Abstract
Population structure, including population stratification and cryptic relatedness, can cause spurious associations in genome-wide association studies (GWAS). Usually, the scaled median or mean test statistic for association calculated from multiple single-nucleotide-polymorphisms across the genome is used to assess such effects, and ‘genomic control' can be applied subsequently to adjust test statistics at individual loci by a genomic inflation factor. Published GWAS have clearly shown that there are many loci underlying genetic variation for a wide range of complex diseases and traits, implying that a substantial proportion of the genome should show inflation of the test statistic. Here, we show by theory, simulation and analysis of data that in the absence of population structure and other technical artefacts, but in the presence of polygenic inheritance, substantial genomic inflation is expected. Its magnitude depends on sample size, heritability, linkage disequilibrium structure and the number of causal variants. Our predictions are consistent with empirical observations on height in independent samples of ∼4000 and ∼133 000 individuals.
Keywords: genome-wide association study, genomic inflation factor, polygenic inheritance
Introduction
Genome-wide association studies (GWAS) have led to the discovery of hundreds of genetic variants that are associated with complex diseases and traits.1 In total, however, the identified variants explain only a fraction of total risk or phenotypic variance, resulting in the so-called ‘missing heritability'.2, 3 One explanation is that most complex diseases and traits are caused by a large number of variants, the effects of which are too small to pass a stringent genome-wide significance level.3 Therefore, large sample sizes are required and many collaborations have been established to achieve this, resulting in published meta-analyses for a range of diseases and traits.4, 5, 6, 7, 8
One standard quality-control measure for GWAS and meta-analysis is genomic control (GC).9, 10, 11 The concept behind this method is that apart from a small number of SNPs that show a true association with the trait or disease, the test statistics for other SNPs should follow the distribution under the null hypothesis of no association between a SNP and the trait. However, artificial differences in allele frequencies due to population stratification, cryptic relatedness and genotyping errors will affect all SNPs and so the test statistics will be inflated across the whole genome.12, 13, 14 For instance, the mean and median χ2 value over all SNPs will be inflated by these artificial differences above their expectations under the null hypothesis of 1.0 and 0.455. This inflation can be detected and corrected for when testing for alleles that are associated with disease. The genomic control method was first proposed before GWAS, when it was hypothesised that the genetic architecture of complex traits was likely to consist of a small number of causal variants (in, eg, candidate genes) comprising a small proportion of the genome, and that a small number of non-associated null SNPs could be chosen to reflect most of the genome that was not associated with the trait. Before large-scale GWAS being conducted, this method was examined in the studies with hundreds of stratified individuals13, 15 and soon became a standard approach to quantify and adjust for population structure. In the first wave of GWAS, the genomic inflation factors observed in GWAS with thousands of individuals were usually <1.1, which were usually interpreted to be due to subtle population structure.16 Much larger inflation factors have been observed in GWAS with large sample size especially when pooling a number of GWAS into a meta-analysis.4, 5 For example, the GIANT meta-analysis of height observed a genomic inflation factor of 1.42 even after GC-correction in each of the participating studies.5
The logic of GC relies on the fact that only a small fraction of the SNPs show a true association with the disease. However, published results from GWAS clearly indicate that there are many causal variants for a particular disease or trait. We therefore addressed the question of what genomic inflation would be expected under polygenic inheritance. We used analytical derivations and simulation studies to quantify the expected mean (λmean) and median (λmedian) of a χ2-statistic for association from a GWAS under polygenic inheritance, in the absence of population structure and genotyping errors. We show that the predicted genomic inflation factors are consistent with those observed in practice.
Methods
Prediction of genomic inflation factors in quantitative trait and case–control association studies
In association analysis of a quantitative trait (QT), the non-centrality parameter (NCP) of χ2-statistic for a causal variant is
where N is the sample size and q2 is the proportion of phenotypic variance explained by a causal variant. Therefore, the NCP for a SNP that is in linkage disequilibrium (LD) with the causal variant is17, 18
where r is the correlation coefficient between the SNP and the causal variant due to LD.
