Abstract
Studies of mammalian splicing factors are often focused on small nuclear ribonucleoproteins or regulatory RNA-binding proteins, such as hnRNP (heterogeneous nuclear ribonucleoprotein) and SR proteins (serine/arginine-rich proteins); however, much less is known about the contribution of DExD/H-box proteins or RNA helicases in mammalian pre-mRNA splicing. The human DEAH-box protein DHX16 [also known as DBP2 (DEAD-box protein 2)], is homologous with Caenorhabditis elegans Mog-4, Schizosaccharomyces pombe Prp8 and Saccharomyces cerevisiae Prp2. In the present study, we show that DHX16 is required for pre-mRNA splicing after the formation of a pre-catalytic spliceosome. We found that anti-DHX16 antiserum inhibited the splicing reaction in vitro and the antibody immunoprecipitated pre-mRNA, splicing intermediates and spliceosomal small nuclear RNAs. Cells that expressed DHX16 that had a mutation in the helicase domain accumulated unspliced intron-containing minigene transcripts. Nuclear extracts isolated from the dominant-negative DHX16-G724N-expressing cells formed splicing complex B, but were impaired in splicing. Adding extracts containing DHX16-G724N or DHX16-S552L mutant proteins to HeLa cell nuclear extracts resulted in reduced splicing, indicating that the mutant protein directly inhibited splicing in vitro. Therefore our results show that DHX16 is needed for human pre-mRNA splicing at a step analogous to that mediated by the S. cerevisiae spliceosomal ATPase Prp2.
Keywords: DExD/H-box protein 16 (DHX16), pre-mRNA splicing, Saccharomyces cerevisiae Prp2, spliceosome
INTRODUCTION
Nuclear pre-mRNA splicing consists of two simple transesterification reactions, but requires one of the most complex macromolecular complexes, the spliceosome, to complete them [1]. The spliceosome is composed of five snRNAs (small nuclear RNAs) and nearly 100 proteins, many of which are either associated with the snRNAs or bound to the intron or exon sequences [2]. The dynamic interactions among the snRNAs, proteins and pre-mRNA substrate are mediated at least in part by DExD/H-box proteins, which are RNA helicases or RNA-dependent ATPases [3]. Although the role of DExD/H-box proteins has been characterized in yeast [4], much less is known about their contribution to human pre-mRNA splicing.
Eight DExD/H-box proteins are required for pre-mRNA splicing in Saccharomyces cerevisiae: Prp2, Prp5, Prp16, Prp22, Prp28, Prp43, Sub2 and Brr2 [5]. Their mammalian counterparts are called DDXs for DExD-box or DHXs for DExH-box proteins [6]. Proteins in this family have a well-conserved helicase domain, a conserved C-terminal domain and a divergent N-terminal domain [7]. Owing to the high conservation, assigning functional orthologues among different species based solely on sequence alignments can be difficult.
Human DHX16 was initially identified as DBP2 (DEAD-box protein 2) in a search for human DEAH-box RNA helicases [8]. The gene resides on chromosome 6p21.3 in the MHC region, which is linked to several malignant and autoimmune diseases. The helicase domain of DHX16 has >72% identity to Schizosaccharomyces pombe Cdc28/Prp8, a protein involved in cell-cycle progression and pre-mRNA splicing [9], and human DHX16 can rescue a temperature-sensitive S. pombe prp8-1 mutant, albeit inefficiently [8], supporting the idea that DHX16 is a functional homologue of Cdc28/Prp8. Another orthologue, Caenorhabditis elegans Mog-4, exhibits 53.9% identity with DHX16. Mog-4 plays a role in the hermaphrodite sperm/oocyte switch, and knockdown of Mog-4 is embryonic lethal [10]. A function in splicing has not been proposed for Mog-4, but Mog-4 is needed for a posttranscriptional repression of the fem-3 sex-determining gene through an interaction with the 3′-UTR (untranslated region) of fem-3 [11]. The S. cerevisiae orthologue for DHX16 is proposed to be Prp2 [10], as DHX16 is found in spliceosome preparations from HeLa cells [12–15]; however, a BLAST search on the S. cerevisiae database (http://www.yeastgenome.org) identified Prp22 as the likely orthologue, whereas the UniGene database (http://www.ncbi.nlm.nih.gov/UniGene) identified Prp43. Therefore the role of DHX16 in human pre-mRNA splicing still needs to be defined.
In the present study, we investigated the contribution of DHX16 in human pre-mRNA splicing. We used an anti-DHX16 antibody to show that DHX16 associates with the spliceosome, is needed for the first transesterification and did not completely dissociate from the spliceosome, even after the second transesterification. We generated several DHX16 helicase-domain-mutants and show that they inhibit splicing of minigene transcripts in vivo and in vitro. Our results indicate that DHX16 contributes to human pre-mRNA splicing before the first transesterification and shares characteristics with S. cerevisiae Prp2.
EXPERIMENTAL
Plasmids
The open reading frame of DHX16 was amplified from pBluescript II SK+ KIAA0577 [16] by PCR using high-fidelity Pfu-Turbo polymerase (Stratagene) with primers containing an EcoRI and an XhoI site (Supplementary Table S1 at http://www.BiochemJ.org/bj/429/bj4290025add.htm) and inserted into pCR (Invitrogen). The K428A, D520A, H523D, S552L and G724N mutations were introduced into DHX16 either by using a site-directed mutagenesis kit (Stratagene) or by following the protocol described by Kunkel et al. [17]. FLAG–DHX16 fusion proteins were constructed by inserting a FLAG tag (MDYKDDDDK) before the second codon of DHX16 via a PCR strategy. The FLAG–DHX16 fragments were cloned between the EcoRI and XhoI sites of pcDNA3.1 (Invitrogen). EGFP (enhanced green fluorescent protein)–DHX16 was made by inserting the EcoRI/XhoI fragment of DHX16 between the EcoRI and SalI sites of pEGFP-C2 (Clontech) such that EGFP was placed at the N-terminus of DHX16. All constructs were sequenced for verification. FLAG–DHX16-G724N (used in Figure 6) contained an additional E484A mutation outside the conserved helicase motif; however, the E484A mutation did not have an observable effect in the in vitro or in vivo splicing assays. pL53In contains a minigene with an intron (MoBiTec).
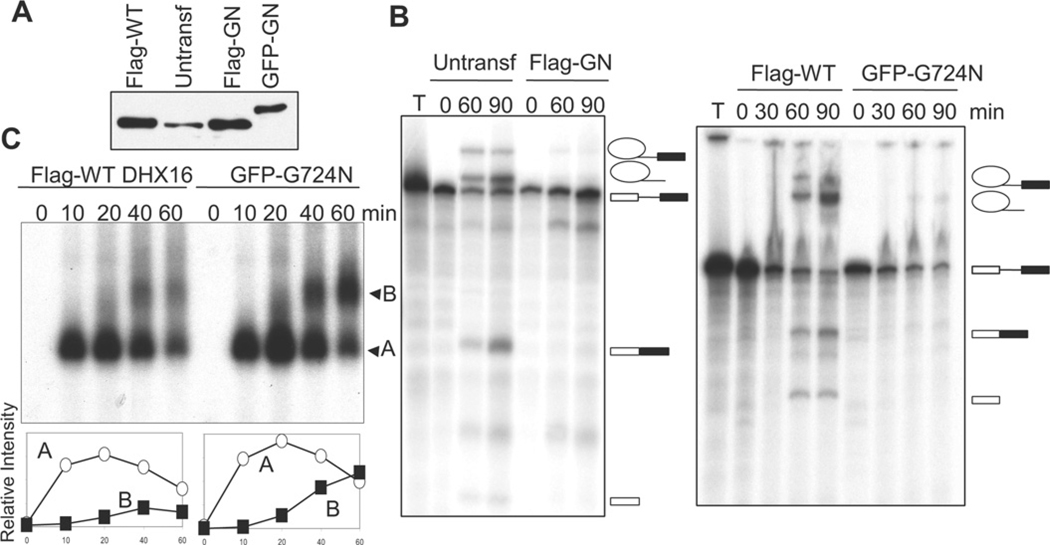
Figure 6. Nuclear extracts from mutant DHX16-expressing cells assemble spliceosomal complex B, but are defective in splicing in vitro.
