SUMMARY
In higher eukaryotes, miRNAs and siRNAs guide translational inhibition, mRNA cleavage, or chromatin regulation. We found that the antisense overlapping gene pair of Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH), a stress-related gene, and SRO5, a gene of unknown function, generates two types of siRNAs. When both transcripts are present, a 24-nt siRNA is formed by a biogenesis pathway dependent on DCL2, RDR6, SGS3, and NRPD1A. Initial cleavage of the P5CDH transcript guided by the 24-nt siRNA establishes a phase for the subsequent generation of 21-nt siRNAs by DCL1 and further cleavage of P5CDH transcripts. The expression of SRO5 is induced by salt, and this induction is required to initiate siRNA formation. Our data suggest that the P5CDH and SRO5 proteins are also functionally related, and that the P5CDH-SRO5 gene pair defines a mode of siRNA function and biogenesis that may be applied to other natural cis-antisense gene pairs in eukaryotic genomes.
INTRODUCTION
Despite the preponderance of large intergenic spaces, eukaryotic genomes contain many overlapping genes. For example, 4%–9% of human genes, up to 22% of Drosophila melanogaster genes, and approximately 10% of Arabidopsis thaliana genes are in convergent overlapping gene pairs, also known as natural cis-antisense gene pairs (Boi et al., 2004; Wang et al., 2005; Jen et al., 2005). Although their functional significance is unclear, one intriguing possibility is that overlapping transcripts in an antisense orientation form double-stranded RNAs that may be processed into small RNAs.
Small 21- to 25-nt, noncoding RNAs are important regulators of gene expression in both plants and animals (Carrington and Ambros, 2003; Bartel, 2004; He and Hannon, 2004). These small RNAs can be divided into two classes: micro-RNAs (miRNAs) and short interfering RNAs (siRNAs). miRNAs are found in a number of multicellular eukaryotes (Ambros, 2003; Carrington and Ambros, 2003; Bartel, 2004) and are generated from longer hairpin precursors by the Ribonuclease III-like enzyme Dicer. After incorporation into an Argonaute-containing RNA Induced Silencing Complex (RISC) (Hannon, 2002; Meister and Tuschl, 2004), base pairing between miRNAs and complementary target mRNAs guides sequence-specific translational inhibition or transcript cleavage (Bartel, 2004). miRNAs have a well-documented role in allowing developmental regulation of multigene families (Palatnik et al., 2003; Carrington and Ambros, 2003; Bartel, 2004).
siRNAs differ from miRNAs in that they are generated from long double-stranded RNAs. siRNAs were first identified in plants undergoing posttranscriptional gene silencing (PTGS; Hamilton and Baulcombe, 1999), and subsequently diverse sets of endogenous siRNAs have been found in plants and animals (Llave et al., 2002; Sunkar and Zhu, 2004; Ambros, 2003; Aravin et al., 2003; Tang et al., 2003; Reinhart et al., 2002). The biogenesis of small RNAs in plants is especially complex. In Arabidopsis, there are four Dicer-like (DCL) proteins (Schauer et al., 2002), six predicted RDRs (Mourrain et al., 2000), and ten predicted Argonautes (Morel et al., 2000). DCL1 is required for the production of ~21-nt miRNAs and trans-acting siRNAs (tasiRNAs) (Park et al., 2002; Peragine et al., 2004; Vazquez et al., 2004b). Allen et al. (2005) demonstrated that an initial DCL1-dependent, miRNA-guided cleavage of tasiRNA primary transcripts sets the 21-nt phase for accurate tasiRNA formation. DCL4 is responsible for the processing of 21-nt tasiRNAs (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). Generation of tasiRNAs also involves RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and SUPPRESSOR OF GENE SILENCING 3 (SGS3) (Peragine et al., 2004, Vazquez et al., 2004b).
DCL2 is involved in the production of some viral siRNAs (Xie et al., 2004) and may substitute for DCL3 or DCL4 when they are absent (Gasciolli et al., 2005). DCL3 and RDR2, as well as the RNA polymerase IV encoded by NRPD1A/SILENCING DEFECTIVE 4, cooperate in generating heterochromatin-associated 24-nt siRNAs from various retroelements and transposons, 5S ribosomal RNA genes, endogenous direct and inverted repeats, and transgenes containing direct repeats (Chan et al., 2004; Xie et al., 2004; Zilberman et al., 2004; Herr et al., 2005; Onodera et al., 2005). The appearance of these siRNAs has been correlated with DNA and histone methylation at the homologous chromatin (Hamilton et al., 2002; Volpe et al., 2002; Zilberman et al. 2004; Xie et al., 2004).
Proline metabolism and particularly Δ1-pyroline-5-carboxylate (P5C), an intermediate in proline synthesis and catabolism, have a key role in stress response and accumulation of reactive oxygen species (ROS). P5C can also promote apoptosis, and this may be mediated, at least partly, through ROS accumulation. In yeast, disruption of PUT2, which encodes Δ1-pyroline-5-carboxylate dehydrogenase (P5CDH), an enzyme that catabolizes P5C, leads to decreased cell growth and ROS accumulation (Nomura and Takagi, 2004; Deuschle et al., 2001). In plants as well, exogenously applied P5C increases ROS production, reduces growth, and induces a number of stress-responsive genes (Deuschle et al., 2004; Hellmann et al., 2000; Iyer and Caplan, 1998). Inactivation of P5CDH causes hypersensitivity to proline and P5C (Deuschle et al., 2004). Also, P5CDH is induced at pathogen infection sites where programmed cell death and ROS accumulation occur (Ayliffe et al., 2002). These lines of evidence, along with the long-studied role of proline accumulation in plant and microbial stress tolerance (Zhu, 2002) indicate that proline and P5C metabolism are likely to be highly regulated processes impacting ROS accumulation, stress response, and cell death across a range of eukaryotes.
We report here that Arabidopsis P5CDH and SRO5, an overlapping gene of unknown function in the antisense orientation, generate both 24-nt and 21-nt siRNAs. We have termed these siRNAs nat-siRNAs because they are derived from natural antisense transcripts. Upon induction of SRO5 by salt stress, a 24-nt SRO5-P5CDH nat-siRNA is produced by a biogenesis pathway requiring DCL2, RDR6, SGS3, and NRPD1A. Cleavage of the P5CDH transcript guided by the 24-nt nat-siRNA sets the phase for accurate production of further 21-nt P5CDH nat-siRNAs by DCL1. The nat-siRNAs downregulate the expression of P5CDH by causing mRNA cleavage. Downregulation of P5CDH leads to proline accumulation that is important for salt tolerance but also causes increased ROS production, which is normally counteracted by the SRO5 protein. Thus, the SRO5-P5CDH nat-siRNAs together with the P5CDH and SRO5 proteins are key components of a regulatory loop controlling ROS production and stress response. The nat-siRNA-mediated crossregulation of P5CDH and SRO5 mRNAs and the functional relationship of these two proteins suggest a regulatory model that may be applied to other cis-antisense gene pairs.
RESULTS
Salt Stress Induces an Endogenous siRNA from a Pair of Antisense Genes
Previously, we cloned miRNAs and putative siRNAs from Arabidopsis plants treated with various abiotic stresses (Sunkar and Zhu, 2004). One putative siRNA (clone # P96-F02) matched the overlapping region between the 3′ end of the P5CDH ORF and the 3′UTR of an unknown gene on the opposite strand (At5g62520), which has recently been designated SRO5 (Figure 1A). These two genes generate convergent transcripts that overlap by 760-nt, and the cloned siRNA sequence matched a 21-nt region (1873–1893) of the SRO5 3′UTR and was complementary to the corresponding region of the P5CDH ORF.