Under the assumption that the causal variants have not been genotyped on the current commercial SNP arrays, the mean of χ2-statistics (λmean) in GWAS is
where m is the number of causal variants, sj is the number of SNPs in LD with the j-th causal variant, n is the total number of SNPs, qj2 is the variance explained by the j-th causal variant and rjk2 is the LD r2 between the j-th causal variant and the k-th of the SNPs that are in LD with it.
Let s0 be the number of SNPs that are completely in linkage equilibrium with the causal variants so that their test statistics are distributed as χ12. In the absence of population structure or technical artefacts, the χ2-statistics of all the SNPs will be a mixture of s0 null SNPs and n−s0 non-null SNPs (distributed as non-central χ2) with a cumulative probability function of
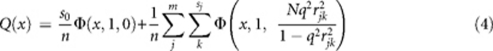 |
where Φ(x, 1, θ) is the cumulative probability of non-central χ2-distribution with NCP of θ.
The median of χ2-statistics (λmedian) is defined as x=c so that Q(c)=0.5. The genomic inflation factor with respect to the median of χ2-statistics is λQTmedian=c/median(χ21)
For a case–control (CC) association study, we assume an underlying threshold-liability model of disease and a multiplicative model of genotype relative risk (GRR). If GRR is small, the variance explained on an underlying liability scale for a genetic variant is19
where p is the allele frequency of the variant, and i=z/K with K being the disease prevalence and z being the height of the standard normal curve at the truncating point pertaining to a probability of K.
Therefore, in a CC association study, the NCP for a causal variant is20
where v is the proportion of cases in the sample. Therefore, the NCP of a SNP in LD with the causal variant in a case–control study is
The mean of χ2-statistics from a genome-wide CC association study is
Analogous to that in the QT association study, the cumulative probability function of χ2-statistics in a case–control study is
and λCCmedian=c/median(χ21) when P(c)=0.5.
Assume that the causal variants have a property that is similar to random SNPs in terms of allele frequency spectrum and LD structure. We randomly sampled m SNPs across the genome to mimic m causal variants. For each ‘causal variant', we searched SNPs for LD within a d Mb region in either direction. Let y denote the genotype code for the causal variant and x for a SNP nearby. We tested for LD between the SNP and causal variant by simple regression, y=b0+b1x+e. We accepted a SNP in LD with the causal variant if the regression P-value <0.05. Obviously, there is a multiple-test problem, but it is unnecessary to correct for it because any SNP in significant LD with the causal variant will be inflated in single-SNP-based association tests.
For a QT, given the heritability (h2) and sample size, we sampled q2 for m causal variants from an exponential distribution with mean of m/h2 and weighted each q2 by h2/Σq2 to constrain the sum of weighted q2 to be h2. Further, we predicted λmeanQT and λmedianQT by equations (3) and (4). For a CC study, given disease prevalence, h2 (heritability of liability on the underlying scale), sample size and number of cases, we predicted λmeanCC and λmeanCC by equations (8) and (9). When m becomes large, it is very likely that some SNPs will be in LD with multiple causal variants. In that case, we calculated s0 as the number of SNPs that were not in LD with the causal variants rather than by using the equation  because otherwise we will underestimate s0 and violate the definitions of equations (4) and (9), that is, Q(x → ∞) and P(x → ∞) would be >1. We then calculated the variance explained by a SNP in LD with s causal variants by
because otherwise we will underestimate s0 and violate the definitions of equations (4) and (9), that is, Q(x → ∞) and P(x → ∞) would be >1. We then calculated the variance explained by a SNP in LD with s causal variants by 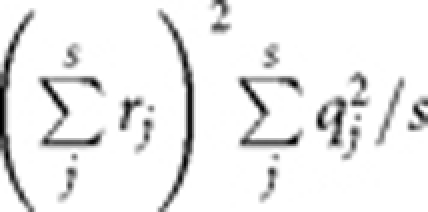 where we summed r rather than r2 because the effects of two causal variants could be either in the same direction or opposite direction. We repeated the procedure 100 times and calculated the mean and SD of the predicted λmean and λmedian.
where we summed r rather than r2 because the effects of two causal variants could be either in the same direction or opposite direction. We repeated the procedure 100 times and calculated the mean and SD of the predicted λmean and λmedian.