Nuclear extracts were isolated from exponentially growing, untransfected HEK-293 cells (untransf), as well as from HEK-293 cells transfected for 72 h with FLAG-tagged wild-type DHX16 (Flag-WT), FLAG-tagged mutant DHX16-G724N (Flag-GN) or EGFP-tagged mutant DHX16-G724N (GFP-GN). (A) Western blot of equal volumes of the extracts probed with the anti-DHX16 antibody. (B) Splicing reactions were carried out with the indicated extracts and for the indicated times and the RNAs from the reactions were resolved on a polyacrylamide/urea gel. T, pre-mRNA substrate that was not incubated with extracts. Pre-mRNA substrate from pRG1 was used for the reactions shown in the left-hand panel and pre-mRNA from AdML was used for those in the right-hand panel. RNA molecules are indicated to the right of the blot and are as described for Figure 2(B). (C) Splicing reactions were carried out using AdML pre-mRNA substrate in extracts form FLAG-tagged wild-type-DHX16- or EGFP-tagged mutant-DHX16-G724N-expressing cells for the indicated times and resolved on a non-denaturing gel. Spliceosomal complexes A and B are indicated. The graphs below the autoradiograph show the amount of spliceosome complex at each time point; complex A (○); complex B (■).
Cell lines and transfection
HeLa, HEK (human embryonic kidney)-293 and HEK-293T cells [HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40)] were cultured in DMEM (Dulbecco’s modified Eagle’s medium; Irvine Scientific) supplemented with 10% (v/v) FBS (fetal bovine serum, HyClone) under a 5% CO2 atmosphere at 37°C. Cells (8 × 105) were transfected with 2–3 µg of plasmid DNA by Lipofectamine™ 2000-mediated transfection (Invitrogen) on a dish, or in suspension and then plated. Calcium phosphate-mediated transfection of HEK-293T cells (60–70% confluent) was performed on plates. Briefly, a 500 µl solution containing 250 mM CaCl2 and 20 µg of plasmid DNA was added dropwise with constant mixing to an equal volume of buffer (50 mM Hepes, pH 7.1, containing 280 mM NaCl and 1.5 mM Na2HPO4). After a 30 min incubation at room temperature (22°C), the mixture was added dropwise to the medium covering the cells and the medium was changed after 18 h.
Antibody production, Western blotting and immunostaining
A peptide (CPKKIGKTREELG) from the C-terminus of DHX16 was synthesized in the Peptide Synthesis Facility at City of Hope, conjugated to keyhole limpet haemocyanin (Pierce), and injected into two rabbits. The animal experiments were performed by certified personnel at the City of Hope Animal Resources Center following the guidelines approved by the Institutional Animal Care and Use Committee (IUCUC no. 93023). The antibody was precipitated from the rabbit serum in saturated ammonium sulfate and resuspended in PBS [18]. To purify the peptide-specific anti-DHX16 antibody, the precipitated antibody was applied to a column of Affi-Gel 10 (Bio-Rad Laboratories) conjugated to the peptide used for immunization. The bound antibody was eluted with 100 mM glycine, pH 3, collected in 1 M Tris/HCl, pH 8, and dialysed against PBS.
For total protein extraction, cells were lysed in RIPA buffer [10 mM sodium phosphate, pH 7.2, 150 mM NaCl, 1% (v/v) NP-40 (Nonidet P40), 1% (w/v) sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 1 mM DTT (dithiothreitol), 1 mM PMSF and 100 units/ml benzonase] or in TNE buffer [10 mM Tris/HCl, pH 8, 1% (v/v) NP-40, 150 mM NaCl and 1 mM EDTA] on ice for 20 min and cleared by centrifugation as described previously [19].
Protein lysates (10–20 µg) were resolved by SDS/PAGE (8% or 10% gels) and proteins were transferred on to PVDF membranes (Millipore). Antibodies used for Western blotting were: rabbit anti-DHX16, anti-TOPOII (topoisomerase type II)-β (Sigma–Aldrich), anti-tubulin (Sigma–Aldrich) and anti-FLAG–M2 (Sigma–Aldrich) antibody, and mouse anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Ambion) antibody. SuperSignal West Pico chemiluminescent substrate (Pierce) was used for detection of HRP (horseradish peroxidase)-conjugated secondary antibodies. To re-probe, the antibody from the first probing was stripped from the membrane in 0.2 M glycine/HCl, pH 2.5 and 0.1% SDS.
For immunostaining, HeLa cells were grown on coverslips and fixed with 4% (w/v) paraformaldehyde for 20 min on ice. After two washes in PBS, cells were incubated with the primary antibody for 1 h on ice. The anti-SC35 antibody was a mouse monoclonal antibody and the anti-DHX16 antibody was a rabbit polyclonal antibody. The secondary antibody used was anti-(mouse Ig)–Cy2 (carbocyamine) or anti-(rabbit-Ig)–Cy5 (indocarbocyamine). DAPI (4′,6-diamidino-2-phenylindole; 1 µg/ml) was added to the final PBS wash (for a 5 min duration). Images were collected via fluorescence microscopy.
Preparation of nuclear and cytoplasmic fractions
HeLa cells (1.2 × 107 cells) were harvested, washed with PBS, and resuspended in one PCV (packed cell volume) of buffer A (10 mM Hepes/KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl and 1 mM DTT) on ice for 15 min. The swollen cells were passed through a 25-gauge needle five times and the cell lysate was centrifuged at 20000 g for 20 s; the supernatant was saved as the cytoplasmic fraction. The pellet was washed with 1 PCV of buffer A, resuspended in a 2/3 PCV of buffer C [20 mM Hepes/KOH, pH 7.9, 25% (v/v) glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 420 mM NaCl, 0.5mM PMSF and 1 mM DTT] at 4°C for 30 min and centrifuged at 20000 g for 5 min; the supernatant was recovered as the nuclear fraction [20].
Whole-cell extract and nuclear extract for in vitro splicing assays
HEK-293T cells were transfected with plasmid DNA on 10-cm-diameter plates by calcium-phosphate-mediated transfection. At 36 h after transfection, cells were harvested (500 g for 1 min), washed in PBS and resuspended in 200 µl of ice-cold buffer E [20 mM Hepes/KOH, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% (v/v) glycerol and 1 mM DTT]. Cells were lysed by sonication for 20 s (in 5 s pulses followed by 30 s rests) at 30% amplitude in a Fisher Scientific Model 500 Ultrasonic Dismembrator. The lysate was centrifuged (20000 g for 20 min at 4°C) and the supernatant was saved as whole-cell extract [21].
