Abstract
Background
Identifying genes with essential roles in resisting environmental stress rates high in agronomic importance. Although massive DNA microarray gene expression data have been generated for plants, current computational approaches underutilize these data for studying genotype-trait relationships. Some advanced gene identification methods have been explored for human diseases, but typically these methods have not been converted into publicly available software tools and cannot be applied to plants for identifying genes with agronomic traits.
Methodology
In this study, we used 22 sets of Arabidopsis thaliana gene expression data from GEO to predict the key genes involved in water tolerance. We applied an SVM-RFE (Support Vector Machine-Recursive Feature Elimination) feature selection method for the prediction. To address small sample sizes, we developed a modified approach for SVM-RFE by using bootstrapping and leave-one-out cross-validation. We also expanded our study to predict genes involved in water susceptibility.
Conclusions
We analyzed the top 10 genes predicted to be involved in water tolerance. Seven of them are connected to known biological processes in drought resistance. We also analyzed the top 100 genes in terms of their biological functions. Our study shows that the SVM-RFE method is a highly promising method in analyzing plant microarray data for studying genotype-phenotype relationships. The software is freely available with source code at http://ccst.jlu.edu.cn/JCSB/RFET/.
Introduction
Among all kinds of environmental stresses (abiotic and biotic) in worldwide agriculture, drought is a major abiotic stress factor with significant impact on agricultural production. While resistance to biotic stresses is sometimes associated with monogenic traits, abiotic stresses are typically associated with multigenic traits, making it more difficult to study [1]. Hydropenia can trigger a cascade of physiological and metabolic activities in plants so the tolerance and susceptibility to drought are very complex. Among all the plants, Arabidopsis thaliana is the most popular model organism used in studying drought tolerance of plants, as it is a typical glycophyte, and many other xerophytes or desiccation-tolerant plants are similar to glycophytes in the drought-resistant mechanism. Hence, we focus on Arabidopsis to explore the drought-resistant genes in this study.
Recently, several research groups have investigated drought-mediated changes in gene expression using microarrays [2]–[5]. Microarray datasets typically include several thousands to tens of thousands of genes with relatively a small number of samples, but many genes are irrelevant or redundant for the purpose of this study. Biologically, there are often tens to hundreds of genes significantly associated to a trait like drought resistance. Hence, it is important to develop computational methods to mine these genes based on microarray data.
Prediction of genes associated with a trait can be formulated as a feature selection problem where key features (genes) of microarray data are indicative of a trait. Various feature selection techniques in handling gene expression data have been proposed. In particular, three types of classification-based methods were developed, i.e., filtering methods, wrapper methods, and embedded methods [6]. Feature selection using embedded SVM evaluation criterion to assess feature relevance is a typical and successful method [7]. Several other studies have provided alternative methods. For example, Support Vector Machine-Recursive Feature Elimination (SVM-RFE) was applied to train SVM for obtaining the weight of each feature and removing the one with the smallest weight iteratively [7]–[11]. This algorithm is superior to the “naïve” ranking with only one time RFE [7], [12]. However, this study [13] is the only study to apply the SVM-RFE method for identifying genes of an agronomic trait. Instead, current methods typically use a simple t-test for identifying important genes relevant to a trait [14], [15]. This could be problematic for integrating data from various sources. Furthermore, many researchers currently study genotype-trait relationships in an ad hoc fashion, e.g. by manually tuning various parameters that are relatively poorly understood [16]. These issues may result in vast amounts of plant microarray data being underutilized for studying genotype-phenotype relationships. One of the reasons behind these issues is lack of publicly available advanced tools. For example, while the SVM-RFE method is highly promising in analyzing microarray data, there was no software tool available.
In this study, we developed a systematic tool by improving the SVM-RFE method for identifying trait-specific genes using microarray data. The tool characterizes drought-resistant genes in Arabidopsis thaliana and is generally applicable to study genotype-phenotype relationships using gene expression data from microarray or RNA-Seq. Furthermore, the tool is freely available with source code at http://ccst.jlu.edu.cn/JCSB/RFET/.