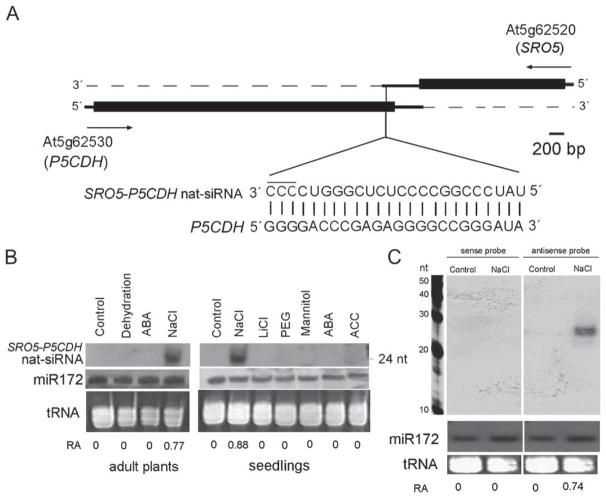
Figure 1. Salt Stress Induces a 24-nt nat-siRNA from the SRO5-P5CDH cis-Antisense Overlapping Genes.
(A) Genomic structure of SRO5 (At5g62520) and P5CDH (At5g62530) genes. Arrows indicate the direction of transcription. Thick and thin solid lines represent ORF and UTR regions, respectively. Sequence of the 24-nt SRO5-P5CDH nat-siRNA is aligned with P5CDH mRNA. The three nucleotides marked by a line on top were deduced from RNA blot analysis and confirmed by primer extension assays (Table S1).
(B) Accumulation of the SRO5-P5CDH nat-siRNA is specifically induced by salt stress. (Left panel) Twenty-day-old soil grown plants were exposed to the following stress or hormone treatments: dehydration (withholding watering for 10 days); ABA (spraying with 100 μM ABA with samples collected 24 hr later); and NaCl (watering with 300 mM NaCl for 2 days). (Right panel) Ten-day-old seedlings exposed to the following treatments in liquid media for 6 hr: Control (nutrient medium), 150 mM NaCl, 15 mM LiCl, 30% (w/v) PEG-8000, 250 mM Mannitol, 100 μM ABA, or 10 μM ACC.
(C) Northern blot of small RNA extracted from control or salt-treated (150 mM, 6 hr) seedlings. The sense and antisense probes are described in the text.
Using an oligonucleotide probe complementary to the 21-nt siRNA, we could not detect any siRNAs in Arabidopsis plants grown under normal conditions (Figure 1B); however, a 24-nt siRNA (referred to hereafter as 24-nt SRO5-P5CDH nat-siRNA) was detected in NaCl-treated adult plants and seedlings. Occasionally, a very weak signal at 21-nt could also be seen in the salt-stressed plants (not shown); this signal likely corresponds to the cloned 21-nt siRNA. No 24-nt SRO5-P5CDH nat-siRNA signal was found in plants exposed to several other stress or homone treatments (Figure 1B), demonstrating that the induction of 24-nt SRO5-P5CDH nat-siRNA is highly specific to NaCl stress.
To define the sequence of the 24-nt SRO5-P5CDH nat-siRNA, we hybridized the small RNA blots with oligonucleotide probe A (5′-GGGGACCCGAGAGGGGCCGGGATA-3′) and oligonucleotide probe B (5′-GACCCGAGAGGGGCCG GGATAGGG-3′) (the sequences in bold letters are complementary to the 21-nt cloned siRNA). Probe A but not B detected the 24-nt SRO5-P5CDH nat-siRNA from salt-stressed plants (data not shown). This suggests that the 24-nt SRO5-P5CDH nat-siRNA sequence is likely 5′-UAUCCCGGCCC CUCUCGGGUCCCC-3′ (Figure 1C). This sequence was confirmed by a modified small RNA cloning approach (Table S1).
Although siRNAs are generated in the double-stranded form, often only one of the strands is incorporated into RISC and can be detected (Schwarz et al., 2003; Khvorova et al., 2003). Twenty-four nucleotide SRO5-P5CDH nat-siRNA was detected with an antisense (relative to SRO5) probe but not with a sense probe (Figure 1C), suggesting that the sense strand as shown in Figure 1A is stable and the other strand is rapidly degraded. Furthermore, examination of the 24-nt SRO5-P5CDH nat-siRNA sequence indicates that the sense strand sequence conforms to the asymmetry rule for siRNA stability (Schwarz et al., 2003).
24-nt SRO5-P5CDH nat-siRNA Is Produced by a Unique Biogenesis Pathway
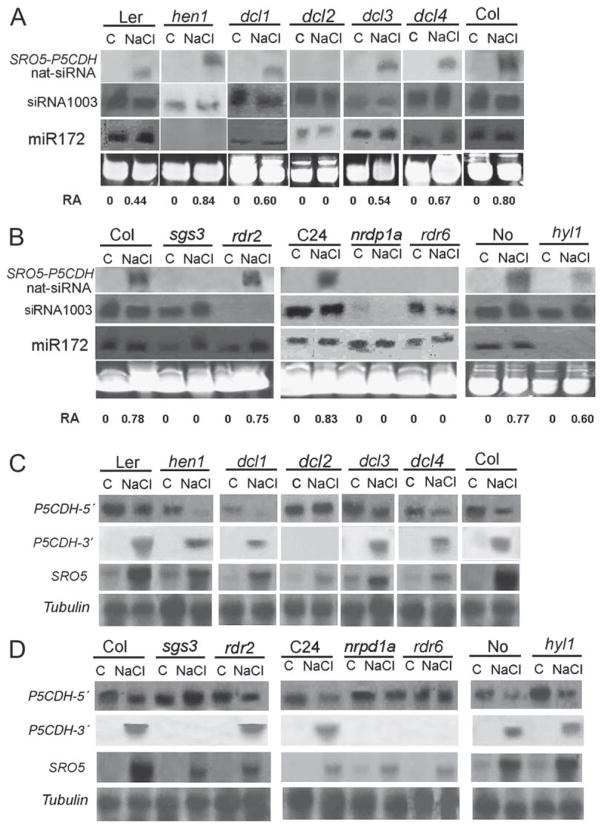
To define the components required for the formation of 24-nt SRO5-P5CDH nat-siRNA, it was examined in salt stressed plants of small RNA biogenesis mutants. The 24-nt SRO5-P5CDH nat-siRNA was detected in salt-treated wild-type plants of all ecotypes tested such as Landsberg (Ler), Columbia, C24, and Nossen (Figure 2). However, it was not detected in the hen1 mutant. Instead, a larger-sized signal was found in hen1 plants (Figure 2A). The 24-nt SRO5-P5CDH nat-siRNA was present in dcl1, dcl3, and dcl4 but not in dcl2. The sgs3, nrpd1a, and rdr6 mutations also blocked the accumulation of 24-nt SRO5-P5CDH nat-siRNA (Figure 2B). In contrast, the rdr2 mutation did not have any effect on 24-nt SRO5-P5CDH nat-siRNA, although it abolished the accumulation of siRNA 1003. The HYL1 gene has been shown to be required for the accumulation of several miRNAs (Han et al., 2004; Vazquez et al., 2004a), including miR172, and the level of 24-nt SRO5-P5CDH nat-siRNA was reduced in the hyl1 mutant plants (Figure 2B). These results suggest that accumulation of the 24-nt SRO5-P5CDH nat-siRNA is dependent on DCL2, RDR6, SGS3, and NRPD1A. This pathway is different from those producing heterochromatin-related siRNAs or tasiRNAs, suggesting that the SRO5-P5CDH nat-siRNA defines a new class of siRNAs.
Figure 2. Accumulation of SRO5-P5CDH nat-siRNA Requires DCL2, RDR6, SGS3, NRPD1A and Is Dependent on Induction of SRO5 mRNA by Salt Stress.