Standard HeLa nuclear extracts were prepared from 5 × 109 HeLaS3 cells (National Cell Culture Center) as described previously [22,23]. For small scale extractions, 5 × 107 cells were transfected with 80 µg of plasmid using Lipofectamine™ 2000, harvested 72 h after transfection and subjected to nuclear extract preparation [20].
In vitro splicing and spliceosomal complex assays
BamHI-digested pRG1 (1.5 µg), encoding a modified MINX (miniature wild-type) pre-mRNA as described previously [24], was transcribed in a 25 µl reaction containing: 2 mM m7G(5′)ppp(5′)G cap analogue, 125 µM ATP, 125 µM CTP, 125 µM GTP, 10 µM UTP, 10 mM DTT, 20 units of SP6 RNA polymerase, 30 µCi of [α-32P]UTP (800 Ci/mmol) and buffer (New England Biolabs). BamHI-digested AdMLPar plasmid [25] was transcribed in: 2.0 mM m7G(5′)ppp(5′)G cap analogue, 0.5 mM ATP, 0.5 mM CTP, 0.1 mM GTP, 0.1 mM UTP, 10 mM DTT, 2 units/µl T7 polymerase, 0.4 units/µl RNasin, 1.2 µCi/µl [α-32P]UTP and buffer (New England Biolabs). Transcripts were subjected to electrophoresis on a 6% polyacrylamide/urea gel, eluted from the gel slice in 0.5 M NH4OAc, 0.1% SDS and 1 mM EDTA, and ethanol-precipitated with 10–20 µg of glycogen.
Labelled transcript (10000–20000 c.p.m.) was used in a 25-µl splicing reaction [26] containing: 40–50% extract, 0.5 mM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate, 20 mM Hepes/KOH, pH 7.6, 2.6% (v/v) polyvinyl alcohol and 0.4 units/µl RNasin at 30°C. The reaction was stopped by adding a solution of 0.3 M NaOAc, pH 5.2, containing 0.1% SDS. After phenol extraction, RNA was ethanol-precipitated with 10–20 µg of glycogen, resuspended and resolved on a 13% polyacrylamide/8 M urea gel (19:1). The gel was imaged by autoradiography or with a PhosphorImager (Typhoon 9000; GE Healthcare), and analysed with ImageQuant TL software (GE Healthcare).
For complex assays, splicing reactions were stopped by adding 0.1 volumes of 10× heparin loading dye [6.5 mg/ml heparin sulfate, 40% (v/v) glycerol, 0.5% Bromophenol Blue, 0.5% Xylene Cyanol, 1 × TBE (45 mM Tris/borate and 1 mM EDTA)] and resolved on a 2% low-melting agarose gel containing 50 mM Tris and 50 mM glycine for approx. 6 h at 70 V [27]. Gels were fixed, dried and imaged by autoradiography or with a PhosphorImager.
Antibody inhibition and immunodepletion of DHX16 from nuclear extract
To test the effect of the anti-DHX16 antibody on splicing, antiserum or pre-immune serum (16 µg of protein equivalent) was added to the splicing mixture without pre-mRNA and incubated at 30°C for 20 min. 32P-labelled pre-mRNA was then added to initiate the reaction. For the peptide-rescue experiment, 15 µg of antiserum or pre-immune serum was incubated at 30°C for 20 min with 600 ng of peptide before being added to the splicing mixture.
To deplete DHX16, 15 µl of anti-DHX16 antibody in TBS (Tris-buffered saline; 50 mM Tris/HCl, pH 7.5, containing 150 mM NaCl) plus 0.01% NP-40 and 0.5 mg/ml BSA was incubated for 2 h at 4°C with 25 µl of Protein A–Sepharose beads (Amersham Bioscience). The beads were then washed with TBS followed by buffer D [20 mM Hepes/KOH, pH 7.9, 20% (v/v) glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF and 1 mM DTT]. The antibody-bound beads were incubated for 2–3 h at 4°C with 100 µl of HeLa nuclear extract and then removed by centrifugation (1000 g for 1 min).
Immunoprecipitation and Northern blotting
Antibody or serum was incubated for 1 h at 4°C with Protein A–Sepharose (Sigma–Aldrich) and washed with immunoprecipitation buffer (20 mM Hepes, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT and 0.05% NP-40). An equal volume of heparin (1 mg/ml) was added to the splicing reaction mixtures (25 µl) and incubated for 5 min at 30°C. The mixture was incubated for a further 2 h at 4°C with 20 µl of Protein A–Sepharose beads coated with antibody, washed with immunoprecipitation buffer and eluted with peptide (50 µl of 0.6 mg/ml) at 4°C for 40 min. RNA was isolated from the eluate by phenol extraction and ethanol precipitation.
For Northern blot analysis, isolated RNA was separated on a 8% polyacrylamide/7.5 M urea gel and transferred on to membrane (NytranN; S&S) (1 mA/cm2, 90 min, in 12 mM Tris/HCl, pH 6.8, containing 6 mM NaOAc and 0.3 mM EDTA) [28] using a Trans-Blot transfer cell (Bio-Rad Laboratories). RNA on the membrane was hybridized at 65°C overnight with 32P-labelled antisense RNA of U1, U2, U4, U5 and U6 transcribed from plasmids pSPU1, pSPU2, pSPU4, pSPU5 and pSPU6 respectively by SP6 RNA polymerase. The membrane was washed with 0.1 × SSC (1 × SSC 0.15 M NaCl/0.015 M sodium citrate)/0.5% SDS at 65°C for 1 h and the signals were detected by autoradiography or with a PhosphorImager.
RNA isolation and RT (reverse transcription)–PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen). RNA was incubated for 15 min at 37°C with 5 units of DNase I (Roche) and then chloroform-extracted. cDNA was synthesized by RT of 5 µg of RNA using random hexamers (New England Biolabs) and Superscript II (Invitrogen). A 0.01 × volume of the RT reaction was used for PCR (95°C for 5 min, then 28–30 cycles of 20 s at 95°C, 20 s at 62°C and 30 s at 72°C, followed by a final extension at 72°C for 7 min) with the indicated primers (Supplementary Table S1).