Materials and Methods
Data source
GEO (http://www.ncbi.nlm.nih.gov/geo/) currently contains 1664 datasets with 90 GDSs, 299 platforms and 1275 series for Arabidopsis thaliana. Among these datasets, we used GSE10670 concerning global expression profiling of wild type and transgenic Arabidopsis plants in response to water stress published on September 1, 2008, and last updated on March 15, 2009. From GEO this is the largest gene expression dataset available up to date for studying plant drought resistance. The data was generated from the Affymetrix platform GPL198 Arabidopsis ATH1 Genome Array. It consists of 22,810 probes and each probe corresponds to one gene. In all we identified 22 samples from GSM269812 to GSM269833, including the wild type (WT), two independent transgenic lines (T6 and T8), and the vector control line (C2), which was used as an additional control. In all related experiments, the relative water content (RWC) of wild type and transgenic leaves during a period of dehydration was monitored. At day 7 when the transgenic plants were still at an RWC of >85%, the wild type and vector control plants were at an RWC of ∼50–60% [17]. Hence, we first chose experimental samples just on T6 and T8, which were expected to reveal drought tolerant genes. Table 1 gives the specific description of the data used, and the class label (0/1) is according to the different stress conditions. Table 2 presents the susceptibility genotype (WT and C2) data, which were used to discard the tuning genes as described later.
Table 1. Resistant samples description.
| N. | Sample | Transgenic line | Stress condition | Rep* | Class label+ |
| 1 | GSM269814 | T6 | well watered | 1 | 1 |
| 2 | GSM269815 | T6 | drought | 1 | 0 |
| 3 | GSM269816 | T8 | well watered | 1 | 1 |
| 4 | GSM269817 | T8 | drought | 1 | 0 |
| 5 | GSM269822 | T6 | well watered | 2 | 1 |
| 6 | GSM269823 | T6 | drought | 2 | 0 |
| 7 | GSM269824 | T8 | well watered | 2 | 1 |
| 8 | GSM269825 | T8 | drought | 2 | 0 |
| 9 | GSM269830 | T6 | well watered | 3 | 1 |
| 10 | GSM269831 | T6 | drought | 3 | 0 |
| 11 | GSM269832 | T8 | well watered | 3 | 1 |
| 12 | GSM269833 | T8 | drought | 3 | 0 |
*: Rep is the number of biological replications.
: Class label is used to indicate well watered (1) and the drought (0), respectively.
Table 2. Susceptible samples description.
| N. | Sample | Genotype | Stress condition | Rep | Class label |
| 1 | GSM269812 | WT | well watered | 1 | 1 |
| 2 | GSM269813 | WT | drought | 1 | 0 |
| 3 | GSM269818 | C2 | well watered | 1 | 1 |
| 4 | GSM269819 | C2 | drought | 1 | 0 |
| 5 | GSM269820 | WT | well watered | 2 | 1 |
| 6 | GSM269821 | WT | drought | 2 | 0 |
| 7 | GSM269826 | C2 | well watered | 2 | 1 |
| 8 | GSM269827 | C2 | drought | 2 | 0 |
| 9 | GSM269828 | WT | well watered | 3 | 1 |
| 10 | GSM269829 | WT | drought | 3 | 0 |
Table caption follows Table 1.