(A and B) Northern analysis of small RNAs from various mutants and wild-type controls. One hundred micrograms of low-molecular-weight RNA was loaded per lane. The blots were hybridized with an oligonucleotide probe corresponding to the sequence of the 24-nt nat-siRNA. The number below the blot indicates relative abundance (RA) of the nat-siRNA with respect to the loading control miR172 or siRNA 1003 when miR172 is not present.
(C and D) Northern blot analysis of P5CDH and SRO5 mRNAs. Twenty-five micrograms of total RNA was probed with P5CDH 5′- or 3′-specific probes or SRO5 probe. The P5CDH 5′ probe includes the 50 bp 5′UTR and the first 400 bp of ORF. The P5CDH 3′ probe corresponds to the last 200 bp of the 3′UTR and detects a cleavage fragment of approximately 700 bp. The P5CDH 3′ probe also hybridized with the ~1.2 kb SRO5 mRNA (not shown). The SRO5 probe detects a transcript of approximately 1.2 kb.
Salt Stress Regulates SRO5 and P5CDH Transcript Levels and P5CDH mRNA Cleavage
To understand the mechanism of salt stress induction of the SRO5-P5CDH nat-siRNA, we investigated whether the expression of SRO5 or P5CDH is regulated by salt stress. SRO5 was not expressed in plants grown under normal conditions, but its expression was induced by NaCl treatment (Figures 2C and 2D). In contrast, P5CDH expression was reduced by NaCl treatment (Figures 2C and 2D). These results suggest that, upon salt treatment, the SRO5 and P5CDH mRNAs can form a dsRNA that is then processed by DCL2 to generate the 24-nt SRO5-P5CDH nat-siRNA.
A possible role of the 24-nt SRO5-P5CDH nat-siRNA is to cause cleavage of P5CDH mRNA. In addition to detecting a full-length P5CDH transcript, a P5CDH 3′UTR probe also hybridized with a smaller band corresponding to the size of the putative 3′ cleavage product. This putative 3′ cleavage product accumulated only in NaCl-treated plants (Figures 2C and 2D). Importantly, in dcl2, sgs3, nrpd1a, and rdr6 mutants where the 24-nt SRO5-P5CDH nat-siRNA was not produced, there was less or no decrease in P5CDH full-length transcript level, and the 3′ cleavage product was not detected (Figures 2C and 2D). Interestingly, in hen1 plants, salt stress still reduced the level of full-length P5CDH transcript and caused accumulation of the 3′ cleavage product. This suggests that the larger-sized siRNA in hen1 mutant plants was still functional in causing P5CDH mRNA cleavage. Taken together, these results suggest that salt stress triggers the expression of SRO5, leading to dsRNA formation and consequently generation of 24-ntSRO5-P5CDHnat-siRNA, which then downregulates P5CDH transcript levels through mRNA cleavage.
Cleavage Site Mapping of P5CDH mRNA Reveals Phased Processing of 21-nt SRO5-P5CDH nat-siRNAs
To verify that the 24-nt SRO5-P5CDH nat-siRNA indeed targets P5CDH mRNA for endonucleolytic cleavage, we carried out 5′ RACE assays using mRNA from salt-treated plants to map potential cleavage sites. Among several cleavage sites found in the P5CDH mRNA in wild-type plants, one site is between nucleotides 12 and 13 of the 24-nt SRO5-P5CDH nat-siRNA (Figure 3). This differs from other ARGONAUTE-catalyzed RNA cleavage sites which occur between nucleotides 10 and 11 (Allen et al., 2004). No cleavage products were found when the assay was performed with mRNA isolated from salt stress treated tissue of rdr6 or sgs3 plants or with mRNA from unstressed wild-type plants (data not shown). The results support our hypothesis that the 24-nt SRO5-P5CDH nat-siRNA can direct P5CDH mRNA cleavage.
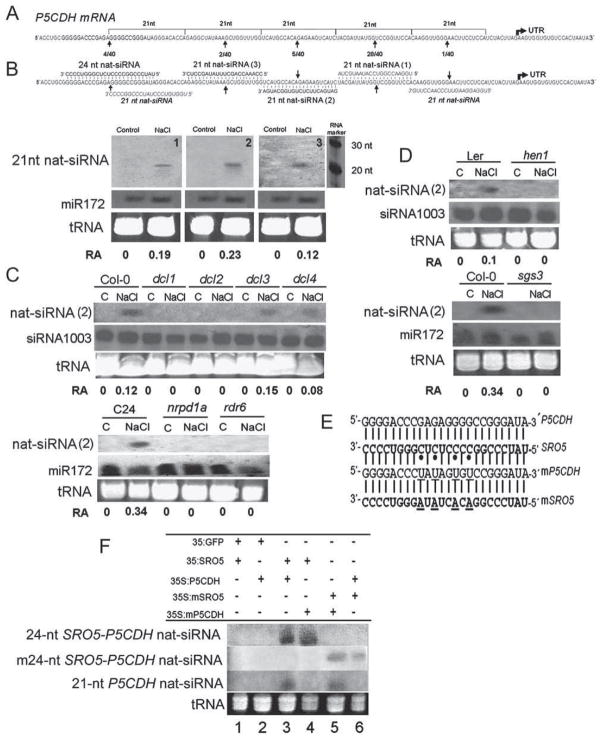
Figure 3. Cleavage Site Mapping of P5CDH mRNA and the Detection of 21-nt nat-siRNAs.
(A) P5CDH mRNA cleavage sites detected by RLM-5′ RACE. Numbers indicate the frequency of cleavage at each site. The positions of predicted 21-nt nat-siRNAs are indicated.
(B) Diagram showing the sequences of 24-nt and 21-nt nat-siRNAs aligned with the P5CDH mRNA (upper panel) and detection of the 21-nt nat-siRNAs (lower panel). Sequences in italics are 21-nt nat-siRNA predicted but not confirmed by Northern blots. One hundred micrograms of low-molecular-weight RNA per lane was probed with oligo probes, 1, 2, and 3, complementary to 21-nt nat-siRNAs numbered. The number below the blot indicates relative abundance (RA) of the nat-siRNA with respect to the loading control miR172.
(C and D) Accumulation of 21-nt nat-siRNA number 2 in different mutants. The number below the blot indicates relative abundance (RA) of the nat-siRNA with respect to the loading control miR172 or siRNA 1003.
(E) Alignment of the 24-nt nat-siRNA region in wild-type and mutated (mSRO5 andmP5CDH) versions of SRO5 and P5CDH used for transient expression in N. benthamiana. Two mutations were introduced on either side of the site of 24-nt nat-siRNA-directed P5CDH cleavage.
(F) Coexpression of various combinations of wild-type and mutant SRO5 and P5CDH constructs in N. benthamiana. As a control, SRO5 and P5CDH were also coexpressed with an unrelated GFP transcript. Small RNA blots were prepared from leaves collected 3 days after infiltration and probed with oligoes complementary to wild-type 24-nt SRO5-P5CDH nat-siRNA, the mutated 24-nt SRO5-P5CDH nat-siRNA sequence (m24-nt nat-siRNA), and a mixture of three 21-nt P5CDH nat-siRNAs (probes 1, 2, and 3; Figure 3B).
Surprisingly, the 5′ RACE assay also revealed four additional cleavage sites in the P5CDH mRNA (Figure 3A). One of these was the most frequent cleavage site (28 out of 40). These additional cleavage sites are 21-nt apart and in-phase with each other. We hypothesized that cleavage guided by the 24-nt SRO5-P5CDH nat-siRNA may set the phase for processing of P5CDH transcript by RDR into dsRNA and then dicing by a different DCL, which generates new 21-nt P5CDH nat-siRNAs. We designed three oligonucleotide probes in an attempt to detect these predicted 21-nt P5CDH nat-siRNAs (1, 2, and 3; Figure 3B). Each of these probes detected a 21-nt P5CDH nat-siRNA in salt-stressed plants. Analysis of one of these siRNAs (probe 2) in the same mutants analyzed above showed that generation of 21-nt P5CDH nat-siRNAs was dependent on DCL1, DCL2, HEN1, RDR6, SGS3, and NRPD1A (Figures 3C and 3D).
