–442. doi: 10.1136/hrt.74.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Ho SY, Anderson RH, et al. The diverse cardiac morphology seen in hearts with isomerism of the atrial appendages with reference to the disposition of the specialised conduction system. Cardiol Young. 2006;16:437–454. doi: 10.1017/S1047951106000382. [DOI] [PubMed] [Google Scholar]
- Wren C, Macartney FJ, Deanfield JE. Cardiac rhythm in atrial isomerism. Am J Cardiol. 1987;59:1156–1158. doi: 10.1016/0002-9149(87)90866-6. [DOI] [PubMed] [Google Scholar]
- Kakura H, Miyahara K, Sohara H, et al. Isolated levocardia associated with absence of inferior vena cava, lobulated spleen and sick sinus syndrome. A case report. Jpn Heart J. 1998;39:235–241. doi: 10.1536/ihj.39.235. [DOI] [PubMed] [Google Scholar]
- Wessels MW, De Graaf BM, Cohen-Overbeek TE, et al. A new syndrome with noncompaction cardiomyopathy, bradycardia, pulmonary stenosis, atrial septal defect and heterotaxy with suggestive linkage to chromosome 6p. Hum Genet. 2008;122:595–603. doi: 10.1007/s00439-007-0436-x. [DOI] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MT, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]