Data preprocessing stage
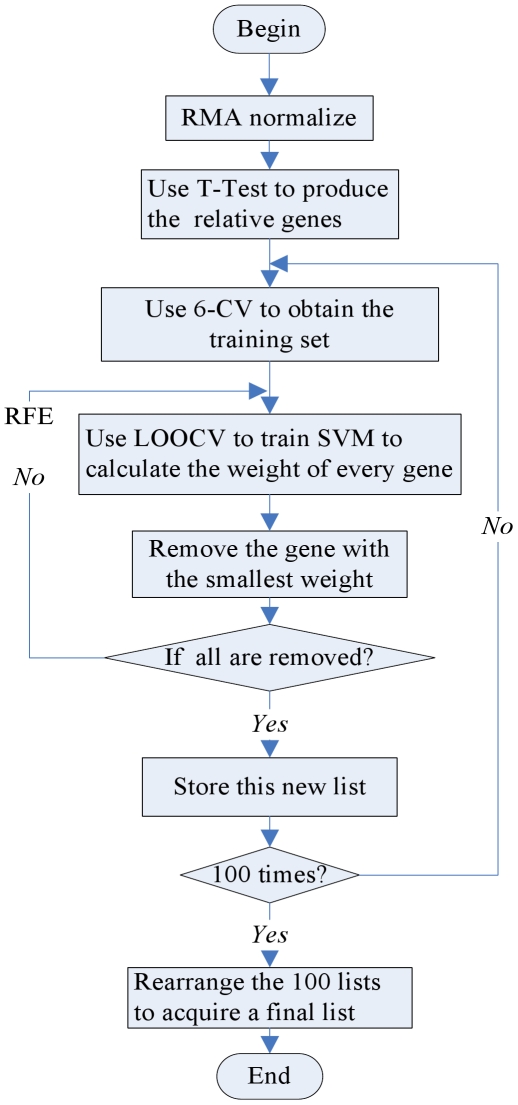
We applied a quantile-based [18] RMA (Robust Multi-chip Averages) method for normalizing microarray data. The RMA feeds probes data stored in Affymetrix CEL into a stochastic model to estimate gene expression and converts the probe data to gene expression data. We conducted the RMA analysis using Bioconductor (http://www.bioconductor.org/), which is an open-source tool for bioinformatics using the R statistical programming language.
T-test method for preliminary selection
For tens of thousands of genes, it would be of high complexity to use the SVM-RFE directly. Hence, we first employed a t-test [19] to filter out unlikely genes involved in drought tolerance. In our preliminary selection we assigned 0.001 as the p-value threshold, resulting in 736 genes, which are still too many for agronomic studies.
Using SVM-RFE method for gene selection
RFE is an iterative procedure for SVM classifier. A cost function  computed on training samples is used as an objective function. Expanding
computed on training samples is used as an objective function. Expanding 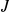 in Taylor series to the second order using the OBD algorithm [20], and neglecting the first order term at the optimum of
in Taylor series to the second order using the OBD algorithm [20], and neglecting the first order term at the optimum of 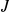 , yield:
, yield:
| (1) |
Here 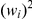 was used as the ranking criterion and we used LIBSVM (a library for Support Vector Machines) [21] with a linear kernel. We present below an outline of the SVM-RFE in the linear kernel. For more details about this method, see Guyon et al.
[7].
was used as the ranking criterion and we used LIBSVM (a library for Support Vector Machines) [21] with a linear kernel. We present below an outline of the SVM-RFE in the linear kernel. For more details about this method, see Guyon et al.
[7].
Algorithm SVM-RFE.
Inputs:
Training samples (microarray datasets)
Class labels (1 for well watered or 0 for drought)
Initialize:
Surviving genes
Gene-ranking list
Limit training samples to good genes
Train the classifier
Compute the weight from each selected gene:
where k indicates the k-th training pattern.
Compute the ranking criterion for the i-th gene
Mark the gene with the lowest ranking
Renew the gene-ranking list
Eliminate the gene with the lowest ranking
Repeat until 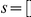
Output:
A gene-ranking list  .
.
We trained the classifier, computed the ranking for the 736 genes obtained and then removed the gene with the lowest ranking. We repeated the process until all the genes were removed. This iterative process is a sequence backward selection (SBS) procedure and at last the method produces a gene-ranking list with weights from high to low.
