Abstract
Receptor tyrosine kinases (RTKs) activate pathways mediated by serine/threonine (Ser/Thr) kinases such as the PI3K (phosphatidylinositol 3-kinase)-Akt pathway, the Ras-MAPK (mitogen-activated protein kinase)-RSK pathway, and the mTOR (mammalian target of rapamycin)-p70 S6 pathway that control important aspects of cell growth, proliferation, and survival. The Akt, RSK, and p70 S6 family of protein kinases transmit signals by phosphorylating substrates on a RxRxxS/T motif. Here, we developed a large-scale proteomic approach to identify over 200 substrates of this kinase family in cancer cell lines driven by the c-Met, epidermal growth factor receptor (EGFR), or platelet-derived growth factor receptor a (PDGFRα) RTKs. We identified a subset of proteins with RxRxxS/T sites for which phosphorylation was decreased by RTKIs as well as by inhibitors of the PI3K, mTOR, and MAPK pathways and determined the effects of siRNA directed against these substrates on cell viability. We found that phosphorylation of the protein chaperone SGTA (small glutamine-rich tetratricopeptide repeat-containing protein alpha) at Ser305 is essential for PDGFRα stabilization and cell survival in PDGFRα-dependent cancer cells. Our approach provides a new view of RTK and Akt-RSK-S6 kinase signaling, revealing many previously unidentified Akt-RSK-S6 kinase substrates that merit further consideration as targets for combination therapy with RTKIs.
Introduction
In virtually all epithelial tumors, growth factor receptor activity is deregulated by activating mutations, genomic amplification, and autocrine loops (1). Accumulating evidence from mouse models and human drug response suggests that signals emanating from the activated tyrosine kinase domain of growth factor receptors are required for tumor initiation and maintenance (2–4). This dependence of tumor cell survival upon the driving oncogene has been called “oncogene addiction” and demonstrates the acute sensitivity of cancer cells to inhibition of the pathways driving their proliferation, growth, and survival (4, 5). However, the complexity of the pathways and multiplicity of kinases activated downstream of RTKs has made it difficult to identify the key substrates that mediate oncogene dependence.
Three core signaling pathways activated downstream of oncogenic RTKs are the Ras-Raf-MAPK (mitogen-activated protein kinase)-RSK (ribosomal S6 kinase) pathway involved in cell proliferation (6, 7), the mTOR (mammalian target of rapamycin)-p70 S6 kinase pathway involved in nutrient sensing and cell growth (8, 9), and the PI3K (phosphatidylinositol 3-kinase)-Akt pathway involved in metabolic and cell survival signaling (10). Each of these pathways activates members of the AGC (cAMP-dependent, cGMP-dependent, and protein kinase C) family of serine/threonine (Ser/Thr) kinases, including Akt, RSK, and p70 S6 kinase, that phosphorylate substrates at the basophilic motif RxRxxS/T (R= arginine, S = serine, T = threonine and x = any amino acid) (11). Although inhibition of these three pathways often correlates with the beneficial effects of tyrosine kinase inhibitors (for instance, the induction of cell death), the downstream targets of these inhibitors remain largely unidentified.
To characterize the cell circuitry activated downstream of Akt, RSK, and p70 S6 kinase, we first developed antibodies that recognized and selectively immunoprecipitated phosphorylated substrates of Akt, RSK, and p70 S6 kinase. Analysis of arginine-rich phosphopeptides by tandem mass spectrometry (MS/MS) is complicated by “neutral loss”, in which arginine residues destabilize nearby phosphorylated amino acids, resulting in the preferential loss of phosphate during the conventional peptide backbone fragmentation necessary for MS/MS based identification. Here, we used two different approaches to overcome neutral loss, electron transfer dissociation (ETD) (12) and two-step protease-based collision-induced dissociation (CID) analysis (13).
Developing appropriate antibodies and overcoming neutral loss enabled us to use a large-scale phosphoproteomic approach to investigate Akt-RSK-S6 kinase signaling downstream of oncogenic EGFR, c-Met, and PDGFRα. Using selective RTK inhibitors (RTKIs), as well as inhibitors specific for the PI3K, mTOR, or MAPK signaling pathways (“pathway inhibitors”), we identified over 200 substrates and identified new circuitry not previously implicated in RTK signaling, including connections to metabolic activity, cell cycle control, transforming growth factor (TGF)-Smad signaling, and regulation of protein stability. Using short interfering RNA (siRNA) screens, we also identified a subset of molecules that participate in a regulatory loop to stabilize RTKs.
Results
Monoclonal Antibodies Directed against Phosphorylated Akt, RSK, and p70 S6 Kinase Substrates
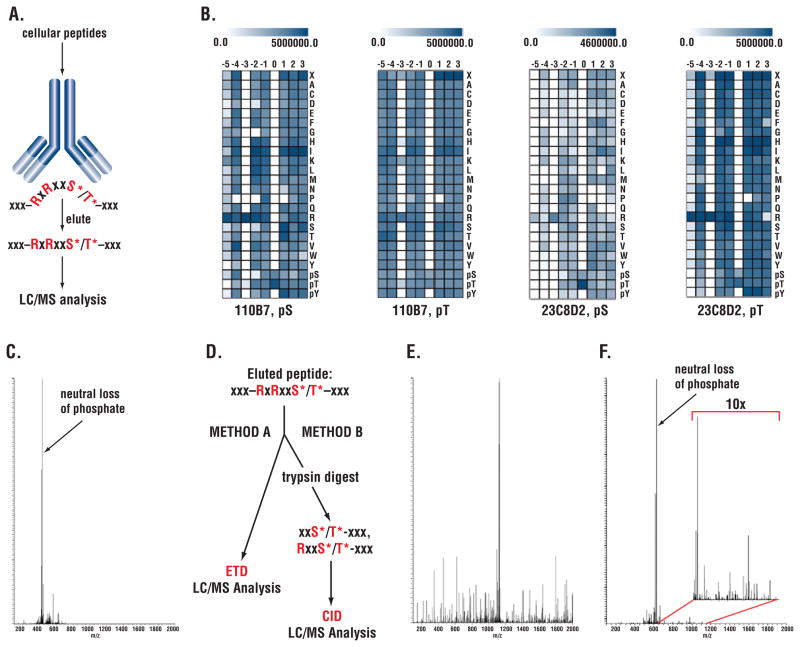
We used an approach described previously (14) to develop rabbit monoclonal antibodies directed against a peptide library of the form RxRxxS*/T*, where phosphorylated Ser (S*) or Thr (T*) are fixed at the 0 position and R is fixed at the −5 and −3 positions (Fig. 1A and Materials and Methods). Antibody specificity was determined by SpotELISA using arrays of phosphopeptide libraries where S* or T* was fixed at the 0 position (Fig. 1B). Results for the two antibodies used in this study, clone 110B7 and 23C8D2, are shown in Fig. 1B. Both antibodies were specific for R at the −3 position. Clone 23C8D2 was also specific for R at the −5 position, whereas clone 110B7 preferred R at this position but was less selective.