RESULTS
DHX16 localizes in the nucleus
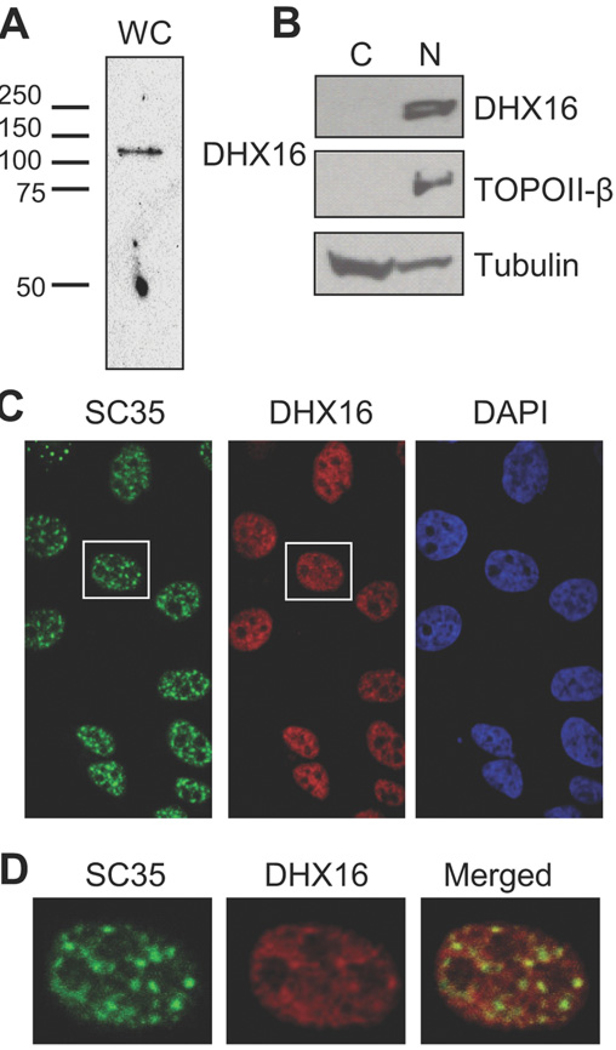
To determine the cellular localization of DHX16, we produced rabbit polyclonal antibodies against the DHX16 C-terminal peptide, PKKIGKTREELG. An approx. 120 kDa protein was detected by peptide-purified antibodies in Western blot analysis of whole cell lysates from HEK-293 cells (Figure 1A), consistent with the 119 kDa calculated molecular mass of DHX16. We then used the antibody to determine the cellular localization of DHX16 in cell fractions and fixed cells. We separated HeLa cell lysates into nuclear and cytoplasmic fractions and performed Western blot analysis. DHX16 was detected in the nuclear fraction, as was the nuclear protein DNA TOPOII-β, whereas tubulin, as expected, was found mostly in the cytoplasmic fraction (Figure 1B). We also stained fixed HeLa cells with antibodies against DHX16 and SR protein SC35, which is a splicing factor and widely used nuclear speckle marker; the anti-SC35 antibody revealed nuclear speckles, where many spliceosomal proteins localize, whereas anti-DHX16 staining was predominantly in the nuclei (Figure 1C). Further examination of the SC35 and DHX16 immunostaining revealed that the two signals overlapped, but the DHX16 staining was more broadly distributed in the nucleoplasm (Figure 1D). We also made an EGFP–DHX16 construct and found the fusion protein localized to the nucleus (results not shown), similar to the observations reported previously [8]. Therefore DHX16 appears to localize to the nucleoplasm and nuclear speckles, consistent with its proposed role in pre-mRNA splicing.
Figure 1. Cellular localization of DHX16.
(A) Western blot analysis of HEK-293 whole-cell lysates (WC) using a peptide-purified antibody against a C-terminal peptide of DHX16. Positions of molecular mass markers in kDa are indicated. (B) HeLa cells were fractionated into cytoplasmic (C) and nuclear (N) fractions, and a 3 × 106 cell-equivalent of each fraction was analysed by Western blotting using anti-DHX16, anti-TOPOII-β and anti-tubulin antibodies. (C) HeLa cells immunostained with anti-SC35 (green) and anti-DHX16 (red) antibodies, and DAPI (blue) for detection of nuclei. (D) Magnified images of the cell outlined by a white rectangle in (C), shown with anti-SC35 antibody staining (green), anti-DHX16 antibody staining (red) and the merged image.
Anti-DHX16 antibody inhibits pre-mRNA splicing in HeLa nuclear extracts
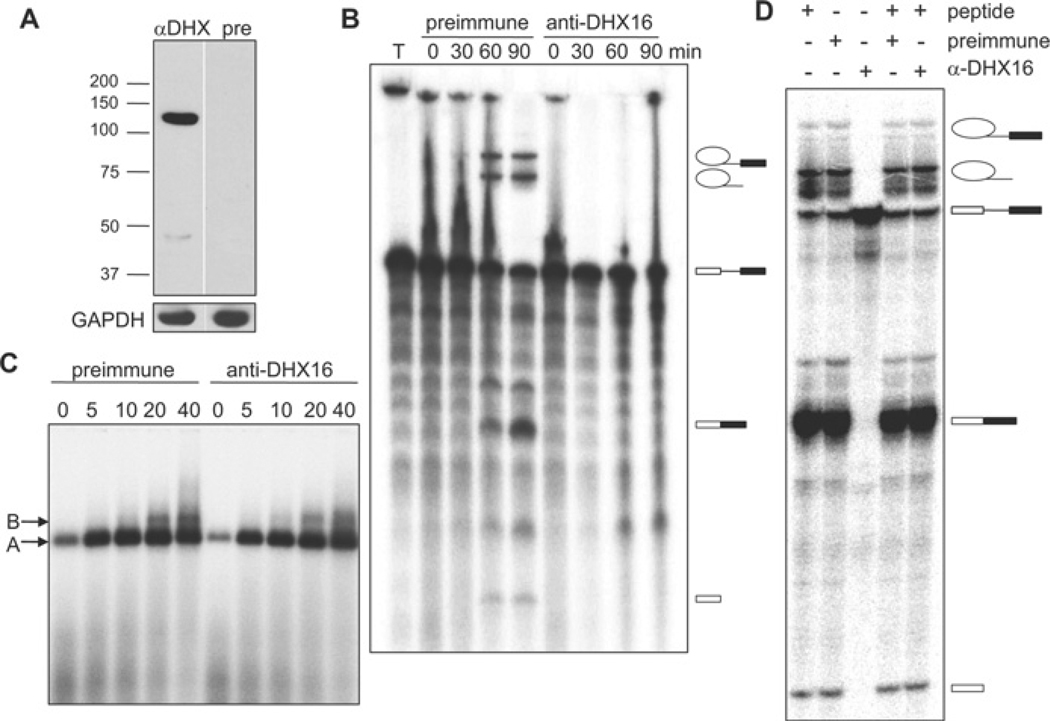
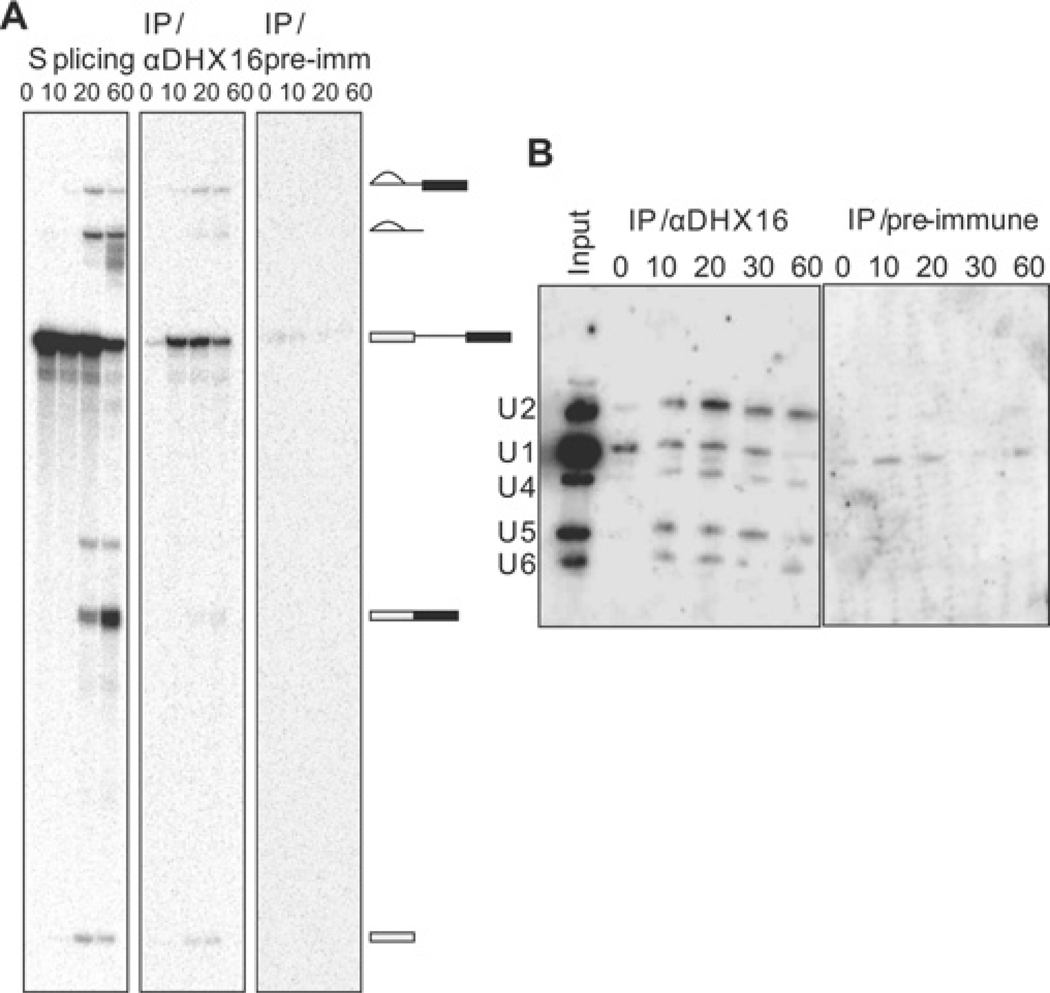
To test whether DHX16 is required for splicing reactions in vitro, we added the anti-DHX16 antiserum to HeLa cell nuclear extracts. Western blot analysis showed that the antiserum reacted with the 120 kDa protein in the HeLa extract, whereas the pre-immune serum did not (Figure 2A). Reaction mixtures were assembled in the presence of anti- or pre-immune serum and 32P-labelled MINX pre-mRNA was added to the reactions to allow splicing to take place for 0, 30, 60 or 90 min. RNA was then extracted and analysed by denaturing