To improve prediction accuracy we conducted several rounds of bootstrapping in the SVM-RFE procedure and in each round one sorted list was produced. However, there is a shortage of experimental samples that are needed to train the SVM. This brings up two issues: one is how to generate training and test sets, and the other is how to combine the weights of each gene in each sorted list. To address these two issues, we developed the following two solutions:
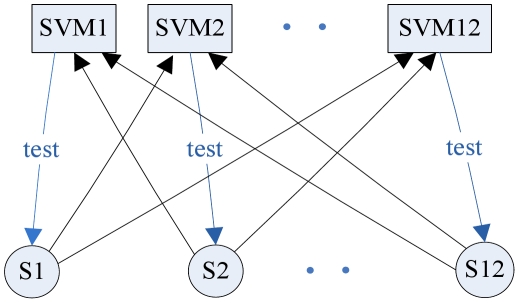
In order to make good use of limited data for predicting drought-resistant genes in Arabidopsis, the generation of training set is a key factor. The dataset was randomly split into n subsets of approximately equal size, then one subset was removed, and the remaining samples formed the training set. Each time a different subset was selected in such a way that all the samples had an equal chance to be selected as the training data. We call it n-CV, where n could be equal to 12, 6, 4, or 3, respectively. In the following examples, there are 12 samples from transgenic lines described in Table 1, and a 6-CV was used for each subset with two samples. Then the Leave One Out Cross Validation (LOOCV) was used for training SVM (see Figure 1). After that we performed 100 times of 6-CV and each 6-CV ran 6 times of the SVM-RFE procedure. The computational complexity of each n-CV is n*(g*O(s3)), where s is the number of samples and g is the number of genes [22]. The computational time on the training stage primarily depends on the number of genes given the small number of training samples. Hence, the computational complexity can be easily handled by filtering out unlikely genes involved. The 100 times 6-CV training took around 90 minutes to get the results on a desktop computer with Intel Core 2 Duo E6750 and DDR2 2GB memory.
- We acquired 100 sorted lists, and designed a re-ranking measure to take occurrence and ranking of every gene into account to form a final ranking list of all genes:
where p is the number of times for CV,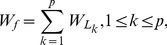
(2) 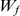 is the sum of one gene's weights in p experiments, and
is the sum of one gene's weights in p experiments, and 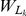 is the weight in the k-th occurrence in sorted lists.
is the weight in the k-th occurrence in sorted lists. 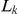 shows the ranking of one feature in the k-th list.
shows the ranking of one feature in the k-th list.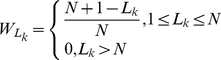
(3)
Figure 1. LOOCV for twelve samples.
Here, we have top 10 (N) genes and 100 (p) SVM-RFE trainings. Thus, we obtain a final list containing the optimal genes sorted by 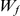 in decreasing order. The algorithm flowchart is shown in Figure 2.
in decreasing order. The algorithm flowchart is shown in Figure 2.
Figure 2. Algorithm flowchart for identifying drought-resistant genes.
Results and Discussion
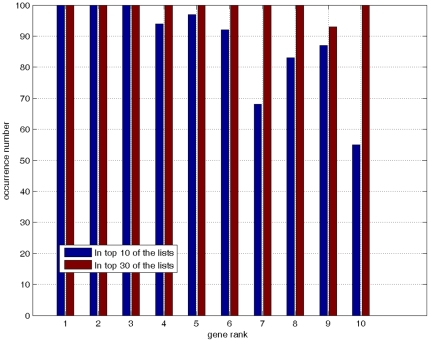
By applying the SVM-RFE method in analyzing the drought-resistant genotype based on the microarray data, we selected 10 genes in the final list according to 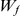 in Eq. 2, as summarized in Table 3. Figure 3 shows the occurrence of these genes in top-10 list and also in top-30 list when conducting 100 times of 6-CV. It shows that the occurrence of these genes in the top list is very high and consistent. We checked the functional annotations using GO (http://www.geneontology.org/), KEGG pathways (http://www.genome.jp/kegg/) and the literatures [2], [23]–[28], and obtained the related information shown in Table 3. From Table 3 it can be seen that most of the genes have no specific molecular function annotations. This is not surprising as the water stress tolerance is a complex trait, which resulted from sophisticated coordination of physiological and biochemical alterations at the cellular and molecular levels [24].
in Eq. 2, as summarized in Table 3. Figure 3 shows the occurrence of these genes in top-10 list and also in top-30 list when conducting 100 times of 6-CV. It shows that the occurrence of these genes in the top list is very high and consistent. We checked the functional annotations using GO (http://www.geneontology.org/), KEGG pathways (http://www.genome.jp/kegg/) and the literatures [2], [23]–[28], and obtained the related information shown in Table 3. From Table 3 it can be seen that most of the genes have no specific molecular function annotations. This is not surprising as the water stress tolerance is a complex trait, which resulted from sophisticated coordination of physiological and biochemical alterations at the cellular and molecular levels [24].