In the sections above, we showed how the genomic inflation factors can be predicted on the basis of the LD structure estimated from random SNPs and the heritability. When m is large (ie, q2 is small), equations (3) and (8) are approximately equal to
where s̄ is the average number of SNPs that are in LD with the causal variants (mimicked by a set of random SNPs) with average r2 of 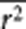 . Since s̄ and
. Since s̄ and 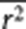 are correlated, in practice, we use
are correlated, in practice, we use 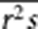 instead of
instead of  .
.
Samples and genotyping
We selected 3925 unrelated individuals (3248 adults and 677 16-year olds) from several GWAS conducted at the Queensland Institute of Medical Research (QIMR).21 All the samples had measured or self-reported height and were genotyped on the Illumina 370K or 610K SNP arrays (Illumina Inc., San Diego, CA, USA). All the samples were collected with informed consent and appropriate ethical approval. A total of 294 831 autosomal SNPs were retained for analysis after stringent quality control. Principal component analysis of ancestry showed that all of these 3925 individuals are of European descent (see ref. 21 for details of the data and quality control procedures). The phenotypes were corrected for age and sex, and standardised to z-scores in the adult and adolescent cohorts separately.
Simulation schemes
We performed simulation studies based on the observed genotype data of 3925 individuals and ∼295K SNPs. We randomly sampled m SNPs as causal variants and generated the effect of each causal variant (b) from a standard normal distribution. We calculated the genetic value of each individual by 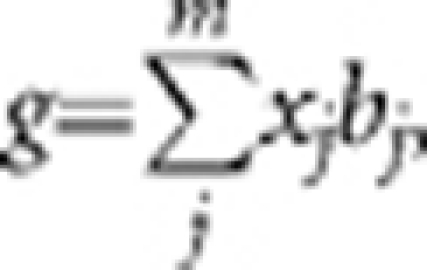 where x is coded as 0, 1 or 2 for genotype qq, Qq or QQ (allele is arbitrarily called Q or q), respectively. We generated residual effects (e) from N (0, var(g) (1−h2)/h2) and calculated the simulated phenotype by y=g+e.
where x is coded as 0, 1 or 2 for genotype qq, Qq or QQ (allele is arbitrarily called Q or q), respectively. We generated residual effects (e) from N (0, var(g) (1−h2)/h2) and calculated the simulated phenotype by y=g+e.
For CC studies, we generated the disease liability in the same way as above. We ranked the individuals by liability and assigned the top 1000 individuals as cases and the remaining individuals as controls.
We used different settings of heritability (h2=0.2, 0.4 and 0.8) and number of causal variants (m=1, 10, 50, 100, 500 and 1000). For each setting, we repeated the simulation 100 times, randomising the positions of causal variants in each simulation replicate. We performed association analyses of the simulated data in PLINK22 and calculated mean and median of χ2-statistics with exclusion of the causal variants.
Results
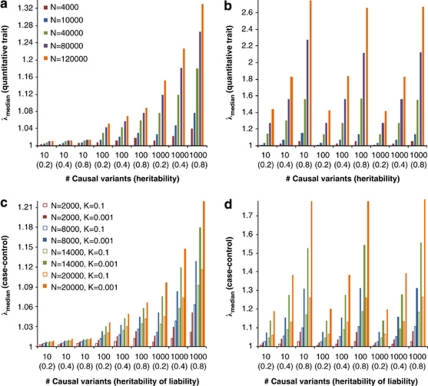
Under the assumption of polygenic inheritance of a quantitative trait and disease liability, we derived analytical equations to predict the genomic inflation factor in GWAS for QT and CC study. We show that in the absence of population structure, the genomic inflation factor, either λmean or λmedian, is not expected to be unity, but is a function of sample size, LD structure, number of causal variants (m) and heritability (h2) for both QT and CC association studies. For the CC study, it depends further on disease prevalence and the proportion of cases in the sample. When m>10, λmean is independent of the number of causal variants and depends only on the heritability, LD structure in the genome and the experimental sample size.