We also looked for, but failed to find, siRNAs deriving from sequences upstream of and in-phase with the 24-nt SRO5-P5CDH nat-siRNA-directed cleavage site by using a mix of 21-nt oligonucleotide probes complementary to the upstream sequences. Additionally, we hybridized the small RNA blots with 24-nt oligonucleotide probes designed to detect other potential 24-nt siRNAs produced in-phase with (either downstream or upstream of) the 24-nt SRO5-P5CDH nat-siRNA(probe sequences used are described in TableS2). These probes failed to detect any siRNA signal (not shown), suggesting that only one 24-nt SRO5-P5CDH nat-siRNA accumulates. Since these 24-nt probes are completely out of phase with the 21-nt P5CDH nat-siRNAs, the result also suggests a lack of siRNAs not in-phase with the 24-nt SRO5-P5CDH nat-siRNA-directed cleavage site, although the experiment cannot rule out the existence of other out-of-phase siRNAs.
The combined data suggest that the 21-nt P5CDH nat-siRNAs were generated by DCL1, and their production was dependent on production of the 24-nt SRO5-P5CDH nat-siRNA by DCL2. To directly test whether formation of the 24-nt SRO5-P5CDH nat-siRNA is required for the formation of 21-nt nat-siRNAs, we transiently expressed combinations of wild-type and mutated P5CDH and SRO5 transcripts (mP5CDH and mSRO5) in Nicotiana benthamiana leaves. mP5CDH and mSRO5 were mutated to introduce four mismatches with the complementary wild-type sequence in the region corresponding to the 24-nt SRO5-P5CDH nat-siRNA (Figure 3E). However, mP5CDH and mSRO5 were perfectly complementary to each other at the 24-nt SRO5-P5CDH nat-siRNA site. Small RNA blots from tissue coexpressing these transcripts were probed with oligonucleotides complimentary to the wild-type and m24-nt SRO5-P5CDH nat-siRNA and with a mixture of 21-nt P5CDH nat-siRNAs (probes 1, 2, and 3; Figure 3B).
The results of this experiment support the hypothesis that 24-nt SRO5-P5CDH nat-siRNA is produced first and required for the formation of 21-nt P5CDH nat-siRNAs. When wild-type P5CDH and SRO5 transcripts were coexpressed in N. benthamiana leaves, both 24-nt and 21-nt SRO5-P5CDH nat-siRNAs were produced (Figure 3F, lane 3). Expression of either gene in combination with the unrelated GFP transcript did not produce any nat-siRNAs (Figure 3F, lanes 1 and 2), demonstrating that both the P5CDH and SRO5 transcripts must be present to generate nat-siRNAs. When wild-type SRO5 was coexpressed with mP5CDH, wild-type 24-nt SRO5-P5CDH nat-siRNA was still produced from the SRO5 transcript but no 21-nt P5CDH nat-siRNAs could be detected (Figure 3F, lane 4). This demonstrated that the initial 24-nt SRO5-P5CDH nat-siRNA could be produced from the long, double-stranded SRO5-P5CDH transcripts despite the mismatches. However, this wild-type 24-nt SRO5-P5CDH nat-siRNA was presumably unable to guide cleavage of mP5CDH due to the mismatches and thus no 21-nt nat-siRNAs could be formed. A similar result was observed from coexpression of P5CDH and mSRO5 (Figure 3F, lane 6). Coexpression of mP5CDH and mSRO5 led to the production of both m24-nt and 21-nt nat-siRNAs (Figure 3F, lane 5). Thus, while initial processing of the SRO5-P5CDH double-stranded RNA is tolerant of mismatches, subsequent production of 21-nt nat-siRNAs is dependent on perfect complementarity between the 24-nt SRO5-P5CDH nat-siRNA and the P5CDH transcript.
Role of SRO5-P5CDH nat-siRNAs in Proline Metabolism and Salt-Stress Tolerance
To determine the physiological role of the salt stress-induced SRO5-P5CDH nat-siRNAs, we compared the levels of salt stress-induced proline accumulation in various mutant plants. In dcl2, sgs3, rdr6, and nrpd1a, which lacked SRO5-P5CDH nat-siRNAs and cleavage of the P5CDH transcript, proline accumulation was not significantly induced by salt stress or was induced to a lesser extent than in the corresponding wild-type (Figure 4A). This result is consistent with their inability to downregulate P5CDH under stress, thereby causing continued proline catabolism and reduced proline accumulation. In contrast, the dcl1 and rdr2 mutants, which were able to degrade P5CDH mRNA, had wild-type levels of proline accumulation under salt stress (Figure 4A). The wild-type level of proline accumulation in dcl1 indicates that although the 21-nt P5CDH nat-siRNAs were not produced, the 24-nt SRO5-P5CDH nat-siRNA alone was sufficient to cause downregulation of P5CDH (Figure 3C). Whether or not the 21-nt P5CDH nat-siRNAs are required to downregulate P5CDH under other stress conditions will be of interest for future experiments.
Figure 4. Proline Accumulation and Salt and Proline Sensitivity in Mutants Affected in SRO5-P5CDH nat-siRNA Accumulation.
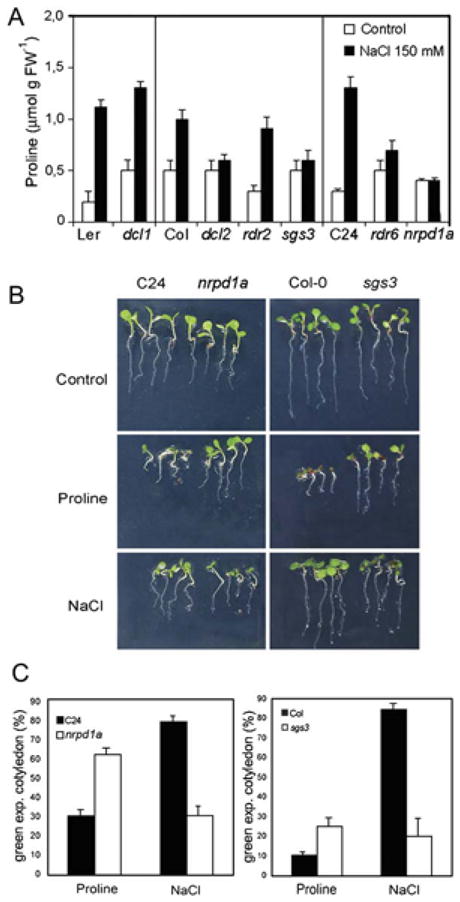
(A) Proline content in 10-day-old seedlings with or without 6 hr of exposure to 150 mM NaCl. Values represent means ± SD from three independent experiments.
(B) Response of sgs3 and nrpd1a to NaCl and exogenous proline. Seedlings were germinated and grown for 7 days in 75 mM NaCl or 5 mM proline for Col-O and sgs3, and 75 mM NaCl or 2 mM proline for C24 and nrpd1a. Photos show representative seedlings.
(C) Tolerance to NaCl or exogenous proline quantified as the percentage of 7-day-old seedlings with green expanded cotyledons. Values represent means ± SD from three independent experiments.
