Abstract
Epigenetic states are responsive to developmental and environmental signals, and as a consequence a eukaryotic cell can have many different epigenomes. In this issue of Cell, Lister et al. (2008) present the floral epigenome of Arabidopsis using next-generation sequencing technology to analyze both DNA methylation at single-base resolution and the expression of small RNAs.
In eukaryotic cells, gene activity is controlled not only by DNA sequences but also by epigenetic marks, which can be transmitted to a cell’s progeny during mitosis or meiosis. Although epigenetic regulation is generally mediated by histone modifications, histone variants, and DNA cytosine methylation, it also involves the production and action of noncoding RNAs, particularly small RNAs (Bernstein et al., 2007; Huettel et al., 2006; Henderson and Jacobsen, 2007). An essential step in understanding epigenetic regulation is to have genome-wide, high-resolution maps of these epigenetic features. Just as the study of genetics has been revolutionized by the ability to sequence entire eukaryotic genomes, next-generation sequencing technologies are now making it possible to determine the genome-wide distributions of methylated DNA, histone variants, and histone modifications. In short, epigenomes can now be sequenced. In this issue, Ecker and colleagues (Lister et al., 2008) present a genome-wide map of DNA methylation at single-base resolution in developing floral tissue of the model plant Arabidopsis. Integrating this map with an analysis of the floral transcriptome and small RNA profiles reveals new insights into the global interplay of DNA methylation, small RNAs, and transcription.
Lister and colleagues used bisulfite sequencing to determine the methylome of Arabidopsis floral tissues. In a complementary paper in Nature, Jacobsen and coworkers (Cokus et al., 2008) independently used the same technology to sequence the methylome of adult Arabidopsis plants. Like previous studies that mapped methylation in Arabidopsis using microarrays (Zhang et al., 2006; Zilberman et al., 2007), these new studies find extensive DNA methylation throughout the genome. They show that methylation is high in heterochromatic regions, dispersed in euchromatic regions, and prevalent in the body of genes. The mapping of DNA methylation at single-base resolution also reveals that local sequence context has a strong effect on cytosine methylation. The results have important implications for clarifying the mechanisms of action and characteristics of DNA methyltransferase enzymes, and also contribute to a better understanding of the evolution of gene promoters and other regulatory sequences.
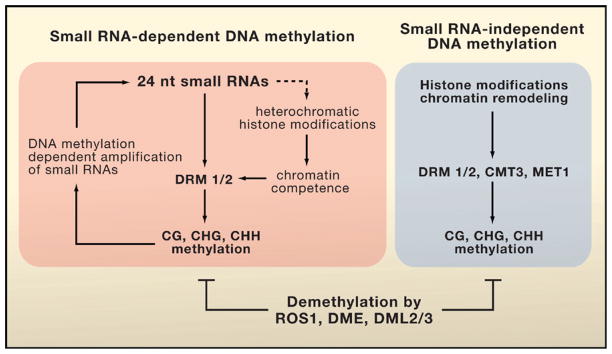
Both of the new studies compared the methylomes of wild-type plants to those of plants lacking individual DNA methyltransferases (DNMTs). De novo methylation in Arabidopsis is carried out by the methyltransferases DRM1 and DRM2, which are orthologs of the DNMT3 family in mammals. In contrast, maintenance of CG and CHG (where H is C, T, or A) methylation is mediated by the DNMT1-like enzyme MET1 and the plant-specific enzyme CMT3, respectively (Henderson and Jacobsen, 2007). Because it is presumed that asymmetric CHH methylation cannot be maintained during DNA replication (Huettel et al., 2006), the de novo methyltransferases DRM1 and DRM2 are expected to be the main (or sole) enzymes responsible for CHH methylation. The new studies, however, show that met1 and cmt3 loss-of-function mutations have even greater effects in reducing the overall level of CHH methylation in the genome than does the loss of both drm1 and drm2. This result suggests that MET1 and CMT3 may also be important for de novo methylation at many genomic regions (Figure 1). In addition, maintenance of methylation at CG and CHG sites may be required for efficient CHH methylation by DRM1 and DRM2.
Figure 1. DNA Methyltransferases and Demethylases in Arabidopsis.