gel electrophoresis. The anti-DHX16 antiserum inhibited splicing as neither the spliced products nor intermediates were detected, whereas splicing occurred in the presence of pre-immune serum (Figure 2B). As yeast extracts lacking active Prp2 can assemble a pre-catalytic spliceosome [29], we also analysed the effect of the anti-DHX16 antiserum on spliceosome formation by native gel electrophoresis. The reaction mixtures containing HeLa nuclear extract and antiserum were incubated for 0, 5, 10, 20 or 40 min, and resolved on a native gel. The formation of splicing complexes A and B was not affected by the antiserum (Figure 2C), even though the splicing reaction was inhibited. To verify the specificity of splicing inhibition, we pre-incubated the antiserum with the peptide antigen prior to addition to the splicing reaction, which reversed the splicing inhibition (i.e. splicing occurred) (Figure 2D, α-DHX containing lanes).
Figure 2. Anti-DHX16 antibody inhibits splicing but not spliceosome formation in vitro.
(A) Western blot of HeLa nuclear extract with anti-DHX16 antiserum (αDHX) or pre-immune serum (pre). The lower panel shows the same blot probed with an anti-GAPDH antibody as a loading control. Positions of molecular mass markers in kDa are indicated. (B) Time course of in vitro splicing of pre-mRNA from pRG1 incubated with HeLa nuclear extracts in the presence of pre-immune or anti-DHX16 antiserum. Lane ‘T’ shows the pre-mRNA transcript only (i.e. not incubated with the extract). RNA molecules are indicated to the right of the gel and are, from top to bottom, IVS (intervening sequence or intron)–exon 2, intron, pre-mRNA substrate, spliced exons and exon 1. (C) Native agarose gel electrophoresis of aliquots of the reactions described in (B). Splicing complexes A and B are indicated. (D) Denaturing gel electrophoresis of RNA from the splicing reactions with HeLa nuclear extracts pre-incubated with anti-DHX16 antiserum, pre-immune serum or peptide as indicated. The splicing reactions were carried out with pRG1 pre-mRNA for 60 min. RNA molecules are indicated as in (B).
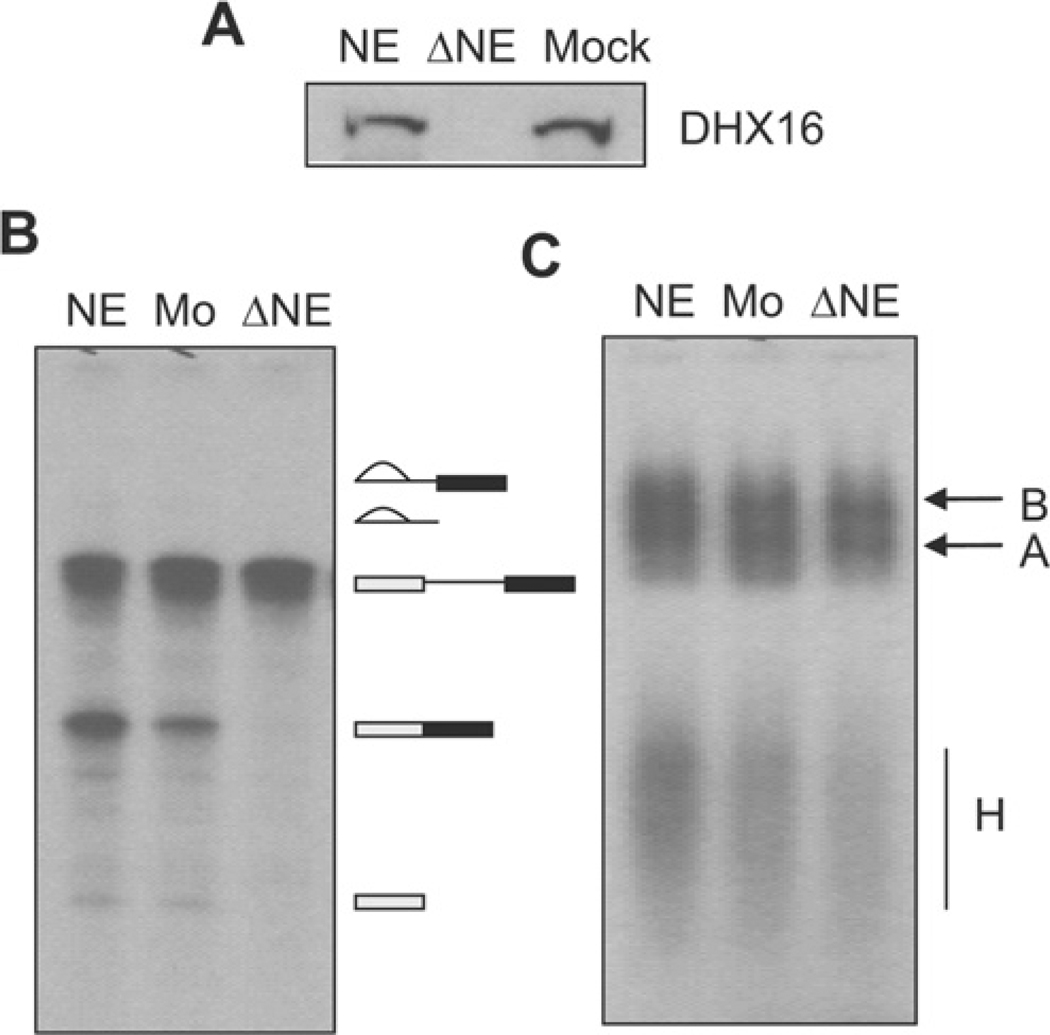
To test further the requirement of DHX16 for splicing, we also analysed the splicing reaction in DHX16-depleted nuclear extract. HeLa nuclear extracts were incubated with or without the peptide-purified anti-DHX16 antibody conjugated to Protein A beads, and splicing was analysed on denaturing gels and formation of splicing complexes was analysed by non-denaturing gels. Western blotting showed that DHX16 was depleted from the extract (Figure 3A, ΔNE). An in vitro splicing reaction with AdML (adenovirus major late) pre-mRNA substrate and the immunodepleted extract exhibited no splicing activity (Figure 3B), but could still assemble spliceosomal complex B (Figure 3C). Taken together, these results indicate that DHX16 is involved in pre-mRNA splicing in vitro and that HeLa nuclear extracts assembled a catalytically inactive spliceosomal complex when DHX16 was inactivated or removed by an anti-DHX16 antibody.