Figure 1. Dual strategies to address neutral loss using monoclonal antibodies recognizing the phosphorylated RxRxxS*/T* motif.
(A) Immunoaffinity purification of phosphopeptides using antibodies recognizing the RxRxxS*/T* motif with x being any amino acid. (B) Specificity of RxRxxS*/T* antibodies using SpotELISA on synthetic peptide libraries. Scale at top indicates ELISA reading; columns represent position relative to phospho-Ser/Thr at 0 position; rows represent fixed amino acid residue at the position. (C) CID spectrum of S6 peptide QIAKRRRLS*SLRASTSKSE showing unproductive neutral loss peak. (Q: Glutamine; I: Isoleucine; A: Alanine; K: Lysine; R: Arginine; L: Leucine; S: Serine, S*: phospho-Serine; T: Threonine; E: Glutamate. (D) Dual strategy to address neutral loss. (E) ETD spectrum of the S6 peptide shown in panel C. (F) CID spectrum of truncated S6 peptide after tryptic digest LS*SLRASTSKSE following IAP. The inset of 10x enlarged image shows efficient backbone fragmentation of the peptide.
MS/MS Analysis of RxRxxS*/T* Substrates and Neutral Loss
Because of neutral loss, immunoaffinity purification of cellular peptides using either clone 110B7 or 23C8D2 followed by MS/MS to identify phosphopeptides resulted in few identifications of peptides containing the full RxRxxS*/T* motif (15, 16). A typical spectrum of a ribosomal protein S6 Ser236 containing peptide is shown as an example (Fig. 1C).
In many cases, analysis using ETD to fragment the peptide backbone (Fig. 1D left side) yielded more productive spectra (compare Fig. 1E and 1C, same peptide). In an alternative approach (13)(Fig. 1D right side), peptides obtained after digestion with GluC, LysC, or chymotrypsin were first immunopurified with the 110B7 antibody, which recognizes both the full RxRxxS*/T* motif and the partial RxxS*/T* motif, and then digested with trypsin to cleave off R at −5 and −3 positions. The truncated peptides were then analyzed by mass spectrometry using CID. Because trypsin cleaves after R, most of these peptides had lost part or all of the R residues in the motif. Although neutral loss of phosphate was still substantial (Fig. 1F), it was usually not as severe as in the presence of nearby R residues, and sufficient peptide backbone fragmentation was obtained (Fig. 1F, insert). This method enabled the identification of phosphorylation sites that were not accessible to ETD analysis, which requires high charge states mostly associated with peptides of considerable length (12, 17). Analysis of tryptic peptides that were directly immunopurified with the 110B7 antibody yielded fewer identifications. These tryptic peptides resulted from incomplete protease digestion, perhaps due to the nearby S*/T* residue, so the peptide still contained a full or partial motif necessary to bind antibody. We next used these two approaches to identify proteins phosphorylated at the RxRxxS/T motif downstream of oncogenic RTKs.
Large-Scale Substrate Identification using ETD and Two-step Protease-Based CID Analysis
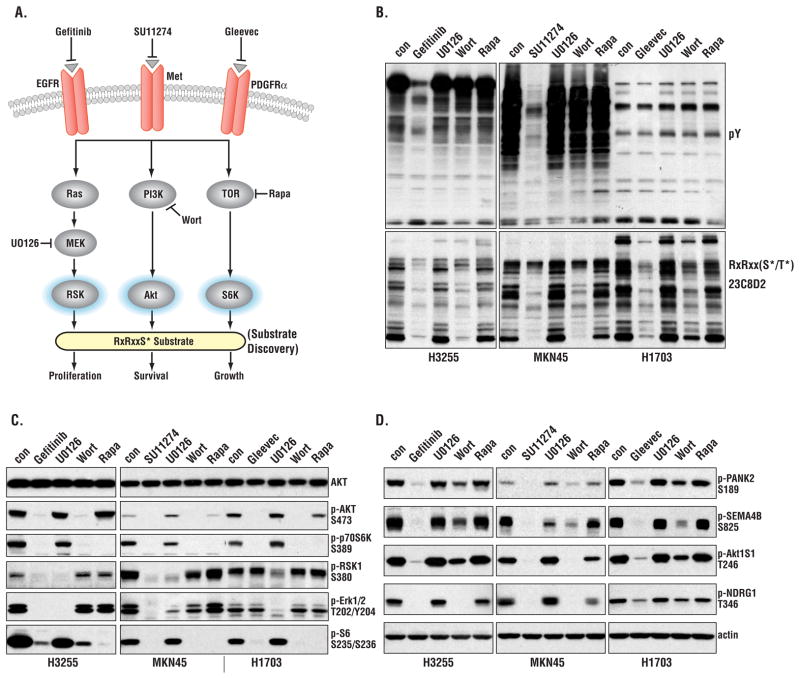
We studied three epithelial cancer cell lines, each of which depends on a different RTK for growth and survival: H1703, a non-small cell lung cancer (NSCLC) cell line driven by PDGFRα (18), MKN-45, a gastric cancer cell line driven by c-Met (19, 20), and H3255, a NSCLC cell line driven by EGFR (21). Fig. 2A shows the overall strategy used to identify substrates downstream of the PI3K-Akt, MAPK-RSK, and mTOR-p70 S6 kinase pathways using selective RTK inhibitors or inhibitors of each of the three pathways. As expected, each RTK inhibitor blocked downstream tyrosine phosphorylation as well as the phosphorylation of many proteins reactive with the RxRxxS*/T* antibody (Fig. 2B). Wortmannin inhibits the PI3K-Akt pathway and noticeably blocked phosphorylation on the RxRxxS/T motif in all three cell lines, whereas the MEK (mitogen-activated or extracellular signal-regulated protein kinase kinase) inhibitor U0126 and rapamycin were less effective at decreasing RxRxxS/T phosphorylation. This suggests that the majority of phosphorylations on RxRxxS/T occur downstream of PI3K-Akt. Control experiments (Fig. 2C) showed the expected selectivity of the drugs used to inhibit the Akt, RSK, and p70 S6 kinase pathways. Gefitinib, which inhibits EGFR, and SU11274, which inhibits c-Met, decreased Akt, RSK1, S6K, and extracellular signaling-regulated kinase (ERK) phosphorylation in H3255 and MKN45 cells, respectively. Gleevec, which inhibits PDGFRα, decreased Akt and S6K phosphorylation in H1703 cells, but not that of ERK or RSK (Fig. 2C).