We also examined the proline and salt-stress sensitivity of nrpd1a and sgs3, two mutants that do not have strong pleiotropic phenotypes and grow well under nonstressed conditions. Consistent with previous observations (Hellmann et al., 2000), high levels of exogenous proline inhibited seedling growth of the C24 and Col wild-types (Figures 4B and 4C). The nrpd1a and sgs3 mutants were clearly more tolerant of exogenous proline (Figures 4B and 4C). This proline tolerance may be caused by a reduced cleavage of P5CDH mRNA leading to greater P5CDH activity and reduced P5C levels in the proline-treated seedlings. Conversely, these mutants were less tolerant of salt stress, which is consistent with their inability to control proline catabolism and accumulate protective amounts of proline.
To further examine the biogenesis of SRO5-P5CDH nat-siRNAs and their role in proline and salt-stress tolerance, we identified two homozygous Arabidopsis mutant plants with a T-DNA insertion in the ORF of SRO5 or P5CDH (Figure 5A). Northern blot analysis showed that expression of P5CDH and SRO5 was abolished in the respective mutants (Figure 5B). Neither mutant could produce the 24-nt SRO5-P5CDH nat-siRNA (Figure 5C) or 21-nt P5CDH nat-siRNAs (data not shown) under salt stress. In the sro5 knockout mutant, P5CDH full-length transcript accumulated to a higher level and was not reduced by salt stress (Figure 5B). These results further demonstrate that production of SRO5-P5CDH nat-siRNAs is dependent on the presence of the P5CDH and SRO5 antisense transcripts, and that SRO5-P5CDH nat-siRNAs are responsible for the down-regulation of the P5CDH mRNA. Interestingly, SRO5 mRNA was detected in the p5cdh knockout mutant even without salt stress (Figure 5B). This indicates that either the p5cdh knockout causes certain physiological stress (e.g., oxidative stress) that can induce SRO5 expression, or that SRO5 transcription or transcript stability is normally suppressed by the P5CDH gene through an unknown mechanism.
Figure 5. SRO5-P5CDH nat-siRNA Accumulation and Salt and Proline Responses in sro5 and p5cdh Knockout Mutants.
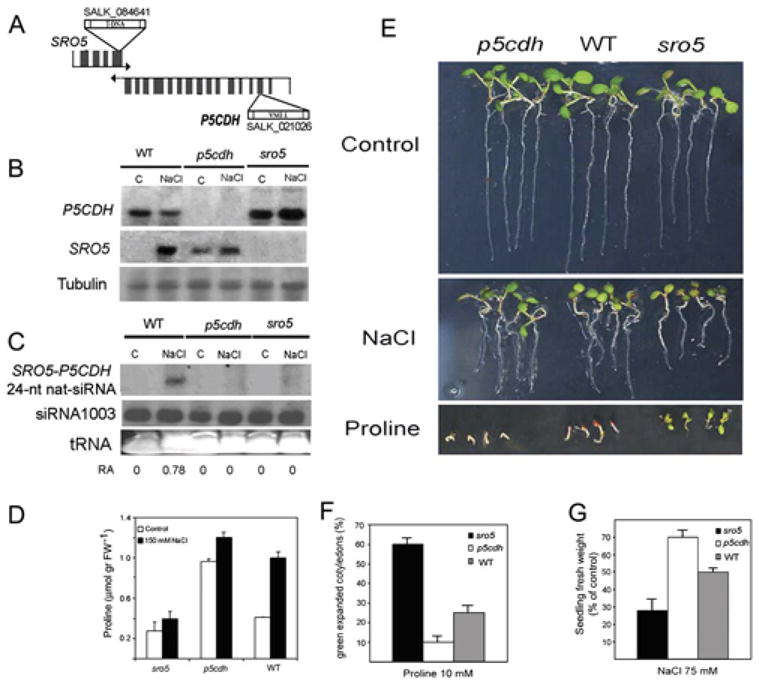
(A) Diagram of T-DNA insertion mutants isolated.
(B) Northern blot of P5CDH and SRO5 mRNAs in wild-type, p5cdh, and sro5 T-DNA lines. RNA (20 μg per lane) from 10-day-old seedling with or without 6 hr exposure to 150 mM NaCl were probed with P5CDH 5′ probe or SRO5 probe.
(C) Northern blot of small RNAs. Eighty micrograms of low-molecular-weight RNA extracted from the same samples as in (B) was blotted and probed with an oligoprobe complementary to the 24-nt SRO5-P5CDH nat-siRNA. The number below the blot indicates relative abundance (RA) of the nat-siRNA with respect to the loading control siRNA 1003.
(D) Proline accumulation in wild-type, p5cdh, and sro5 T-DNA lines after 6 hr exposure to 150 mM NaCl. Data are means ± SD (n = 10).
(E) Phenotypes of seedlings grown on 75 mM NaCl or 5 mM proline for 7 days.
(F) Proline response quantified as the percentage of seedlings with green expanded cotyledons. Data are means ± SD (n = 10).
(G) NaCl response quantified as seedling fresh weight relative to control not treated with NaCl. Data are means ± SD (n = 10).
The p5cdh mutant accumulated more proline than wild-type under control conditions and slightly more under salt stress (Figure 5D). The mutant was slightly more tolerant to NaCl stress under our conditions, which is possibly related to its higher proline accumulation (Figures 5E and 5F). As reported previously (Deuschle et al., 2004), the p5cdh mutant was more sensitive than wild-type to exogenous proline (Figures 5E and 5F). In contrast, the sro5 mutant accumulated less proline under salt stress (Figure 5D) and was more sensitive to NaCl stress (Figures 5E and 5G) and more tolerant of exogenous proline (Figures 5E and 5F). These phenotypes are all consistent with the increased expression of P5CDH in sro5.
Altered ROS Sensitivity and Accumulation in sro5 Indicates a Functional Link between the SRO5 and P5CDH Proteins
Our results suggest that nat-siRNA regulation of P5CDH is dependent on the SRO5 mRNA. However, SRO5 is a coding gene, and we hypothesized that the SRO5 protein may also be functionally related to P5CDH. It has been shown that P5CDH downregulation leads to the accumulation of its substrate, P5C (Deuschle et al., 2004). P5C itself or glutamate semialdheyde, which is in spontaneous equilibrium with P5C, causes the accumulation of high levels of ROS (Deuschle et al., 2001; Nomura and Takagi, 2004). Therefore, the SRO5 protein may function in counteracting this ROS accumulation in a manner that balances the effects of reduced P5CDH activity.
We found that sro5 plants were more sensitive to H2O2-mediated oxidative stress (Figure 6A). Also, there was substantially more accumulation of ROS, particularly H2O2, in salt-stressed sro5 and p5cdh seedlings (Figures 6B and 6C). The increased ROS level in p5cdh under salt stress was expected based on previous observations that P5C can cause ROS accumulation (Deuschle et al., 2001; Nomura and Takagi, 2004). The observation that sro5 had even greater ROS accumulation and sensitivity than p5cdh suggests a role for SRO5 protein in counteracting the increased ROS production caused by decreased P5CDH activity, either by blocking ROS production or increasing ROS detoxification.
Figure 6. SRO5 Is Required for Oxidative Stress Response.