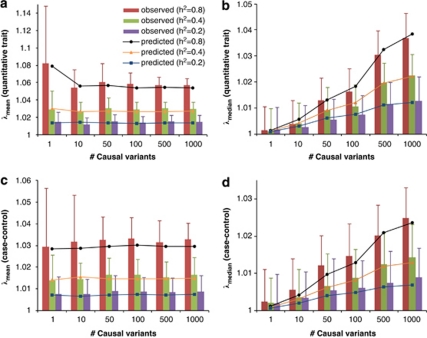
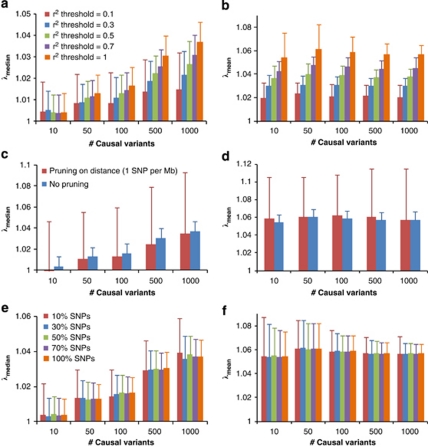
We demonstrate our method using a data set of 3925 unrelated individuals and 294 831 SNPs selected from several GWAS at the QIMR.21 We validated the analytical equations by simulations based on the actual genotype data. Results show that both λmean and λmedian increase with h2 (Figure 1), decrease when pruning SNPs for LD, but do not change when selecting SNPs at random or based on physical distance (Figure 2). Conditional on h2, λmean is approximately constant, but λmedian increases with m, as predicted by theory. The reason is that when m increases, more SNPs (in LD with the causal variants) will depart from the null distribution (χ12) so that the median of χ2-statistics will deviate more from the expected median of (χ12), whereas the effect of each locus decreases as constrained by the heritability, so that the mean test statistic remains the same. Given h2 and m, we predicted λmean and λmedian by theory, but conditional on the observed LD structure. The LD structure is important because there are many SNPs in LD with each causal variant and so many SNPs have an inflated χ2 and this increases the mean and to a less extent the median. We used the LD between SNPs as a proxy for the LD between SNPs and causal variants. In general, the predicted λmean and λmedian agree well with those observed from simulations (Figure 1). For a particular data set, when m is large (eg, m>10), λmean depends only on trait heritability.
Figure 1.
Genomic inflation factor observed in simulation versus that predicted by theory. Data are simulated based on real genotypes of 3925 individuals and 294 831 SNPs with different numbers of causal variants (m=1, 10, 50, 100, 500 and 1000) and heritabilities (h2=0.2, 0.4 and 0.8). Each column represents the average of λmean (a and c) or λmedian (b and d) observed from 100 simulations. Error bars are SD. Each marked line represents the predicted λmean or λmedian averaged over 100 prediction replicates given m and h2. For case–control studies (c and d), h2 refers to heritability of liability on the underlying scale.
Figure 2.
Genomic inflation factor for pruned (or selected) SNPs in simulation study. GWAS for quantitative trait is simulated based on real genotypes of 3925 individuals and 294 831 SNPs with heritability of 0.8 and with different numbers of causal variants (10, 50, 100, 500 and 1000). Each column represents an average of λmean (b, d and f) or λmedian (a, c and e) observed from 100 simulations. Error bars are SD. In (a and b), SNPs are pruned for LD using PLINK22 with threshold r2 value of 0.1, 0.3, 0.5 and 0.7. In (c and d), SNPs are pruned based on physical distance so that any pair of SNPs are at least 1 Mb away from each other. In (e and f), 10, 30, 50 and 70% SNPs are randomly sampled from all of the SNPs.