Figure 2. Effects of RTKIs and pathway inhibitors on Akt-RSK-S6 kinase signaling.
(A) Strategy of probing signaling pathways with different inhibitors. Wort, wortmannin; Rapa, rapamycin. (B) Immunoblotting of three cancer cell lines treated with the indicated inhibitors. (C) Immunoblotting showing the effect of inhibitors on Akt, RSK, S6, and ERK phosphorylation. (D) Confirmation of MS/MS results by immunoblotting wih phosphospecific antibodies. Actin was used as loading control.
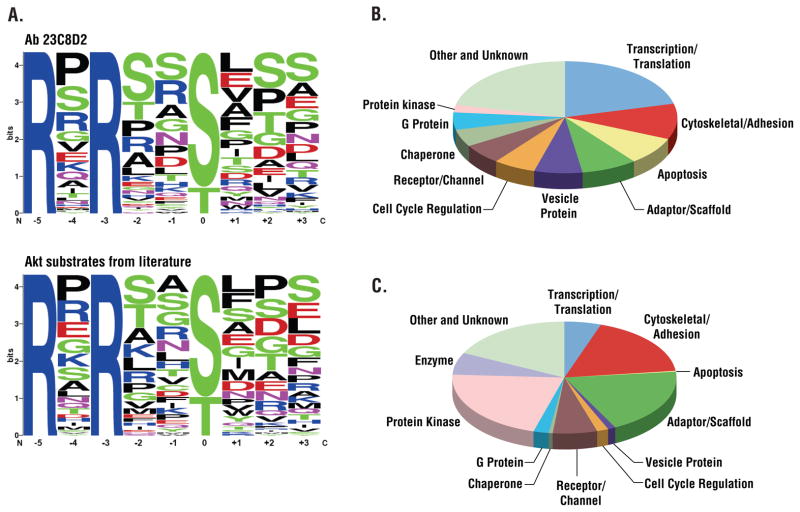
Lysates obtained from the three cell lines were digested with the protease GluC. Peptides from each sample were separately immunoaffinity-purified with the 110B7 or 23C8D2 antibodies and subjected to ETD mass spectrometric analysis. We identified 183 sites with the full RxRxxS*/T* motif, 81 of which (44%) had not previously been identified. In other studies we used the CID approach with the two-step protease digestion. Overall, using both ETD and CID, we identified over 1200 partial or full RxRxxS*/T* sites from the three cancer cell lines. Table S1 shows over 500 full (RxRxxS*/T*) motif sites from over 300 proteins and over 700 partial (RxxS*/T*) motifs from over 500 proteins identified from the three cell lines. When these sites were queried against PhosphoSitePlus™ [an on-line resource of post-translational modifications available at www.phosphosite.org (22)], more than 47% (575 sites) appeared to have not previously been identified. Data have been deposited in PhosphoSitePlus™ and are freely available. Table S1 summarizes each of the peptide assignments from all mass spectrometric experiments in this study. Analysis of the sequence context of the phosphorylated sites immunoprecipitated using the RxRxxS*/T* antibody (clone 23C8D2) revealed diverse RxRxxS*/T* sequences (Fig. 3A) similar to those of known Akt substrates (22)
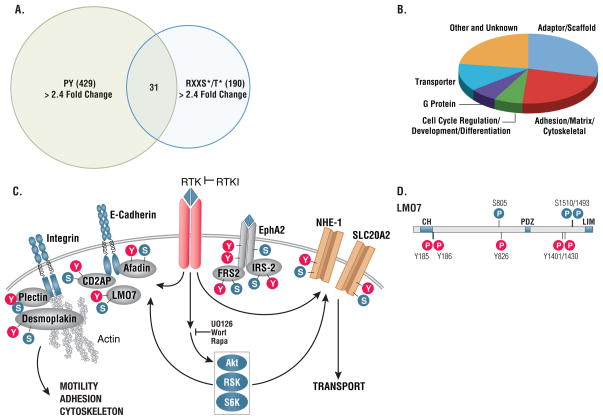
Figure 3. Comparison of sites and proteins identified using RxRxxS*/T* and pY antibodies.
(A) Amino acid distributions in singly phosphorylated phosphoserine peptides identified using the indicated RxRxxS*/T* antibody compared to known Akt substrates from the literature. Phosphopeptide sequences containing the full motif RxRxxS*/T* were aligned with phospho-serine/threonine at the 0 position, and the frequency of each amino acid at each flanking position was calculated and plotted. (B) Protein containing RxRxxS*/T* sites identified with 110B7 antibody arranged by protein type. Only the top 11 classes were shown, the rest were classified as other. (C) Proteins containing pY sites identified using the phosphotyrosine (pY-100) antibody arranged by protein type.