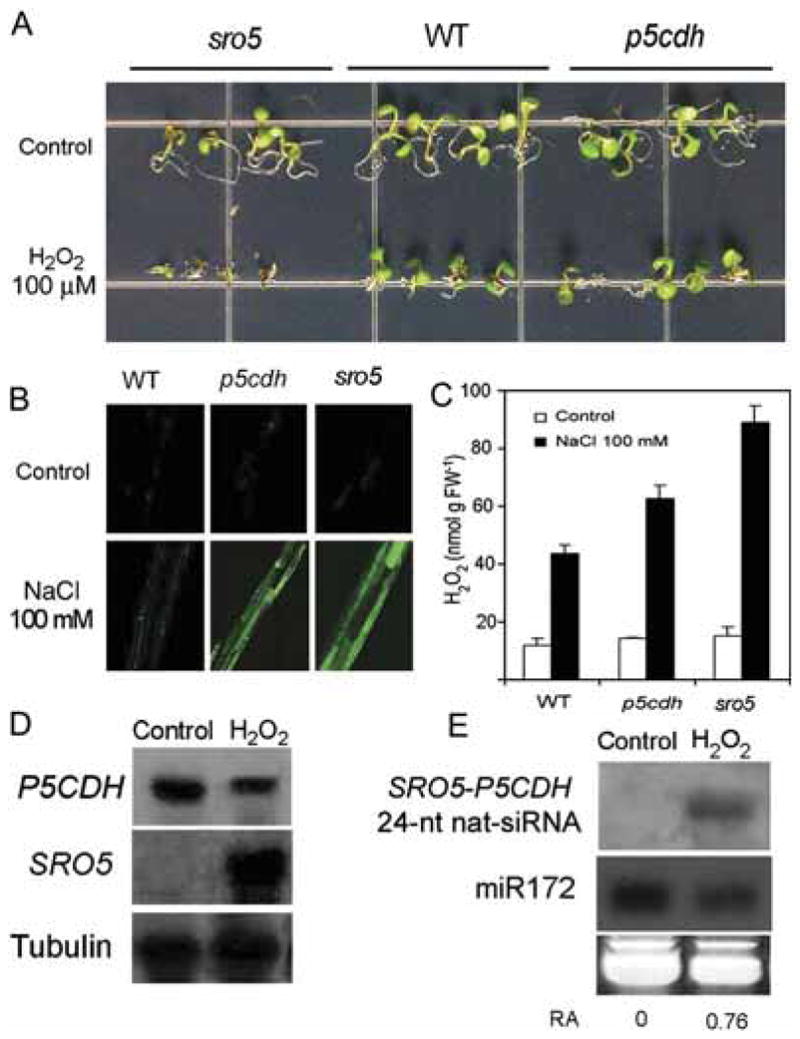
(A) Hypersensitivity of sro5 mutant to oxidative stress induced by 100 μM exogenous H2O2.
(B) ROS staining in seedling roots after exposure to 100 mM NaCl for 12 hr.
(C) H2O2 accumulation quantified in seedlings exposed to 100 mM NaCl for 12 hr. Data are means ± SD (n = 10).
(D) Regulation of P5CDH and SRO5 mRNAs by oxidative stress. Seedlings were treated or not treated (control) with 6 hr exposure to 10 mM H2O2.
(E) Accumulation of 24-nt SRO5-P5CDH nat-siRNA is induced by H2O2. Small RNA (100 μg per lane) from the same tissues used in (D) were probed with an oligonucleotide complementary to the 24-nt SRO5-P5CDH nat-siRNA. The number below the blot indicates relative abundance (RA) of the nat-siRNA with respect to the loading control miR172.
A role for the SRO5 protein in ROS regulation is supported by its intracellular localization. SRO5 is predicted to be a mitochondrial protein, and we confirmed this by observing that transiently expressed SRO5-YFP fusion protein is targeted to the mitochondria (Figure S1). Proline catabolism occurs in the mitochondria, and P5CDH is localized on the matrix side of the inner mitochondrial membrane (Deuschle et al., 2001; Nomura and Takagi, 2004). Also, SRO5 shares partial sequence homology to RADICLE INDUCED CELL DEATH 1 (RCD1). RCD1 is involved in controlling ROS-induced cell death, and rcd1 plants are hypersensitive to ozone, which causes oxidative stress (Ahlfors et al., 2004). Further support for a role of SRO5 protein will require demonstration that an untranslatable SRO5 mRNA can cause P5CDH cleavage but cannot suppress ROS accumulation.
Salt treatment causes oxidative stress, and we also found that application of H2O2 induced the expression of SRO5, induced the 24-nt SRO5-P5CDH nat-siRNA, and decreased P5CDH transcript levels (Figures 6D and 6E). The result suggests that the salt-stress induction of SRO5 and SRO5-P5CDH nat-siRNA formation might be mediated by increased ROS under salt stress.
DISCUSSION
A Fine-Tuned Biological Regulatory Circuitry Involving nat-siRNAs
We have discovered a class of siRNAs, which we call nat-siRNAs, that are central components of a stress regulatory circuit (Figure 7). In this circuit, salt stress induces the expression of SRO5 mRNA, and together with the P5CDH mRNA already present, the SRO5 mRNA forms dsRNA that is processed by DCL2 into 24-nt nat-siRNAs. The accumulation of detectable levels of the 24-nt SRO5-P5CDH nat-siRNAs also requires the presumably amplifying activity of the type IV RNA polymerase NRPD1A (Onodera et al., 2005; Herr et al., 2005), the RNA-dependent RNA polymerase RDR6, and SGS3, which is functionally coupled with RDR6 (Peragine et al., 2004). One of the 24-nt SRO5-P5CDH nat-siRNAs is stable and targets the P5CDH mRNA for cleavage. Initial cleavage of P5CDH mRNA guided by this 24-nt SRO5-P5CDH nat-siRNA establishes a phase for subsequent production of 21-nt nat-siRNAs by DCL1 and further degradation of P5CDH mRNA. The downregulation of P5CDH reduces proline degradation, thereby enhancing proline accumulation, which is beneficial to salt-stress tolerance. However, reduced P5CDH activity also leads to the accumulation of the toxic metabolic intermediate P5C and ROS accumulation, which is probably counteracted by the SRO5 protein either through a direct detoxification activity or regulatory mechanism. The initial induction of SRO5 mRNA by salt stress may be mediated by oxidative stress that is an inevitable consequence of salt stress. We propose that the insights gained from this characterization of the P5CDH-SRO5 cis-antisense gene pair will be useful in the study of the many other such gene pairs present in eukaryotic genomes.
Figure 7. Diagram of Phased Processing of SRO5-P5CDH nat-siRNAs and Their Role in a Salt-Stress Regulatory Loop.
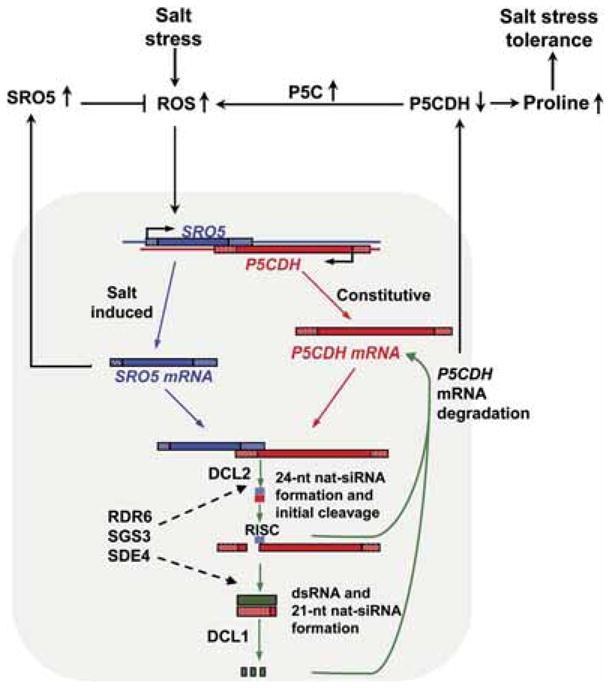
Salt stress induces ROS accumulation which in turn induces the expression of SRO5. This initiates a series of nat-siRNA processing steps (shown in the shaded area of the model) that result in the downregulation of P5CDH. When SRO5 is induced, dsRNA can form by annealing of the SRO5 and P5CDH transcripts (note that poly-A tails of the mRNAs are omitted for clarity), and this initiates DCL2-dependent formation of 24-nt nat-siRNA. This initial cleavage of the P5CDH mRNA causes phased generation of 21-nt nat-siRNAs by a DCL1-dependent mechanism and additional cleavage of the P5CDH transcript. RDR6, SGS3, and NRPD1A are suggested to be involved in the formation of both 24- and 21-nt SRO5-P5CDH nat-siRNAs. The importance of P5CDH downregulation by nat-siRNAs is further illustrated by the functional relationship between the SRO5 and P5CDH proteins (portion of model outside the shaded area). Degradation of P5CDH mRNA decreases P5CDH activity and proline degradation. Decreased proline degradation contributes to salt-induced proline accumulation and salt tolerance but, at the same time, leads to the ROS accumulation caused by buildup of the proline catabolic intermediate P5C. This ROS accumulation is potentially harmful but is also a signal to initiate further stress responses. The SRO5 protein counteracts P5C-induced ROS accumulation, thus completing a regulatory loop that allows fine tuning of ROS accumulation and stress response.