Figure 3. HeLa nuclear extracts immunodepleted of DHX16 assemble spliceosomal complex B but are not active for splicing.
DHX16 was depleted from HeLa nuclear extracts with the anti-DHX16 antibody or mock-depleted with Protein A beads. NE, nuclear extract; ΔNE, DHX16-depleted NE; Mock or Mo, mock-depleted NE. (A) The degree of DHX16 depletion was verified by immunoblot analysis using the anti-DHX16 antibody. (B) Denaturing gel electrophoresis of RNA from the splicing reactions containing the indicated extracts carried out for 60 min. RNA molecules are indicated to the right of the blot and are described as for Figure 2(B). (C) Native agarose gel electrophoresis of spliceosomal complexes in an aliquot of the reactions in (B) treated with 0.65 µg/µl heparin. B, splicing complex B; A, splicing complex A; H, hnRNP complexes.
DHX16 associates with pre-mRNA, splicing intermediates and snRNA during the splicing reaction
To investigate whether DHX16 is associated with the spliceosome, we used an anti-DHX16 antibody to precipitate the protein and its associated complexes during the splicing reaction. HeLa nuclear extracts were incubated with 32P-labelled MINX pre-mRNA under splicing conditions for 0, 10, 20 or 60 min (Figure 4A, left-hand panel). At the indicated times the reaction mixtures were immunoprecipitated with the peptide-purified anti-DHX16 antibody and eluted with the peptide antigen. RNA was isolated from the eluate and analysed on denaturing gels (Figure 4A, middle panel). A set of immunoprecipitation reactions using the pre-immune serum was also carried out for comparison (Figure 4A, right-hand panel). Pre-mRNA was associated with DHX16 from 10 min to 60 min, whereas the excised exon 1 and intron–exon 2 lariat intermediates associated with DHX16 at the 20 and 60 min time points (Figure 4A, middle panel). Small amounts of intron and spliced exons were also present in the immunoprecipitated samples. These results indicate that DHX16 associates with the spliceosome before the first transesterification, remains associated with the spliceosome after the first step, and probably even after the second transesterification, suggesting that human DHX16 may have a function in the later stage of pre-mRNA splicing. Protein MS has shown that DHX16 is present in purified spliceosomal complex B, as well as in complex C (see the Introduction section). Thus this is an interesting difference between human DHX16 and yeast Prp2, because yeast Prp2 activates the spliceosome prior to the first step and is not involved in the later stage of the splicing reaction [30].
Figure 4. Pre-mRNA, splicing intermediates and snRNAs co-immunoprecipitate with DHX16.
Splicing reactions with HeLa nuclear extracts were incubated for the indicated times and immunoprecipitated (IP) with the peptide-purified anti-DHX16 antibody. (A) The left-hand panel shows the splicing reactions with 32P-labelled pre-mRNA (no immunoprecipitation); RNA isolated from 1/15 of a 25 µl splicing reaction was applied in each lane. The middle (α-DHX16) and right-hand panel (pre-immune serum) show 32P-labelled RNA that co-immunoprecipitated with DHX16; 2/3 of the RNA extracted from the immunoprecipitate of a 25 µl splicing reaction was applied in each lane. (B) Northern blot analyses of snRNAs co-immunoprecipitated with DHX16. Splicing reaction mixtures were incubated for the specified time and subjected to immunoprecipitation with the anti-DHX16 antibody (α-DHX16) or pre-immune serum. One-third of the RNA extracted from the immunoprecipitation reaction was run on denaturing gels, blotted and probed with 32P-labelled antisense snRNAs. Input RNA (2/15 of the RNA from the splicing reaction) was also analysed for comparison. The positions of U-snRNAs are indicated to the left of the blot.
To verify that DHX16 was associated with the spliceosome during the splicing reaction, the immunoprecipitate was analysed by Northern blot for the presence of spliceosomal snRNAs (Figure 4B). By comparing the control immunoprecipitation with that using the pre-immune serum (Figure 4B), it was found that the U2, U4, U5 and U6 snRNAs were specifically associated with DHX16 after 10–60 min incubations (Figure 4B, left-hand panel). U1 snRNA was also found in the immunoprecipitates, but it was not clear whether the signal was specific. Nonetheless, the presence of four snRNAs in the DHX16 immunoprecipitates indicated that DHX16 associated with the pre-mRNA or splicing intermediates via the spliceosome.