We compared the distribution of RxRxxS*/T* sites based upon protein function classification to the distribution of tyrosine-phosphorylated proteins identified in the same extracts by sequential immunoprecipitation with antibodies directed against RxRxxS*/T* and antibodies that recognized phosphotyrosine (Fig. 3B and C). Different classes of proteins were targeted by the Akt-like kinases and tyrosine kinases. Targets of Akt-like kinases include proteins involved in transcription, translation, apoptosis, and vesicle transport. In contrast, the main protein classes targeted by tyrosine kinases are protein kinases, adaptor and scaffold proteins, and cytoskeletal proteins. Nuclear proteins were also frequently found to be substrates of Akt-like kinases, whereas tyrosine kinases phosphorylate mostly membrane-proximal protein.
Tyrosine Kinase Activated RxRxxS*/T* Pathways
To quantify changes in RxRxxS*/T* phosphorylation status downstream of RTKs we used SILAC (stable isotope labeling with amino acids in cell culture) and the two-step protease approach combined with Orbitrap and VISTA software-based quantification (23). We obtained lysate from SILAC-labeled (24) MKN-45, H3255, or H1703 cells after no treatment or after their respective treatment with SU11274, gefitinib, or Gleevec. We also prepared SILAC-labeled lysates treated with inhibitors of the PI3K-Akt, MAPK-RSK, and mTOR-p70 S6 kinase pathways from each cell line. Untreated samples (from cells grown in medium containing heavy-isotope arginine and lysine) and inhibitor-treated samples (from cells grown in medium with natural isotopic abundance) were mixed in equal protein amounts.
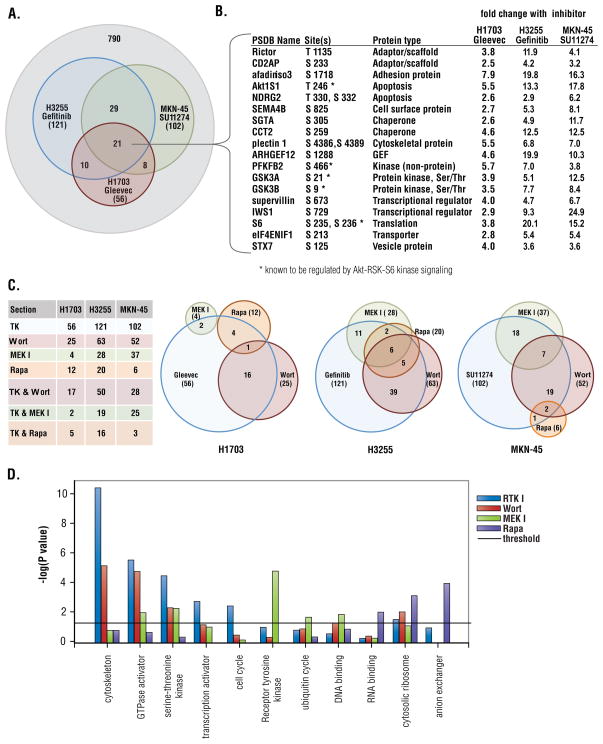
This analysis identified an extensive network of RxRxxS*/T* sites that changed downstream of the activated tyrosine kinase in each of the three cell lines (Table S2). For example, sites on over 100 proteins changed more than 2.4 fold (threshold of significant change, see Materials and Methods) in MKN45 cells treated with the c-Met inhibitor SU11274 and phosphorylation on over 120 proteins decreased upon treatment of H3255 cells with gefitinib (Fig. 4A). Phosphorylation of a core set of 21 proteins decreased downstream of all three RTKs (Fig. 4A and 4B). This core set of proteins included known Akt-RSK-S6-kinase substrates such as glycogen synthase kinase 3A and B (GSK3A/B), ribosomal protein S6, AKT1S1, and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2), a key enzyme involved in glycolysis. Previously unrecognized targets of these RTK pathways such as Rictor, a key component of mTOR complex 2 (mTORC2), and the chaperone SGTA were also identified (Fig. 4B).
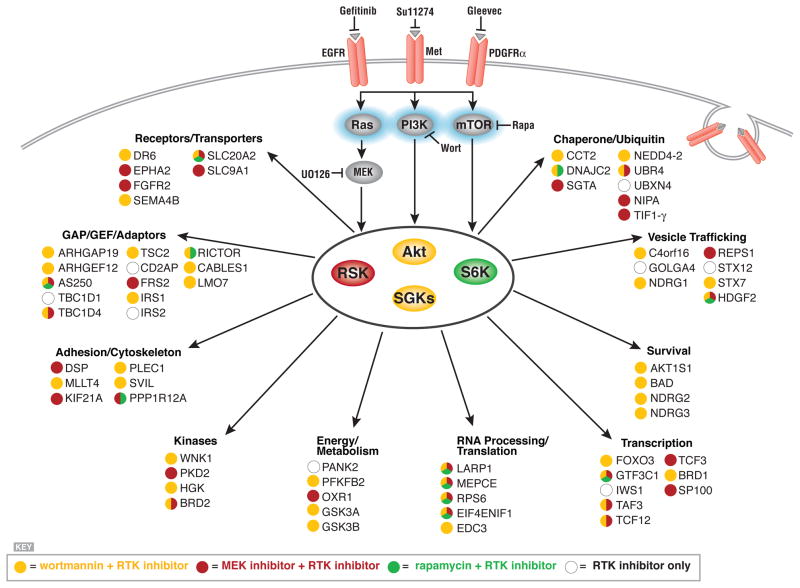
Figure 4. Pathway mapping downstream of EGFR, c-Met, and PDGFRα using RTKIs and pathway inhibitors.
(A) Overlap of RxRxxS*/T* protein phosphorylation inhibited by Gleevec, gefitinib, and SU11274 in H1703, H3255, MKN45 cells, respectively. (B) Proteins whose phosphorylation was decreased by corresponding RTK inhibitors in all three cancer cell lines, phosphorylation sites and fold change in phosphorylation status relative to untreated cells are shown. (C) Venn diagrams of protein phosphorylation decreased by inhibitors in three cancer cell lines. (D) Enrichment of GO terms among the targets of each inhibitor (relative to the total numbers in their respective categories) was determined using the Pathway Studio program by Fisher’s Exact Test. Significance is represented as the −log(P value); the significance threshold is 1.301 = −log (P = 0.05).