DNA methyltransferases can be targeted by both small RNA-dependent and RNA-independent pathways to achieve methylation at CG or non-CG sites. For some loci, DNA demethylases actively remove methylation to prevent silencing. Small RNA production is controlled by DNA methylation in a self-reinforcing positive feedback loop. The dotted line indicates the possibility that small RNAs may direct heterochromatic histone modifications.
Lister et al. also sequenced the transcriptome of Arabidopsis floral tissues, including an extensive analysis of small RNA populations. Comparing this information with the DNA methylome revealed that sequences matching small RNAs are 25 times more likely to be methylated than sequences without small RNA matches. This striking correlation between small RNAs and DNA methylation supports the notion that RNA-directed DNA methylation occurs genome wide (Huettel et al., 2006). RNA-directed DNA methylation was first discovered in plants more than a decade ago and involves a pathway that generates 24 nucleotide small RNAs that are then bound to Argonaute 4 (AGO4). RNA-directed DNA methylation requires the chromatin remodeling factor DRD1 and DRM1/DRM2 to direct the methylation of corresponding genomic DNA (Huettel et al., 2006) (Figure 1). Interestingly, Lister et al. found a tendency for the sense strand (relative to the small RNA) to be methylated. An attractive model to explain strand-specific methylation is that nascent transcripts from the sense strand may hybridize with the small RNAs, thus bringing the effector complex for RNA-directed DNA methylation to the sense strand for methylation.
Yet small RNAs are associated with only one-third of methylated loci (Lister et al., 2008). Thus, the remaining two-thirds of genomic cytosine methylation must be directed by mechanisms that are independent of small RNAs (Figure 1). Some small RNA-independent methylation corresponds to gene body methylation (where methylation is concentrated within the gene rather than at the ends of a gene). The function of gene body methylation remains enigmatic because its loss in the met1 mutant does not substantially affect transcript levels (Zhang et al., 2006; Lister et al., 2008). However, loss of gene body CG methylation in the met1 mutant is often compensated for by an increase in CHG methylation (Cokus et al., 2008; Lister et al., 2008), suggesting that maintaining gene body methylation is important. Perhaps gene body methylation helps to inhibit cryptic transcription initiation (Zilberman et al., 2007). Alternatively, it might suppress recombination or transposon insertion within genes. For small RNA-independent DNA methylation, DNA methyl-transferase enzymes may be directed to the target sequences by specific histone modifications or by chromatin remodeling.
Although DNA methylation is a relatively stable epigenetic modification, it can be very dynamic and is subject to active demethylation (Zhu et al., 2007). In principle, patterns of DNA methylation can result from the targeted action of methyltransferases or indiscriminate methyltransferase activity that is later reset or trimmed by demethylases (or a combination of these two mechanisms). Whereas the identities of DNA demethylases remain elusive in mammals, in Arabidopsis ROS1 and related DNA glycosylases function as locus-specific demethylases through a base-excision repair pathway (Gong et al., 2002; Zhu et al., 2007; Penterman et al., 2007). Lister et al. sequenced the floral DNA methylome and transcriptome in a mutant deficient in the demethylases. They found hundreds of discrete regions with increased DNA methylation in this mutant, further supporting a crucial role of the demethylases in pruning DNA methylation in transposons and other loci and thereby upholding their expression levels (Zhu et al., 2007). Intriguingly, the methylation levels at some other sites were decreased in the demethylase mutant. This result suggests that these large DNA glycosylase proteins with potential chromatin-interaction domains may be important for maintaining DNA methylation at some sites. Alternatively, the expression of some DNA methyltransferase genes may be reduced in the demethylase mutant as a compensatory response to hypermethylation at certain critical sites. Huettel et al. (2006) reported that in met1 and other mutants with DNA hypomethylation, ROS1 gene expression is significantly reduced. With these kinds of compensatory responses in mind, it is clear that not all of the altered DNA methylation patterns in the met1 mutant can be attributed directly to MET1 enzyme activity.
The pioneering work of Lister et al. and Cokus et al. provides valuable snapshots of portions of the epigenomes of Arabidopsis. The transcriptomes of an organism are continually changing in response to developmental and environmental cues. Similarly, the epigenome is not static and can be molded by developmental signals, environmental perturbations, and disease states. Therefore, many epigenomes will need to be sequenced for a single organism, making epigenome sequencing perhaps even more challenging than genome sequencing.
References
- Bernstein BE, Meissner A, Lander ES. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Cell. 2008 doi: 10.1016/j.cell.2008.03.029. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]