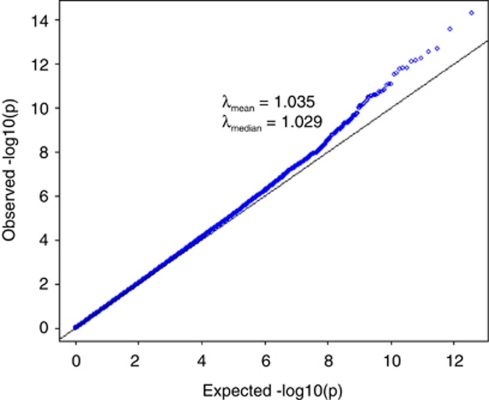
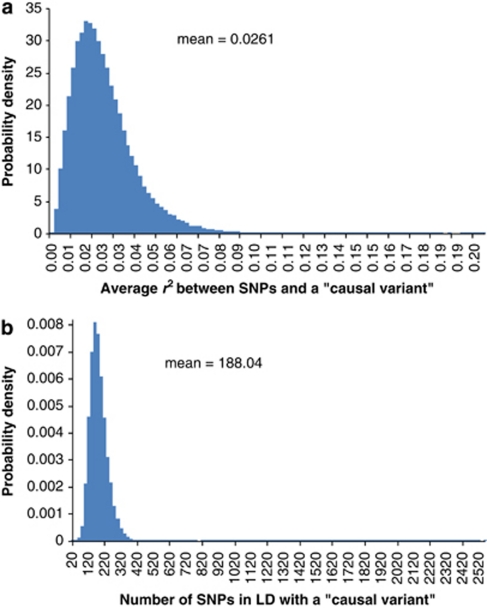
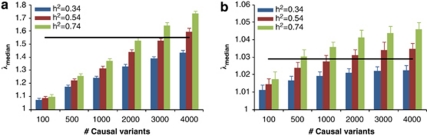
We performed standard GWAS of height using the QIMR data set and observed λmean=1.035 and λmedian=1.029 (Figure 3). We have shown previously that there is no evidence of population structure in this data set (Supplementary Figures 2 and 3 and Supplementary Tables 1 and 2 of ref. 21). We searched for SNPs in LD with 100 000 randomly selected loci and estimated an average of 188 SNPs that are in LD with each locus, with an average r2 of 0.026 (Figure 4). We have previously estimated in this data that 45% of phenotypic variation for height can be explained by ∼295K common SNPs.21 Assuming that the unobserved causal variants are similar to random SNPs with respect to allele frequency and LD, we estimated h2=0.54 (SE=0.1) after adjustment for imperfect LD between the causal variants and SNPs.21 Given h2=0.54, we predicted λmedian to be from 1.028 to 1.035 assuming that the number of causal variants for height ranges from 1000 to 4000 (Figure 5a), consistent with an observed λmedian of 1.029 and with height being highly polygenic.
Figure 3.
Quantile–quantile plot of height association result for QIMR data set (3925 unrelated individuals and 294 831 SNPs). All the SNPs passed stringent quality control and all the individuals are of European ancestry as verified by SNP data. The mean and median of χ2-statistics are 1.035 and 1.029, respectively.
Figure 4.
Histograms of (a) number of SNPs in significant LD with a ‘causal variant' and (b) average r2 between these SNPs and the ‘causal variant'. The ‘causal variants' are mimicked by randomly sampling (without replacement) 100 000 out of 294 831 SNPs across the genome. Simple regression is used to test for SNPs in LD with each ‘causal variant' within 5-Mb distance in either direction.
Figure 5.
Predicted median of χ2-statistics (λmedian) of height association study in (a) the QIMR data and (b) the GIANT meta-analysis. Each column is mean±2SD of 25 prediction replicates. The straight lines are the observed λmedian in real data analyses.
We accessed the test statistics of the discovery set of GIANT meta-analysis (MA) of height with ∼133 000 individuals and ∼2.8-M genotyped and imputed SNPs.5 We excluded ∼636K SNPs with effective sample sizes <126 000 and extracted ∼270K SNPs in common with the QIMR data set. We observed λmean=1.95 and λmedian=1.55. Assuming that the LD structure that underlies the GIANT MA results is similar to that in the QIMR data and h2=0.54, we predicted λmedian for the GIANT MA to be from 1.32 to 1.59 with the assumption of 1000–4000 causal variants (Figure 5b).