Phosphorylation of many sites was also decreased by wortmannin, U0126, or rapamycin (Table S2). Inhibitor analysis allowed a rough mapping of the circuitry downstream of the different RTKs (see Fig. 4C and 4D), consistent with the profile of inhibitor sensitivity (Fig. 2C). For example, in both H3255 and MKN-45 cells, gefitinib and SU11274 blocked signaling through all three pathways (Fig. 2C) and this is confirmed in the large numbers of substrates with phosphorylation status sensitive to these inhibitors (Fig. 4C). In contrast, in H1703 cells, Gleevec inhibited the Akt and S6K pathways but not the MAPK-RSK pathway (Fig. 2C), reflected in the small number of sites for which phosphorylation decreased in response to both Gleevec and the MEK inhibitor U0126 (Fig. 4C).
Overall, roughly half of the RxRxxS*/T* sites showing decreased phosphorylation in response to a RTKI could be mapped to one or more of these three pathways. Phosphorylation of known substrates showed expected patterns of inhibitor sensitivity, for example, phosphorylation of GSK3A/B and AKT1S1 are both wortmannin-sensitive, and phosphorylation of the ribosomal protein S6 was inhibited by both wortmannin and rapamycin.
Functional categories significantly enriched among the different targets sensitive to each inhibitor are shown in Figure 4D. Cytoskeletal proteins, GTPase activators, and Ser/Thr kinases were enriched among proteins showing decreased phosphorylation following treatment with RTKIs and wortmannin. RTKs and proteins involved in ubiquitination were enriched among targets sensitive to inhibition of MEK, and proteins involved in RNA binding, cytosolic ribosome, and anion exchange were enriched among targets sensitive to inhibition of mTOR.
Using phosphospecific antibodies prepared to several identified sites, we confirmed changes observed by mass spectrometry in the phosphorylation status of pantothenate kinase 2 (PANK2, phosphorylated on Ser189), semaphorin-4B (SEMA4B, phosphorylated on Ser825), AKT1S1 (phosphorylated on Thr246), and N-myc downstream regulated 1 (NDRG1, phosphorylated on Thr346) in response to RTK and specific pathway inhibitors (Fig. 2D).
Connections Downstream of Oncogenic RTKs
We ranked substrates by fold change in phosphorylation status after exposure to RTKI or pathway inhibitor in the three cell lines. The proteins showing the greatest change, and their sensitivity to pathway inhibitors, are shown in Fig. 5. These data suggest the existence of previously unexplored connections between the AGC signaling pathways and various cellular processes and proteins. These included such processes as energy metabolism, cell survival and apoptosis, and trafficking, and various Ser/Thr kinases, receptors, adaptors, and scaffolding proteins.
Figure 5. Connections iidentified using RTKIs and pathway specific inhibitors.
Proteins inhibited by two or more RTKIs and the indicated pathway inhibitor(s) are grouped by protein function. Color coding indicates sensitivity to pathway inhibitor.
Sequential immunoprecipitation with antibodies directed against phosphotyrosine and antibodies that recognized RxRxxS*/T* identified proteins showing a greater than 2.5 fold change in phosphorylation status at both tyrosine and RxRxxS/T residues (Fig. 6A, Table S3). These proteins were highly enriched for membrane proximal signaling components, such as adaptors, transporters, and cytoskeleton proteins (Fig. 6B and 6C), and included transporters, such as NHE-1 and SLC20A2, and signaling adaptors, such as IRS2, FRS2, CDAP2, and afadin (Fig. 6C). In many cases the regulated sites of tyrosine and Ser/Thr phosphorylation were in close proximity, suggesting targeting of common functional activities (see LMO7, in Fig. 6D).
Figure 6. Proteins subject to both RxRxxS*/T* and tyrosine phosphorylation.
(A) Overlap of proteins showing greater than 2.4 fold change on RxRxxS/T and tyrosine phosphorylation sites. (B) Functional categorization of proteins phosphorylated on both RxRxxS/T and tyrosine sites downstream of RTKs. (C) Examples of protein types phosphorylated on both RxRxxS/T and tyrosine sites downstream of RTKs.. (D) Tyrosine and RxRxxS/T phosphorylation sites on individual proteins phosphorylated downstream of RTKs are often clustered in close proximity as shown for LOM7 at Ser805 and Tyr826, Ser1493/1501 and Tyr1401/1430.
Functional Assay of Substrates: Cell Viability Screen
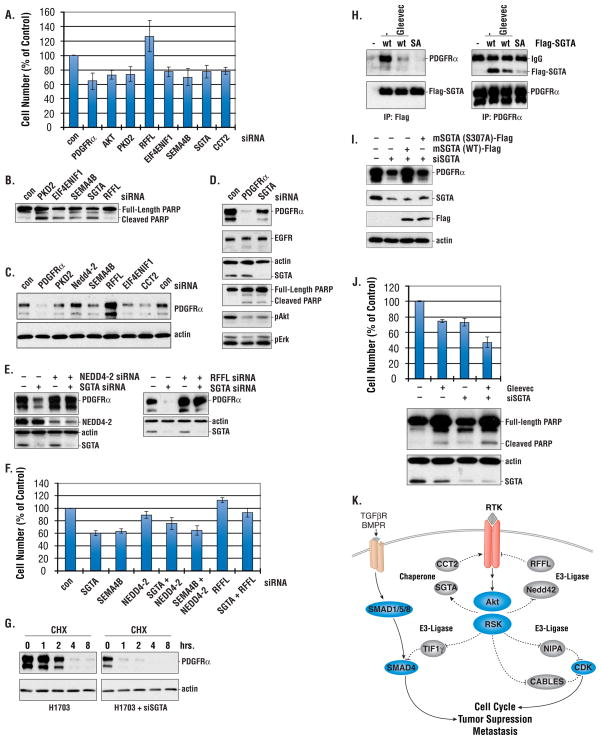
We investigated the roles of 33 proteins in cancer cell viability based on their phosphorylation sensitivity to RTKIs and pathway inhibitors (Table S4). We examined the effects of siRNA-mediated knockdown of these candidates in H1703 cells. PDGFRα siRNA was used as a positive control for cell viability, because inhibition of PDGFRα by Gleevec induces cell death (18). A siRNA scored as positive if the cell number following transfection was > 20% below or above the cell number of the control siRNA. Using this criterion, we identified six proteins that affected cell viability (Fig. 7A and 7B). siRNAs directed against PKD2 (a protein kinase), EIF4ENIF1 (a nuclear import factor), SEMA4B (a membrane protein), and SGTA and CCT2 (both chaperones) scored as decreasing cell viability, as did siRNA knock-down of PDGFRα and Akt. siRNA inhibition of RFFL, a molecule with E3 ligase activity that is involved in endosome sorting, showed increased cell viability.