Table 3. Selected 10 genes related to drought-resistant genotype.
| Rank | Probe ID | Platform ORF | Gene Title | GO: Function | GO: Process | GO: Component |
| 1 | 248352_at | At5g52300 | LTI65(LOW-TEMPERATURE-INDUCED 65) | abscisic acid mediated signaling pathway/response to abscisic acid stimulus/response to cold/response to salt stress/response to water deprivation | ||
| 2 | 247723_at | At5g59220 | Protein phosphatase 2C,putative/PP2C, putative | catalytic activity/protein serine/hreonine phosphatase activity | response to abscisic acid stimulus/ response to water deprivation | chloroplast |
| 3 | 249052_at | At5g44420 | PDF1.2 | defense response | cell wall / endomembrane system | |
| 4 | 265342_at | At2g18300 | basic helix-loop-helix (bHLH) family protein | nucleus | ||
| 5 | 257365_x_at | At2g26020 | PDF1.2b(plant defensin 1.2b) | defense response | cell wall / endomembrane system | |
| 6 | 266743_at | At2g02990 | RNS1(RIBONUCLEASE1); endoribonuclease/ribonuclease | endoribonuclease activity/ribonuclease activity | response to wounding | cell wall/extracellular region/plasma membrane |
| 7 | 258897_at | At3g05730 | hypothetical protein | endomembrane system | ||
| 8 | 266462_at | At2g47770 | benzodiazepine receptor-related | response to abscisic acid stimulus/response to osmotic stress/response to salt stress | Golgi stack/endoplasmic reticulum/membrane | |
| 9 | 248218_at | At5g53710 | hypothetical protein | |||
| 10 | 262347_at | At1g64110 | AAA-type ATPase family protein | ATP binding/nucleotide binding |
Figure 3. The occurrence of selected 10 genes in the top-10 and top-30 lists when conducting 100 times of 6-CV.
The gene order is the same as that in Table 3.
Table 3 reveals some interesting biological features. Both the first and second genes directly respond to water deprivation. The cellular component of the second gene is chloroplast where photosynthesis of plants occurs. As photosynthesis uses carbon dioxide and water releasing oxygen, it is likely that this gene plays some role in water utilization in chloroplast. The third and fifth genes are both related to defense response, whose pathway is known to have cross-talk with drought resistance pathway [29].
The sixth gene in the table responds to wounding and it may help repair the damage of cell caused by water loss. Some other genes in the list may also be related to drought resistance. The eighth-ranked gene responds to osmotic stress and salt stress. Osmotic adjustment is an important physiological mechanism adapting to water stress [24]. Osmotic adjustment can maintain a dynamic balance between damage and repair of cellular components to relieve plants injury and improve plants' ability of stress resistance.
In the column GO: Component of Table 3, all of the 3rd, 5th, 6th, 7th, and 8th genes belong to membrane systems, like endomembrane, plasma membrane, and so on. Membrane system is the key part damaged by drought stress and it is the most sensitive original reaction site against adversity [25]. Membrane, together with associated proteins, provides cells with not only a relatively stable internal environment, but also provides a switch to material transportation, energy exchange and information transmission between cells and the environment. Therefore, these five genes may help adjust osmotic membranes to boost drought resistance. In all we have demonstrated that seven genes may be closely related to water tolerance for Arabidopsis, i.e., the 1st, 2nd, 3rd, 5th, 6th, 7th, and 8th genes in Table 3.
We also repeated the computational process with the susceptibility genotype samples (WT and C2), and obtained the top-10 gene list as described in Table 4. From the relative function annotations, it appears that most of these genes have little relationship with the ability of drought resistance, which may explain why these genotypes do not have capacities of drought resistance.