Helicase domain mutants of DHX16 block splicing of minigene transcripts
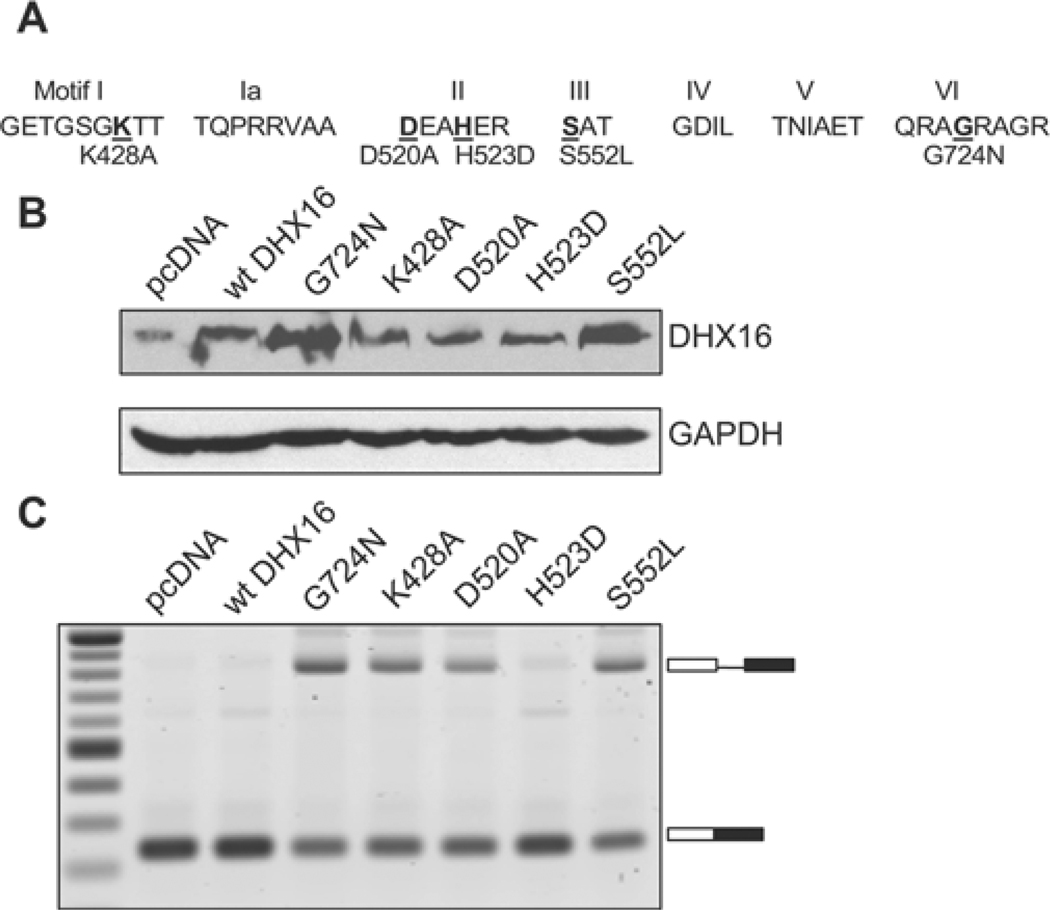
To study further the role of DHX16 in splicing, we created DHX16 mutants by mutating various amino acids in the conserved helicase domain: K428A, D520A, H523D, S552L and G724N (Figure 5A). The latter three mutations are analogous to mutations in yeast Prp2 (H349D, S378L and G551N) that result in diminished ATPase activity and a dominant-negative phenotype [31,32]. We transfected HEK-293 cells with a mutant plasmid, a wild-type plasmid or the vector plasmid, along with an intron-containing minigene plasmid, pL53In. Western blot analysis of the transfected cell lysates, using the anti-DHX16 antibody, indicated expression of DHX16 from the transfected plasmid (Figure 5B). We then isolated total RNA and performed RT–PCR to detect unspliced and spliced minigene RNA. Unspliced pL53In transcript was detected in cells expressing DHX16-K428A, -D520A, -S552L and -G724N, a very small amount of unspliced RNA was detected in cells that expressed DHX16-H523D, and little or no unspliced transcript was detected in cells transfected with the wild-type or vector control (Figure 5B). The weak dominant-negative effect of the H523D mutation was somewhat unexpected, because an equivalent mutation in yeast Prp2 (H349D) is quite effective in inhibiting splicing [32]. Nevertheless, overproducing a DHX16 mutant protein with an altered conserved amino acid in motif I (K428A), II (D520A), III (S552L) or VI (G724N) (Figure 5A) resulted in accumulation of unspliced minigene transcript, suggesting that pre-mRNA splicing, as detected by a model intron-containing minigene, was impaired when DHX16 function was interrupted in vivo.
Figure 5. DHX16 mutants block splicing of a minigene transcript in vivo.
HEK-293 cells were transfected with the pL53In minigene together with a DHX16 plasmid as indicated. (A) Schematic representation of the DHX16 mutants. Seven conserved motifs (I to VI) in the helicase domain are depicted. The mutated amino acid is shown in bold and underlined; the substituted amino acid is shown below with its protein sequence position. (B) Western blot using the anti-DHX16 antibody or anti-GAPDH antibody (control) on protein lysates of cells transfected with the mutant DHX16. (C) RT–PCR with total RNA isolated from the transfected cells. The unspliced and spliced RNAs are indicated on the right (box, exon; line, intron).
Nuclear extracts from mutant DHX16-G724N-expressing cells are impaired in splicing and accumulate spliceosome complex B
To analyse further the effect of the mutant protein on splicing, we prepared nuclear extracts from cells transfected with a mutant or wild-type DHX16 construct. We chose to study the DHX16-G724N mutant because of its robust inhibition of minigene transcript splicing (Figure 5C). HEK-293 cells were transfected with a plasmid carrying FLAG-tagged wild-type DHX16, FLAG-tagged DHX16-G724N or EGFP-tagged DHX16-G724N. Nuclear extracts were then prepared from transfected and untransfected cells. Western blot analysis showed that the transfected wild-type and mutant DHX16 proteins were expressed at a higher level than the native protein (Figure 6A). When assayed for in vitro splicing activity, nuclear extracts derived from the mutant DHX16-expressing cells had much less splicing activity than extracts from untransfected or wild-type DHX16-expressing cells (Figure 6B). The extracts containing DHX16-G724N failed to splice the MINX (Figure 6B, left-hand panel) or AdML (Figure 6B, right-hand panel) pre-mRNA substrates, indicating a general reduction in splicing activity in those extracts.
To analyse formation of the splicing complex, we resolved aliquots of the in vitro splicing reactions carried out in extracts from wild-type-DHX16-transfected cells and EGFP–DHX16-G724N-transfected cells on a native agarose gel (Figure 6C). Both wild-type and mutant extracts formed complexes A and B. The kinetics of complex A formation were similar between the wild-type and mutant extracts, but the mutant extract produced more complex B at later time points (Figure 6C). Therefore nuclear extracts isolated from DHX16-G724N-expressing cells are capable of assembling spliceosomal complex B, but are not effective in carrying out the transesterification reactions. This is consistent with DHX16 having a function after spliceosome formation and prior to the first catalytic step of splicing, similar to yeast Prp2 protein [30].
The weak activity in mutant extracts could be due to inhibition of the splicing reaction by the mutant protein in vitro or a lack of another late splicing factor in the extract (e.g. if there was less mRNA encoding a late splicing factor due to splicing inhibition by DHX16-G724N in the transfected cells). To test this, we isolated whole-cell extracts from cells that expressed mutant DHX16 and mixed them with the standard HeLa nuclear splicing extracts. If splicing activity was reduced beyond a simple dilution effect, we would conclude that the weak splicing activity shown in Figure 6 was caused by dominant inhibition by DHX16-G724N in vitro. The mixing experiment indicated that mutant DHX16 proteins, such as DHX16-G724N and DHX16-S552L, inhibited splicing in trans (see the Supplementary Results section and Supplementary Figure S1 at http://www.BiochemJ.org/bj/429/bj4290025add.htm). Thus the weak splicing activity in vitro (Figure 6) and in vivo (Figure 5) most probably resulted from direct participation of the mutant protein in splicing.