Figure 7. Functional assay of AGC-family substrates.
(A) Cell viability after siRNA transfection. H1703 cells were collected 60 hrs after transfection with the indicated siRNA, stained with Trypan Blue and counted. Numbers are mean ± SD from three different wells. Controls were transfected with scrambled siRNA. Results are representative of three independent experiments. (B) Immunoblotting of H1703 cells transfected with siRNAs. 60 hrs after transfection, cell lysates were made and probed with PARP antibody. (C) Immunoblotting of H1703 cells transfected with siRNAs. Cell lysates were made as in 7b and probed with PDGFRα antibody. (D) Immunoblotting of H1703 cells transfected with PDGFRα and SGTA siRNAs. 60 hrs after transfection, cell lysates were collected and probed with the antibodies indicated. (E) Immunoblotting of H1703 transfected with siRNAs. Cell lysates were made as in 7b and probed with the antibodies indicated. (F) Cell viability of H1703 cells transfected with siRNAs. siRNA transfected cells were collected and counted as in 7a. (G) Half life of PDGFRα in H1703 cells transfected with SGTA siRNA or treated with Gleevec. 48 hrs after siRNA transfection or overnight treatment with Gleevec (1 mM), cells were treated with the protein synthesis inhibitor cycloheximide (CHX, 0.1 mg/ml) for the indicated time and cell lysates were collected and probed with PDGFRα antibody. (H) SGTA S305 phosphorylation is required for SGTA/PDGFRα interaction. Cells were transfected with Flag-tagged wildtype SGTA (WT) or S305A mutant (SA). 32 hrs after transfection, cells were treated with Gleevec (1 uM) for 2.5 hrs and cell lysates were immunoprecipitated with Flag (left panel) or PDGFRα (right panel) and probed with antibodies indicated. (I) SGTA Ser305 is required for PDGFRα stability. SGTA siRNA knockdown cells were complemented with murine SGTA WT, or the S307A mutant. Cell lysates were collected and probed with antibodies indicated. (J). Gleevec sensitivity of H1703 cells transfected with SGTA siRNA. 32 hrs after SGTA siRNA transfection, cell were treated with Gleevec (1 uM) for 24 hrs and cells were counted as in 7a. Results are representative of 2 independent experiments. (K). A model for the effect of putative Akt substrates on PDGFRα stability, cancer cell growth and death.
PDGFRα is essential for H1703 cell survival (18), prompting us to test whether these proteins affect cell viability by regulating PDGFRα abundance. PDGFRα protein level was decreased by SGTA and CCT2 siRNAs (Fig. 7C and 7D); in contrast, PDGFRα abundance increased in cells transfected with siRNA directed against either of the two E3 ligases, RFFL and Nedd4-2, whereas siRNAs directed against PKD2 and SEMA4B had no effect on PDGFR abundance. Although SGTA siRNA reduced PDGFR protein level, decreased phosphorylated Akt and increased PARP cleavage (a marker of apoptosis), it had little effect on EGFR abundance (Fig. 7D). Double knockdown of SGTA in combination with that of either NEDD4-2 or RFFL showed that loss of NEDD4-2 or RFFL overcame the effects of SGTA siRNA on PDGFR (Fig. 7E) and partially restored cell viability (Fig. 7F), suggesting a role of these E3 ligases in regulation of PDGFR abundance. Together, these results highlight the importance of pathways regulating RTK abundance on survival of cancer cells.
Phosphorylation of SGTA at Ser305 regulates PDGFRα stability
To examine the effect of SGTA on PDGFRα protein stability, we measured PDGFRα turnover in H1703 cells transfected with siRNA directed against SGTA. PDGFRα half-life was reduced in SGTA knockdown cells and in cells treated with Gleevec, suggesting that SGTA may stabilize PDGFRα (Fig. 7G). Our MS/MS analysis identified ser305 on SGTA as sensitive to RTKIs (Table S2). To examine the role of SGTA-Ser305 in PDGFRα stability we developed a phosphorylation-specific antibody to Ser305 (fig. S1a). We found that phosphorylation of SGTA-Ser305 was increased by PDGF, an increase that was inhibited by Gleevec (fig. S1b). Western analysis showed that phosphorylation of SGTA-Ser305 was also decreased by SU11274 and U0126 in MKN45 cells (fig. S1c), consistent with the results from MS/MS SILAC analysis. Analysis of mouse embryonic fibroblast (MEF) cells that lacked different AKT isoforms showed that phosphorylation of Ser305 was decreased in MEF AKT2 KO cells, suggesting that AKT2 could be a candidate SGTA Ser305 kinase (fig. S1D).
We next investigated the mechanisms whereby SGTA phosphorylation at Ser305 regulates PDGFRα stability. We found that Flag-tagged wild-type SGTA (Flag-SGTA) coimmunoprecipitated with PDGFRα, and this interaction was inhibited by Gleevec (Fig. 7H). The association between SGTA and PDGFRα was reduced in H1703 cells transfected with a Flag-SGTA S305A mutant, suggesting that phosphorylation of SGTA at Ser305 promotes interaction of SGTA with PDGFRa.