Table 4. Selected 10 related to water-susceptibility genotype.
| Rank | Probe ID | Platform ORF | Gene Title | GO: Function | GO: Process | GO: Component |
| 1 | 262128_at | At1g52690 | late embryogenesis abundant protein, putative / LEA protein, putative | embryonic development ending in seed dormancy | ||
| 2 | 264580_at | At1g05340 | hypothetical protein | biological_process | ||
| 3 | 258499_at | At3g02540 | RAD23-3 (PUTATIVE DNA REPAIR PROTEIN RAD23-3); damaged DNA binding | proteasome binding///ubiquitin binding | nucleotide-excision repair/proteasomal ubiquitin-dependent protein catabolic process | nucleus |
| 4 | 258239_at | At3g27690 | LHCB2.3; chlorophyll binding | chlorophyll binding | photosynthesis/response to blue light/response to far red light/response to red light | chloroplast envelope/chloroplast thylakoid membrane/light-harvesting complex/thylakoid |
| 5 | 266462_at | At2g47770 | benzodiazepine receptor-related | response to abscisic acid stimulus/response to osmotic stress/response to salt stress | Golgi stack/endoplasmic reticulum/membrane | |
| 6 | 258347_at | At3g17520 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | embryonic development ending in seed dormancy | ||
| 7 | 247095_at | At5g66400 | RAB18 (RESPONSIVE TO ABA 18) | cold acclimation/response to 1-aminocyclopropane-1-carboxylic acid/response to abscisic acid stimulus/response to stress/response to water deprivation | ||
| 8 | 247723_at | At5g59220 | protein phosphatase 2C, putative / PP2C, putative | catalytic activity/protein serine/threonine phosphatase activity | response to abscisic acid stimulus/response to water deprivation | chloroplast |
| 9 | 262382_at | At1g72920 | disease resistance protein (TIR-NBS class), putative | transmembrane receptor activity | intrinsic to membrane | |
| 10 | 248352_at | At5g52300 | LTI65 (LOW-TEMPERATURE-INDUCED 65) | abscisic acid mediated signaling pathway/response to abscisic acid stimulus/response to cold/response to salt stress/response to water deprivation |
Our understanding of the functions of these seven genes is far from complete. Compared the top-10 gene list obtained from the resistant genotype with the susceptibility genotype list, 3 genes are the same, which are the 1st, 2nd, and 8th in Table 3, and the 10th, 8th, and 5th in Table 4. We call the same ones tuning genes [16]. Maybe their adaptability to hydropenia is the result of irritable reactions to environmental changes. So by removing the tuning genes from the top-10 gene list with transgenic genotype, the inference is that the real drought-resistant genes should be in the 7 ones in Table 5 (a subset of Table 3). And with the same thought, we analyzed the two top 100 gene lists using the resistant genotype and the susceptibility genotype, respectively (see Table S1 and Table S2 for the detailed information). Comparing the top-100 gene list obtained from the resistant genotype with the susceptibility genotype list, it can be seen that 37 genes overlap (tuning genes). In Table S1, the “Overlap” column indicates the tuning genes. We compared our result against the result published by Huang et al. [30] to look for the overlap of the genes identified to be involved in drought resistance. Our comparison shows that 5 out of the 10 genes from resistant genotype and 6 out of the 10 genes from susceptible genotype are identified to be the same. When we expanded our analysis to include the top 100 genes, 50 genes of the resistant genotype and 42 of the susceptible genotype overlap with the gene lists in the published results. GO term enrichment analysis was also performed for the annotations of the top 100 genes in the resistant and susceptibility genotypes, respectively using Amigo GO Term Enrichment Tool (http://amigo.geneontology.org/cgi-bin/amigo/term_enrichment) as shown in Table 6. All the GO terms identified for both genotypes with a significant p-value less than 0.001 have functional categories related to the drought stress.
Table 5. The new list with the tuning genes removed from the top-10 resistant gene list.
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Probe ID | 249052_at | 265342_at | 257365_x_at | 266743_at | 258897_at | 248218_at | 262347_at |
| Platform ORF | At5g44420 | At2g18300 | At2g26020 | At2g02990 | At3g05730 | At5g53710 | At1g64110 |
Table 6. GO Term Enrichment for resistant and susceptibility genotypes.