Phased Processing of nat-siRNAs
siRNAs are extremely diverse, and their biogenesis is especially complex in plants (Xie et al., 2004; Baulcombe, 2004; Meister and Tuschl, 2004). siRNAs from transposons and other repeat sequences are important for RNA-dependent DNA methylation and heterochromatin formation (Zilberman et al., 2004; Xie et al., 2004). Biogenesis of these siRNAs requires NRPD1A, RDR2, and DCL3 (Xie et al., 2004). The tasiRNAs that target specific mRNAs for cleavage require RDR6, SGS3, DCL1, and DCL4 (Peragine et al., 2004, Vazquez et al., 2004b; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). In contrast, our data suggest that the nat-siRNAs described here are a unique class in which the 24- and 21-nt nat-siRNAs are generated by DCL2 and DCL1, respectively.
Unexpectedly, the production of the 24-nt SRO5-P5CDH nat-siRNA is also dependent on the atypical DNA-dependent RNA polymerase NRDP1A. The fact that formation of the nat-siRNAs but not P5CDH or SRO5 transcript accumulation is dependent on NRPD1A suggests a DNA-independent function of this mysterious protein. The role of RDR6 is also intriguing since it is required not just for the formation of 21-nt nat-siRNAs, where the need to generate a new RNA strand is clear, but, also for the generation of the 24-nt SRO5-P5CDH nat-siRNA, where dsRNA from the SRO5-P5CDH cis-antisense transcripts should already be present. Perhaps RDR6 is required for subsequent amplification of the 24-nt SRO5-P5CDH nat-siRNA generated from the initial antisense transcript pair.
Our results show that initial cleavage of P5CDH mRNA guided by this 24-nt SRO5-P5CDH nat-siRNA sets the phase for accurate processing by DCL1 to yield the 21-nt P5CDH nat-siRNAs. Production of these 21-nt nat-siRNAs is also dependent on RDR6, SGS3, and NRPD1A. It is possible that this dependency is due to the phasing requirement for the 24-nt nat-siRNA. Alternatively, these components may be needed to process the cleaved P5CDH mRNA fragment into dsRNA that is then competent for dicing by DCL1. In the absence of 21-nt nat-siRNA formation, as in dcl1, the 24-nt SRO5-P5CDH nat-siRNA is sufficient to downregulate the P5CDH transcript under the salt-stress conditions used in our experiments. It may be that 21-nt nat-siRNA- guided degradation of P5CDH mRNA serves to ensure P5CDH downregulation. Nonetheless, future investigations to determine whether the 21-nt nat-siRNAs might have additional functions are needed.
Another important question that arises from these experiments is that of how a specific 24-nt SRO5-P5CDH nat-siRNA is initially generated from the relatively long (760-nt) P5CDH-SRO5 overlapping region. This situation may be analogous to the generation of a single, precise miRNA from a hairpin precursor with a long dsRNA-like stem. Alternatively, many different 24-nt siRNAs may be produced from the long overlap, but only one is stable and incorporated into the RISC to guide subsequent mRNA cleavage.
Implications for the Regulation and Function of Genes in Natural cis-Antisense Pairs
Our proposed regulatory model (Figure 7) has implications beyond the regulation of P5CDH and SRO5. In both animals and plants, genome analyses have found thousands of genes in convergent overlapping pairs that can generate complementary transcripts (Boi et al., 2004, Wang et al., 2005; Jen et al., 2005). Various expression profiling approaches, such as genome tiling arrays (Yamada et al., 2003; Bertone et al., 2004) and MPSS (massively parallel signature sequencing; Brenner et al., 2000), have detected widespread antisense transcription throughout genomes. As illustrated by the SRO5-P5CDH example here, these antisense transcript pairs can form dsRNAs that are processed by a Dicer into nat-siRNAs. In the SRO5-P5CDH example, one of the overlapping genes (SRO5) is inducible. This may be a widespread scenario as our preliminary analysis indicates that most of the nearly 2000 genes in convergent overlapping pairs in Arabidopsis are regulated by various environmental or hormonal stimuli (T. Girke and J.-K.Z., unpublished data). Our preliminary survey has detected several other nat-siRNAs from cis-antisense gene pairs, and all of these nat-siRNAs are inducible and detected only under specific abiotic or biotic stress-treatment conditions (our unpublished data). Consistent with their inducible nature, these nat-siRNAs including the nat-siRNAs from the SRO5-P5CDH pair were not identified by recent deep small RNA profiling by the MPSS method, which was carried out with non-stressed plants (Lu et al., 2005). Whether or not these induced nat-siRNAs also downregulate the corresponding noninducible overlapping gene by mRNA cleavage is an important topic for further investigation.
Jen et al. (2005) have argued against dsRNA- or siRNA-mediated RNA cleavage as a mode of regulation of antisense overlapping gene pairs. Their proposal is based largely on gene expression data from DNA microarrays that showed apparent coexpression of cis-antisense gene pairs. We believe that such microarray data offers only an incomplete picture of the regulation of overlapping genes because it may not distinguish full-length transcripts from stable 3′ cleavage products. Thus, we think that the proportion of cis-antisense gene pairs that are genuinely coexpressed remains an open question.
The SRO5-P5CDH example implies a functional link between proteins encoded by antisense overlapping genes such that regulating the two genes in a converse manner is beneficial. Whether or not such a functional link also exists between other pairs of antisense overlapping genes remains to be determined. In most cases, only one gene in the antisense overlapping pair has a defined function or contains known domains; thus, the possibility of a functional link between proteins encoded by overlapping genes offers an approach to identify the physiological function of a number of unknown genes.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Arabidopsis thaliana mutants, rdr2-1, dcl2-1, dcl3-1, and dcl4-1, were a generous gift of Jim Carrington (Center for Gene Research and Biotechnology, Oregon State University). dcl1-9 and hen1-1 were provided by Xuemei Chen. sde1 (rdr6 in this study) and sde4/nrpd1a were kindly provided by David Baulcombe (John Innes Center for Plant Science Research, Sainsbury Laboratory, United Kingdom). sgs3 was kindly provided by Herve Vaucheret (Laboratoire de Biologie Cellulaire, Institut National de la Recherche Agronomique, Versailles, France). hyl1 was a gift from Nina Federoff (The Huck Institute of Life Science, Pennsylvania State University). These mutants were in the Columbia (Col-0), Landsberg erecta (Ler), Nosssen-0 (No), or C24 genetic backgrounds as indicated in the text and figures. T-DNA insertion mutants of P5CDH (salk_021026) and SRO5 (salk_084641) were identified using the SIGnAL website (http://signal.salk.edu) and obtained from ABRC.
Seedlings were grown in half-strength MS (3% w/v sucrose) agar plates under 16/8 hr light/dark cycle of fluorescent light (120 μE/min/m2) at 22°C. After 10 days, the seedlings were treated by immersing the roots in half-strength MS liquid media (3% w/v sucrose) with or without the addition of salt, osmotic agents (PEG, mannitiol), or hormones, at concentrations indicated in the figure legends. Tissue was collected 6 hr after the treatment. In the experiments with adult plants, 20-day-old plants grown in pots and kept in a growth chamber with a 16/8 hr light/dark cycle at 22°C were used.