DISCUSSION
By using antibodies and helicase domain mutants, we have shown that DHX16 contributes to human pre-mRNA splicing prior to first transesterification reaction. A pre-catalytic spliceosome, splicing complex B, was stalled when DHX16 function was inhibited. Although human DHX16 has been suggested to be a S. cerevisiae Prp22 homologue [6], our results indicate that DHX16 is probably a functional orthologue of S. cerevisiae Prp2. As DHX16 complements S. pombe cdc28/prp8 double-mutants that are defective in pre-mRNA splicing [8], those studies [6,8] further argued that S. pombe Cdc28/Prp8 is likely to be a splicing complex B helicase. Thus the requirement of a DEAH-box protein to activate a pre-catalytic spliceosome for the first transesterification reaction in pre-mRNA splicing [30] appears to be evolutionarily conserved.
Similarity between DHX16 and S. cerevisiae Prp2 resides in the helicase and C-terminal domains; there is essentially no similarity in their N-terminal regions. The N-terminal region of DHX16 is enriched in lysine, glutamate and arginine residues, and a similar KER domain enriched in these residues was initially found in CAF-I (chromatin assembly factor I) as a histone-binding domain [33]. Human DHX38 (a homologue of S. cerevisiae PRP16) has an N-terminal region enriched in arginine, glutamate, aspartate and serine residues, which is not found in S. cerevisiae Prp16 [34]. In addition, the N-terminal region of human DHX8 (a homologue of S. cerevisiae Prp22) is enriched in arginine and serine residues, which is not found in S. cerevisiae Prp22 [35]. These unique N-terminal regions may play an important role in human mRNA splicing and may contribute to a potential difference in splicing regulation between humans and S. cerevisiae. In fact, full-length DHX38 cannot rescue S. cerevisiae with a prp16 deletion; only a chimaeric construct containing an S. cerevisiae N-terminus with human helicase and C-terminal domains can rescue the deletion [34]. We found that DHX16 did not complement S. cerevisiae prp2 temperature-sensitive or deletion mutants (M. Kato and R. J. Lin, unpublished work), suggesting that the N-terminal domains of DHX16 and Prp2 are not compatible. Interesting future studies could elucidate the unique function of the KER domain in DHX16 function.
Anti-DHX16 antibody immunoprecipitated splicing intermediates, as well as some spliced products (Figure 4), indicating that DHX16 remained associated with the spliceosome after the first transesterification. However, S. cerevisiae Prp2 exits the spliceosome after the first transesterification [36,37]. It is possible that DHX16 remains on the human spliceosome because it has a more stable interaction with a late spliceosomal factor such as GPKOW (G-patch domain and KOW motifs), which is found in splicing complex C and could be a homologue of yeast Spp2 [15]. Yeast Prp2 interacts with Spp2 and this interaction is essential for splicing [37,38]. We are currently investigating the interaction between DHX16 and GPKOW and whether their interaction plays a role in keeping DHX16 on the spliceosome. Furthermore, it is of interest to determine whether DHX16 plays an additional role in splicing after the first transesterification.
Unspliced intron-containing minigene transcripts were detected when cells overexpressed a dominant-negative DHX16 mutant, either by transient transfection (Figure 5) or by inducible stable transfection (M. Kato and R. J. Lin, unpublished work). This is consistent with a requirement for DHX16 before the first transesterification. We have also identified, by genomic tiling microarray, several endogenous transcripts that were unspliced in mutant-DHX16-expressing cells (M. Gencheva and R. J. Lin, unpublished work). It appears that expressing mutant DHX16 in a human cell results in accumulating intron-containing pre-mRNAs. This is analogous to yeast prp2 mutants accumulating spliceosomes that contained unspliced pre-mRNA in vivo [31,32,39]. Further characterization of DHX16 shall aid in the understanding of how a human spliceosome becomes active in splicing.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Rajesh Gaur, Sailesh Gopalakrishna Pillai, and the Gaur group for help with the in vitro splicing assay, including gifts of HeLa nuclear extracts, pRG1, and pAdMLPar. We thank Joan Steitz for SP6 plasmids carrying human anti-U-snRNA sequences, Takahiro Nagase for pBluscript II SK+KIAA0577, and Xiang-Dong Fu for anti-SC35 antibody. We thank Douglas Black and Peter Stoilov (UCLA), as well as members of the Lin lab, for stimulating discussion and insightful suggestions. We thank Keely Walker for proofreading the manuscript prior to submission.
FUNDING
This work was supported by the National Institutes of Health [grant number GM40639] and by a City of Hope Cancer Center seed grant (to R.J.L.).
Abbreviations used:
- DHX
DExH-box
- DTT
dithiothreitol
- EGFP
enhanced green fluorescent protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GPKOW
G-patch domain and KOW motifs
- HEK
human embryonic kidney
- HEK-293T cells
HEK-293 cells expressing the large T-antigen of SV40 (simian virus 40)
- hnRNP
heterogeneous nuclear ribonucleoprotein
- NP-40
Nonidet P40
- PCV
packed cell volume
- RT
reverse transcription
- snRNA
small nuclear RNA
- SR protein
serine-arginine rich protein
- TBS
Tris-buffered saline
- TOPOII
Type II topoisomerase.
Footnotes
AUTHOR CONTRIBUTION
Marieta Gencheva, Mitsuo Kato and Alain Newo performed experiments and all authors contributed to experimental design, ideas and manuscript writing. Ren-Jang Lin put together the final version of the manuscript for submission.
REFERENCES
- 1.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? BioEssays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 5.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 6.Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 7.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Imamura O, Saiki K, Tani T, Ohshima Y, Sugawara M, Furuichi Y. Cloning and characterization of a human DEAH-box RNA helicase, a functional homolog of fission yeast Cdc28/Prp8. Nucleic Acids Res. 1998;26:2063–2068. doi: 10.1093/nar/26.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puoti A, Kimble J. The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3276–3281. doi: 10.1073/pnas.97.7.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belfiore M, Mathies LD, Pugnale P, Moulder G, Barstead R, Kimble J, Puoti A. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3' untranslated region in Caenorhabditis elegans. RNA. 2002;8:725–739. doi: 10.1017/s1355838202028595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 13.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 16.Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. IX: the complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. Antibodies. [Google Scholar]
- 19.Nayler O, Hartmann AM, Stamm S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 2000;150:949–962. doi: 10.1083/jcb.150.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KA, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka N, Dreyfuss G. Preparation of efficient splicing extracts from whole cells, nuclei, and cytoplasmic fractions. Methods Mol. Biol. 2008;488:357–365. doi: 10.1007/978-1-60327-475-3_23. [DOI] [PubMed] [Google Scholar]
- 22.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 24.Gaur RK, McLaughlin LW, Green MR. Functional group substitutions of the branchpoint adenosine in a nuclear pre-mRNA and a group II intron. RNA. 1997;3:861–869. [PMC free article] [PubMed] [Google Scholar]
- 25.Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayeda A, Krainer AR. Mammalian in vitro splicing assays. Methods Mol. Biol. 1999;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 27.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brow DA, Vidaver RM. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1:122–131. [PMC free article] [PubMed] [Google Scholar]
- 29.Lin RJ, Lustig AJ, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- 30.Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plumpton M, McGarvey M, Beggs JD. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwalds-Gilbert G, Kim DH, Kim SH, Tseng YH, Yu Y, Lin RJ. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA. 2000;6:1106–1119. doi: 10.1017/s1355838200992483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Reed R. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono Y, Ohno M, Shimura Y. Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell. Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SH, Lin RJ. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1993;90:888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL., Jr The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lardelli RM, Thompson JX, Yates JR, III, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.