| Resistant Genotype | GO Term | Aspect | P-value | Sample frequency | Background frequency |
| GO:0050896 response to stimulus | P | 1.49e-04 | 36/99 (36.4%) | 4570/29887 (15.3%) | |
| GO:0006950 response to stress | P | 5.10e-04 | 23/99 (23.2%) | 2221/29887 (7.4%) | |
| GO:0009628 response to abiotic stimulus | P | 6.17e-04 | 18/99 (18.2%) | 1421/29887 (4.8%) | |
| GO:0009725 response to hormone stimulus | P | 1.74e-03 | 14/99 (14.1%) | 935/29887 (3.1%) | |
| GO:0009719 response to endogenous stimulus | P | 4.41e-03 | 14/99 (14.1%) | 1014/29887 (3.4%) | |
| GO:0009611 response to wounding | P | 7.93e-03 | 6/99 (6.1%) | 151/29887 (0.5%) | |
| GO:0042221 response to chemical stimulus | P | 9.68e-03 | 20/99 (20.2%) | 2085/29887 (7.0%) | |
| Susceptibility Genotype | GO:0009628 response to abiotic stimulus | P | 5.47e-04 | 18/100 (18.0%) | 1421/29887 (4.8%) |
| GO:0009266 response to temperature stimulus | P | 4.96e-03 | 9/100 (9.0%) | 407/29887 (1.4%) |
The SVM-RFE algorithm was used in cancer marker gene prediction with abundant datasets. However, there are much fewer microarray samples for finding the drought-resistant genes of Arabidopsis. Hence, we modified the original SVM-RFE method to address this drawback by using bootstrapping and leave-one-out cross-validation. Our study shows that the improved method is effective for identifying drought-resistance genes. Since the sparseness of gene expression data for studying genotype-trait relationships is a common issue, the method provides a framework for handling this issue. The framework has some advantages over some other feature selection methods that require extensive training data, such as random forest [31]. This is by no means a replacement of additional experimental data, but it can effectively utilize the sparse data available to generate useful hypotheses and guide further targeted experimental work.
Although Arabidopsis is a model organism for plant gene function analysis and gene expression studies, few genes related to drought resistance mechanisms are annotated with direct experimental evidences. There is a need to predict additional drought-resistance genes based on gene expression data. The predictions of drought-resistance genes generated by the JU/MU development of the SVM-RFE method-based software provides useful hypothesis for experimentalists to verify. For example, the 3rd, 8th and 10th genes in Table 3 are not annotated as drought-resistance genes, but they are highly likely involved in drought resistance. Perhaps a major challenge in the future is to inquire into the relative contribution of each gene to water tolerance. The JU/MU approach is applicable to the study of plant genes related to other stress resistance and genes associated with any agronomic trait in general.
Supporting Information
Detailed information of top 100 genes from resistant genotype.
(DOC)
Detailed information of top 100 genes from susceptibility genotype.
(DOC)
Acknowledgments
We thank Zhang Chen and Du Wei for helpful discussions and technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National High-Tech R&D Program of China (2009AA02Z307), National Natural Science Foundation of China (61073075, 60903097, and 10872077), U.S. Department of Energy (DE-FOA-0000223) and the U.S. National Science Foundation (1025752). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology. 2005;16(2):123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan A, Lafitte HR, Chen JX, Mansueto L, Bruskiewich R, et al. Gene expression microarrays and their application in drought stress research. Field Crops Research. 2006;97(1):101–110. [Google Scholar]
- 3.Matsui A, Ishida J, Morosawa T, Okamoto M, Kim J-M, et al. Arabidopsis tiling array analysis to identify the stress-responsive genes. Plant Stress Tolerance: Methods and Protocols. 2010;639:141–155. doi: 10.1007/978-1-60761-702-0_8. [DOI] [PubMed] [Google Scholar]
- 4.Kankainen M, Brader G, Törönen P, Palva ET, Holm L. Identifying functional gene sets from hierarchically clustered expression data: map of abiotic stress regulated genes in Arabidopsis thaliana. Nucleic Acids Res. 2006;34(18):e124. doi: 10.1093/nar/gkl694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Ruan J, Ho TH, You Y, Yu T, et al. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21(14):3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- 6.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 7.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46(1–3):389–422. [Google Scholar]