As noted previously, transfection of SGTA siRNA resulted in decreased PDGFRα abundance in H1703 cells (Fig. 7I) and increased cleaved caspase-3 positive cells (fig. S1E). The effect of SGTA siRNA on PDGFRα abundance and cleaved caspase activity were partially reversed by cotransfection with a murine wildtype SGTA construct that is resistant to human SGTA siRNA but not by murine SGTA lacking the Ser305 phosphorylation site (S307A) in mouse.
The effects of SGTA on PDGFRα stability prompted us to investigate whether destabilization of PDGFRα could enhance the sensitivity of H1703 cells to Gleevec. Indeed, transfection of H1703 cells with SGTA siRNA enhanced cell death in response to Gleevec (Fig. 7J). This suggests that destabalization of PDGFRα could further enhance the antitumor effects of Gleevec.
Discussion
A focused view of Akt-RSK-p70 S6 Kinase Signaling
This study provides a large-scale identification and analysis of substrates phosphorylated by the Akt, RSK, and S6 family of protein kinases. Neutral loss has limited the analysis of the basic phosphopeptide substrates characteristic of the AGC family of proteins kinases, restricting our view of this important class of protein kinase substrates. Here we used two different approaches to overcome neutral loss, one involving improved MS/MS peptide fragmentation and the other using proteases to remove basic residues. Our approach largely overcomes the neutral loss problem but has certain limitations, including antibody-dependent phosphopeptide preferences, ETD analysis favoring peptides of high charge state, and the general problem of peptides being either too small or large for MS/MS analysis depending upon the combination of proteases used. Despite these limitations, the method enabled a broad view of signaling activity across multiple AGC kinase pathways and allowed quantitative measurements of pathway activity and the identification of many previously unknown substrates.
Pathway Inhibitors Define Core Networks
We identified and characterized Akt-like substrates in three cancer cell lines, each dependent on a different receptor tyrosine kinases, and identified 200 proteins with phosphorylation sites that decrease more than 2.4 fold following treatment with RTKI. We identified a set of “core substrates” activated by multiple RTKs: phosphorylation of 21 proteins was inhibited by each of the three different RTKIs (Fig. 4A and 4B) and that of 66 proteins was inhibited by at least two different RTKIs (Fig. 5). Phosphorylation of approximately half of the sites affected by RTKIs was also inhibited by blocking either the PI3K, MAPK, or mTOR signaling pathways, indicating the approach enriches proteins acting in these pathways. Among the 200 proteins with phosphorylation that was inhibited by RTKIs, 77 were also inhibited by wortmannin, 40 by the MEK inhibitor U0126, and 21 by rapamycin. Phosphorylation on AGC target sites of nine proteins was inhibited by all three pathways including ribosomal protein S6, EIF4ENIF1, and LARP1. This redundant targeting may render these elements more difficult to inhibit and suggests that understanding this circuitry will be important to the future development of targeted cancer therapeutics.
Oncogenic RTK Signaling: Connections to Akt-like Substrates
This study maps an extensive signaling network operating downstream of the oncogenic receptor tyrosine kinases, EGFR, c-Met and PDGFR involving phosphorylation at basophilic motifs typical of Akt, RSK, and p70 S6 kinase signaling. Our analysis indicates that the majority of the substrates identified function in various related cellular activities (Fig. 5). We identified many inputs from RTKs to receptors, adaptors, scaffolds, and G-proteins acting at the interphase between the cytoskeleton and plasma membrane, suggesting a possible role in control of cell shape and motility. Other prominent targets of RTK signaling included components of pathways regulating protein stability, vesicle transport, translation, transcription, cell survival, and energy metabolism (fig S2). Some of these substrates were previously identified (25) in a study of signaling networks activated by EGF in HeLa cells, confirming the importance of these molecules as effectors of RTK signaling.
We identified multiple inputs from RTK signaling to E3 ubiquitin ligases that connect to cell cycle progression, TGFb/BMP Smad signaling, and protein stability and trafficking. Both the E3 ligase NIPA and the tumor suppressor protein CABLES1 regulate cell cycle progression by targeting cyclins and cyclin dependent kinases (26, 27). Another possible connection occurs through the E3 ligase, TIF1γ, which adds mono-ubiquitin to Smad4, removing it from the pool transducing TGFb and BMP signaling (27). TIF1γ could thus provide a link from RTKs to the TGFb and BMP pathways.
Our approach allows analysis of different protein modifications from the same sample. For instance we used antibodies directed against phosphotyrosine and antibodies directed against RxRxxS*/T* motifs to identify proteins targeted by both tyrosine and Ser/Thr kinases (Table S3). Identification of such proteins suggests that rapid membrane-proximal tyrosine phosphorylation may be followed by Ser/Thr phosphorylations that limits or adjusts signal duration or intensity. This kind of feedback regulation is known to limit insulin signaling through the phosphorylation of IRS1 by p70 S6 kinase at Ser residues (28).
Regulation of RTK Stability
Downregulation of growth factor RTKs plays an important role in terminating RTK signaling (8). Upon ligand binding, RTKs are removed from the cell surface through endocytosis and either recycled back to the plasma membrane or degraded in the lysosome. Although defective RTK endocytosis leads to prolonged RTK signaling and has been linked to malignancy (29), the pathways and molecules targeted remain largely unknown. Based upon our phosphoproteomic network mapping and siRNA screens, two chaperones, SGTA and CCT2 were identified as important for cell viability. Further investigation revealed that SGTA was critical for stabilization of PDGFRα and received inputs from the PI3K and MAPK pathways, which converged on Ser305. Mutational analysis suggested that Ser305 phosphorylation is important for the interaction of SGTA with PDGFRα and for its ability to stabilize PDGFRα. siRNA knockdown of the E3 ligases Nedd4-2 and RFFL overcame the effect of SGTA on PDGFR abundance, suggesting a model in which oncogenic RTKs act through MAPK- and PI3K- dependent pathways to stabilize their own expression by coordinating the balance between chaperone-mediated receptor stabilization and ubiquitin-mediated degradation (Fig. 7K).