Discussion
We have shown by theory, simulation studies and analysis of multiple data sets that a significant inflation of test statistics is to be expected under polygenic inheritance even when there is no population structure. We have provided options in our software tool GCTA23 to estimate LD structure and perform GWAS simulations, and provided an R-script to implement the theoretical predictions as described above (http://gump.qimr.edu.au/gcta/gc). Of course, we are not denying that there may be spurious associations because of population structure for single SNPs,14, 16 but are questioning whether λmean or λmedian is an appropriate statistic to indict and adjust for population structure. In the absence of population structure, λmean reflects the trait heritability and λmedian further reflects the number of causal variants.
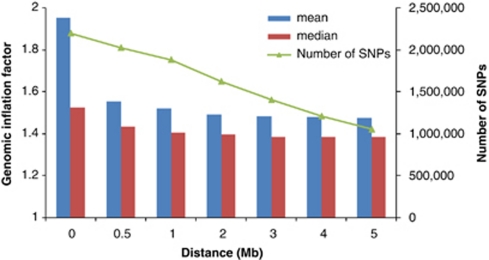
Standard GC theory predicts that the expected value of λmean and λmedian are the same,9, 10 because the distribution of the test statistic is a scaled (χ12). Under polygenic inheritance, however, λmean and λmedian show explicitly different patterns with different sample size, heritability and disease prevalence (Figure 6). Results from the GIANT MA also show a much larger λmean than λmedian, as predicted from the polygenic model. When removing SNPs within d Mb (d=0.5, 1, …, or 5) of the 318 top hits (180 hits at genome-wide false-positive rate of 0.05 and additional 138 hits at genome-wide false discovery rate of 0.05) from ∼2.2-M SNPs in the GIANT MA, λmean decreases from 1.95 to 1.48 and λmedian decreases from 1.53 to 1.39, but they do not converge, consistent with polygenic inheritance (Figure 7). Adjustment for GC in large meta-analyses may therefore be too conservative and reduce the power to detect significant SNP-trait associations.
Figure 6.
Predicted genomic inflation factor for quantitative trait (a and b) and case–control (c and d) association studies. Prediction is based on 294 831 SNPs with different numbers of causal variants and heritabilities (h2), sample size (N) and disease prevalences (K, for case–control study). Each value is an average over 100 prediction replicates. For the case–control study, the number of cases and controls is equal.
Figure 7.
Genomic inflation factor for ∼2.2-M SNPs (with exclusion of ∼636K with effective sample sizes <126 000 from the total ∼2.8 M SNPs) in GIANT meta-analysis for height with ∼133 000 samples. A total of 318 top hits were identified by GIANT meta-analysis (genome-wide false discovery rate of 0.05).5 Any SNP within d Mb distance (d=0.5, 1, …, or 5, x-axis) of the top hits is removed and genomic inflation factor is calculated using all of the remaining SNPs.
In the presence of both population structure and polygenic inheritance (which may be regarded as a general case in practice), we cannot distinguish whether population structure or polygenic inheritance is the major cause of the genomic inflation unless we are able to estimate the proportion of phenotypic variance explained by all the SNPs and that attributed to population structure. It may be possible to discriminate polygenic inheritance from population structure by testing for associations between markers on different chromosomes. Population structure, including the presence of cryptic relationships among individuals in the sample, implies a correlation between alleles on different chromosomes. A genome-wide inflation of the test statistic with little or without such correlation is a strong support for polygenic variation.
Acknowledgments
We thank all three reviewers for helpful comments. We acknowledge funding from the Australian National Health and Medical Research Council (NHMRC Grants 389891, 389892, 613672 and 613601), the Australian Research Council (ARC Grants DP0770096 and DP1093900) and the US National Institute of Health (NIH Grants AA13320, AA13321 and DA12854).
The authors declare no conflict of interest.
Supplementary Material
References
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249 796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DPB, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Zheng G, Freidlin B, Gastwirth JL. Robust genomic control for association studies. Am J Hum Genet. 2006;78:350–356. doi: 10.1086/500054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. The Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Hao K, Li C, Rosenow C, Wong WH. Detect and adjust for population stratification in population-based association study using genomic control markers: an application of Affymetrix Genechip Human Mapping 10 K array. Eur J Hum Genet. 2004;12:1001–1006. doi: 10.1038/sj.ejhg.5201273. [DOI] [PubMed] [Google Scholar]
- WTCCC Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2010;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]