- 8.Lu Y, Gao Y, Cao Z, Cui J, Dong Z, et al. A study of health effects of long-distance ocean voyages on seamen using a data classification approach. BMC Medical Informatics and Decision Making. 2010;10:13. doi: 10.1186/1472-6947-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y, Wilkins D. Improving the performance of SVM-RFE to select genes in microarray data. BMC Bioinformatics. 2006;7(Suppl 2):S12. doi: 10.1186/1471-2105-7-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Tuck DP. MSVM-RFE: extensions of SVM-RFE for multiclass gene selection on DNA microarray data. Bioinformatics. 2007;23(9):1106–1114. doi: 10.1093/bioinformatics/btm036. [DOI] [PubMed] [Google Scholar]
- 11.Johannes M, Brase JC, Fröhlich H, Gade S, Gehrmann M, et al. Integration of pathway knowledge into a reweighted recursive feature elimination approach for risk stratification of cancer patients. Bioinformatics. 2010;26(17):2136–2144. doi: 10.1093/bioinformatics/btq345. [DOI] [PubMed] [Google Scholar]
- 12.Yousef M, Ketany M, Manevitz L, Showe LC, Showe MK. Classification and biomarker identification using gene network modules and support vector machines. BMC Bioinformatics. 2009;10:337. doi: 10.1186/1471-2105-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang JX, Zhang F, Wang Y, Fu Y, Xu D, et al. Salt tolerance genes selection in Oryza Sativa using SVMRFE based on Microarray. 2011. pp. 30–35. 3rd International Conference on Bioinformatics and Computational Biology (BICoB), New Orleans.
- 14.Swindell W, Huebner M, Weber A. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity. 2007;99:143–150. doi: 10.1038/sj.hdy.6800975. [DOI] [PubMed] [Google Scholar]
- 15.Lionneton E, Aubert G, Ochatt S, Merah O. Genetic analysis of agronomic and quality traits in mustard (Brassica juncea). Theor Appl Genet. 2004;109:792–799. doi: 10.1007/s00122-004-1682-0. [DOI] [PubMed] [Google Scholar]
- 16.Maury LL, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nature Reviews Genetics. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 17.Perera IY, Hung CY, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell. 2008;20(10):2876–2893. doi: 10.1105/tpc.108.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 20.LeCun Y, Denker JS, Solla SA. Optimal brain damage. In: Touretzky DS, editor. Advances in Neural Information Processing Systems II. San Mateo, CA: Morgan Kaufman; 1990. pp. 598–605. [Google Scholar]
- 21.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. 2001. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- 22.Tsang IW, Kwok JT, Cheung PM. Core Vector Machines: Fast SVM Training on Very Large Data Sets. Machine Learning Research. 2005;6:363–392. [Google Scholar]
- 23.Czechowski T, Stitt M, Altmann T, KUdvardi M, Scheible W. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiology. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramanjulu S, Bartels D. Drought-and desiccation-induced modulation of gene expression in plants. Plant, Cell and Environment. 2002;25(2):141–151. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- 25.Villadsen D, Rung JH, Nielsen TH. Osmotic stress changes carbohydrate partitioning and fructose-2, 6-bisphosphate metabolism in barley leaves. Functional Plant Biology. 2005;32(11):1033–1043. doi: 10.1071/FP05102. [DOI] [PubMed] [Google Scholar]
- 26.Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Current Opinion in Plant Biology. 2006;9(2):189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Kwon YR, Lee HJ, Kim KH, Hong SW, Lee SJ, et al. Ectopic expression of Expansin3 or Expansinβ1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnology Letters. 2008;30(7):1281–1288. doi: 10.1007/s10529-008-9678-5. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Hua X, Guo J, Qi D, Wang L, et al. Enhanced tolerance to drought stress in transgenic tobacco plants overexpressing VTE1 for increased tocopherol production from Arabidopsis thaliana. Biotechnology Letters. 2008;30(7):1275–1280. doi: 10.1007/s10529-008-9672-y. [DOI] [PubMed] [Google Scholar]
- 29.Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, et al. Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol. 2004;55(3):327–342. doi: 10.1007/s11103-004-0685-1. [DOI] [PubMed] [Google Scholar]
- 30.Huang DQ, Wu WR, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany. 2008;59(11):2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Gao Z. Random forests: an important feature genes selection method of tumor. Acta Biophys Sin. 2009;25:51–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed information of top 100 genes from resistant genotype.
(DOC)
Detailed information of top 100 genes from susceptibility genotype.
(DOC)