Tolerance of salt or exogenous proline was assessed by sowing seeds on half-strength MS agar plates (3% sucrose) containing the concentrations of proline indicated in the text or figure legends. After 7 days, the seedling response (survival or fresh weight) was quantified. For p5cdh and sro5 mutants, the seeds were pre-soaked in MS solution containing 1mM proline for 48 hr before they were subjected to the above tolerance assays.
RNA Analysis
Total RNA was extracted from 10-day-old seedlings with Trizol reagent (Invitrogen). Twenty micrograms of total RNA was separated on 1.2% formaldehyde-MOPS agarose gels and blotted onto Hybond-N+ membranes (Amersham Biosciences). Hybridization was carried out at 65°C using PerfectHyb Plus buffer (Sigma). Probes were labeled with 32P-dCTP using a Ready-To-Go DNA Labeling Kit (Amersham Biosciences). Blots were washed twice in 2× SSC and 0.1% SDS for 20 min at 65°C and once in 1× SSC 0.1% SDS.
For detection of low-molecular-weight RNA, total RNA was purified from at least 1 gram of seedlings using hot-phenol extraction followed by two extractions of phenol/chloroform/isoamyl alcohol (25/24/1). Total RNA were precipitated with ethanol and pellet was dissolved in 750 microliter DEPC-water. High molecular weight RNA were selectively precipitated from the total RNA by the addition of one volume of 20% PEG-1M NaCl (Llave et al., 2002). The resulting low-molecular-weight-enriched RNA (80–100 μg) was then separated by 17% denaturing polyacrylamide gels and electrically transferred to Hybond-N+ membranes. Low-molecular-weight RNA blots were probed with miR172, siRNA1003, and 24 and 21-nt nat-siRNA DNA oligonucleotides complementary to small RNA sequences. The probes were end-labeled with γ32P-ATP using T4 kinase (NEB). Unincorporated nucleotides were removed using G-25 spin columns (Amersham) as per manufacturer’s instructions. Blots were prehybridized for at least 1 hr and hybridized overnight at 38°C using PerfectHyb Plus buffer (Sigma). The blots were washed twice in 2× SSC and 0.2% SDS for 15 min at 38°C and once with 1× SSC 0.1% SDS for 15 min at 38°C. Membranes were exposed to X-ray films and to Typhoon phosphoimager and relative abundance (RA) was estimated using Image Quant software 5.2.
Transient Expression in Nicotiana benthamiana
Site-directed mutagenesis was performed to generate P5CDH and SRO5 genes (mP5CDH and mSRO) mutated in the region complementary to the 24-nt SRO5-P5CDH nat-siRNA. Both mutant and wild-type P5CDH and SRO5 were incorporated into the binary vector pMDC32 with expression under control of the 35S promoter. These constructs were transformed into Agrobacterium tumefaciens strain 3301. Overnight cultures grown in presence of 30 μM acetosyringone were harvested by centrifugation, and cells resuspended in 10 mM MgCl2, 10 mM (pH 5.6), and 150 μM acetosyringone to an OD600 of 0.5. After 2 hr incubation at room temperature, Agrobacterium suspension was infiltrated into expanding leaves of N. benthamiana using a needleless syringe (Llave et al., 2002). Leaves were harvested 3 days after infiltration and small RNA extraction and blotting performed as described above.
RLM 5′ RACE
A modified procedure for RNA ligase-mediated rapid amplification of cDNA ends (RLM 5′RACE) was performed using the GeneRacer Kit (Invitrogen). Total RNA was extracted from 10-day-old seedlings using the TRIZOL method described above. The GeneRacer RNA Oligo adaptor was directly ligated to total RNA (100 ng) without calf intestinal phosphatase treatment. The GeneRacer oligo dT primer was then used for cDNA synthesis. Initial PCR was carried out using the GeneRacer 5′ Primer and P5CDH RACE primer 1 (5′-CCCTCTACCCCTTGACTAAGTCAACG-3′), following the protocol of Llave et al. (2002). Nested PCR was carried out using 1 μl of the initial PCR reaction, the GeneRace 5′ nested primer, and P5CDH RACE primer 2 (5′-CTTATTCCCTCACACGGAAACTACTCG-3′). RACE fragments were cloned and sequenced after gel purification.
Proline and ROS Determination
Proline was quantified using the acid ninhydrin assay (Deuschle et al., 2004). For ROS detection, 7-day-old seedlings were transferred to media containing 100 mM NaCl for 12 hr. The salt-treated and untreated control seedlings were then incubated with 50 μM 5-(and -6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA, Molecular Probes) for 30 min following the methods of Shin and Schachtman (2004). Fluorescence images were obtained with a Leica SP2 confocal microscope.
For quantitative measurement of H2O2 production, 7-day-old seedlings were transferred to media with and without 100 mM NaCl and incubated for 12 hr. Salt-treated and control seedlings were extracted using 20 mM sodium phosphate buffer (pH 6.5) according to the protocol of Shin and Schachtman (2004) and H2O2 quantified using an Amplex red hydrogen/peroxidase assay kit (Molecular Probes) following the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We are grateful to Shou-Wei Ding, Xuemei Chen, and Hailing Jin for helpful suggestions and stimulating discussion and to David Baulcombe, Jim Carrington, Xuemei Chen, Nina Federoff, and Herve Vaucheret for providing seeds of various mutants. We also thank Rebecca Stevenson, Mingtang Xie, and Pradeep Jain for excellent technical assistance and Juan Díaz Pendón for invaluable assistance with the N. benthamiana transient expression experiments. This work was supported by National Institutes of Health grants R01GM59138 and R01GM0707501 and National Science Foundation grant IBN-0212346 to J.-K.Z.
Footnotes
Supplemental data include one figure, two tables, and references and are available with this article online at http://www.cell.com/cgi/content/full/123/7/1279/DC1/.
References
- Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Taurianinen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, et al. Arabidopsis radical-induced cell death1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925– 1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie ZX, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie ZX, Gustafson AM, Carrington JC. micro- RNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Roberts JK, Mitchell HJ, Zhang R, Lawrence GJ, Ellis JG, Pryor TJ. A plant gene up-regulated at rust infection sites. Plant Physiol. 2002;129:169–180. doi: 10.1104/pp.010940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Boi S, Solda G, Tenchini ML. Shedding light on the dark side of the genome: overlapping genes in higher eukaryotes. Curr Genomics. 2004;5:509–524. [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo SJ, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Hellmann H, Daschner K, Binder S, Frommer WB. A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001;27:345–356. doi: 10.1046/j.1365-313x.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, Frommer WB. The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell. 2004;16:3413–3425. doi: 10.1105/tpc.104.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 2000;123:779–789. doi: 10.1104/pp.123.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Iyer S, Caplan A. Products of proline catabolism can induce osmotically regulated genes in rice. Plant Physiol. 1998;116:203–211. [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 2005;6:R51. doi: 10.1186/gb-2005-6-6-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and rniRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and posttranscriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nomura M, Takagi H. Role of the yeast acetyltransferase Mpr1 in oxidative stress: Regulation of oxygen reactive species caused by a toxic proline catabolism intermediate. Proc Natl Acad Sci USA. 2004;101:12616–12621. doi: 10.1073/pnas.0403349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu XL, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by micro- RNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Park W, Li JJ, Song RT, Messing J, Chen XM. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICERLIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004a;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004b;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZX, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZX, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen HM, Shinn P, Palm CJ, Southwick AM, Wu AC, Kim C, Nguyen M, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.