Molecular chaperones have emerged as a promising target in cancer therapy (29). However, the adaptations that sensitize cancer cells to chaperone inhibitors remain unknown. Our network analysis places SGTA downstream of RTK signaling and suggests that SGTA may participate in a regulatory loop acting to enhance cancer cell sensitivity to chaperone inhibitors. Given the importance of SGTA for androgen receptor signaling in prostate cancer (30), the discovery of SGTA as a critical regulator of PDGFR stability could have important therapeutic implications.
In non-small cell lung cancer patients with EGFR mutations, the EGFR inhibitor gefitinib provides substantial therapeutic benefit. However, most patients develop resistance through EGFR mutations that block drug binding or by kinase switching, wherby a different kinase, such as c-Met, substitutes for EGFR growth and survival signaling (31). Robust, but not durable, response to RTKIs highlight the need to inhibit pathway activity at multiple levels (4). Our findings suggest that selective targeting of the chaperone and ubiquitin pathways should act together with RTKIs to further inhibit oncogenic kinase signaling. In support of this idea, we show that siRNA knockdown of SGTA enhances cell death by Gleevec, suggesting that approaches targeting both kinase activity and stability may confer additional clinical benefit.
Materials and Methods
SpotELISA
Synthetic biotinylated phosphopeptides were arrayed in 96 well avidin plates, 0.1 pmol peptide per well. Peptides in each well contained a residue fixed at one position (−5 to +3) relative to the centrally fixed (position 0) phospho-Serine or phospho- Thr residue, besides Arginine fixed at −5 and −3 positions. The peptide plates were blocked by BSA-PBST, and probed with motif antibody 110B7 and 23C8D2 at 4 °C overnight, then the plates were washed and probed with Europium-labeled anti-rabbit lgG. Plates were read at 615nm with a Wallac Victor plate reader.
siRNA transfection, viability assay, and immunoblotting
H1703 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and transfected with siRNA (Dharmacon) using Mirus reagents and protocol. Transfection controls were cells treated with a scrambled control siRNA from Cell Signaling Technology. Viability of cells was measured 60 hrs after transfection by trypan blue staining and counting on a hemocytometer. Lysates for immunoblotting were prepared following the standard protocols of Cell Signaling Technology. In brief, cells were collected in lysis buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 ug/ml leupeptin, protease inhibitors). The cell lysate was sonicated for 2 × 5 second, and cleared by centrifugation for 10 min at 14,000 rpm. Protein concentration is measured using Coomassie protein assay reagent (Pierce). Equal amount of protein lysate (20 – 30 ug) is loaded on SDS-PAGE, probed with antibodies indicated in the figures. All the antibodies used in this study, except for Nedd4-2 (Abcam), were from Cell Signaling Technology. The pSGTA (S305) antibody was raised by immunizing rabbit with a synthetic peptide containing phospho-Serine 305.
Cell culture and drug treatment
Cells were cultured in DMEM medium or RPMI 1640 medium supplemented with 10% fetal bovine serum. For SILAC experiments, cells were grown for at least 5 passages in DMEM medium with either heavy isotope lysine (L-Lysine:2HCl (U-13C6) and arginine (L-Arginine:HCl (U-13C6) (untreated cells) or normal isotope lysine and arginine (inhibitor-treated cells). H3255 cells and MKN-45 cells were starved overnight before harvesting. For drug treatment, cells were treated with the following drugs for 2.5 hrs: RTKI (1 uM), wortmannin (0.1 uM), rapamycin (10 nM), MEK inhibitor U 0126 (10 uM). RTKIs included: Gleevec (PDGFRα inhibitor) for H1703 cells, gefitinib(EGFR inhibitor) for H3255 cells, and SU11274 (Met inhibitor) for MKN-45 cells. Controls were treated with DMSO vehicle.
Phosphopeptide immunoaffinity purification and analysis by LC-MS/MS mass spectrometry
Phosphopeptide immunoaffinity purification from different cell lines was performed as described previously (32). A detailed description of the procedures can be found in Supplemental Data.
Supplementary Material
Phosphorylation of SGTA at S305.
Akt-RSK-S6-kinase networks downstream of RTKs
Summary of all peptide assignments from all mass spectrometric experiments in this study
Proteins with RxRxxS*/T* or RxxS*/T* sites phosphorylation changed by inhibitors
Proteins with RxRxxS*/T* or RxxS*/T* sites and pY sites both phosphorylation changed by RTK inhibitors (overlap).
33 proteins tested in siRNA inhibition experiments.
Acknowledgments
We thank other members of the Cell Signaling Technology, Inc. (CST) for help and fruitful discussions, especially Roberto Polakiewicz, Ting-Lei Gu, and Klarisa Rikova. We thank Claudia Hoffman, Rachel Poor, Kim Anderson, and David Comb for graphic assistance.
Footnotes
Author contributions: A.M., Y.L., A.G., J.V., J.M., K.S., J.Z., A.P., J.M.R. performed experiments; Y.W., J.K., P.H. performed bioinformatics analysis; L.C., S.G. J.R. and M.J.C. designed experiments, analyzed data and wrote the paper.
Accession numbers: Phosphoproteomics data has been deposited with the PhosphoSitePlus protein modification database, http://ms.phosphosite.org:5080/.
Reference and Notes
- 1.Gazdar AF, Minna JD. Deregulated EGFR signaling during lung cancer progression: mutations, amplicons, and autocrine loops. Cancer Prev Res (Phila Pa) 2008;1:156–160. doi: 10.1158/1940-6207.CAPR-08-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 6.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 7.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 8.Bache KG, Slagsvold T, Stenmark H. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 2004;23:2707–2712. doi: 10.1038/sj.emboj.7600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 10.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villén J, et al. Manuscript in preparation. [Google Scholar]
- 14.Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakiewicz RD, Comb MJ. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 15.Leitner A, Foettinger A, Lindner W. Improving fragmentation of poorly fragmenting peptides and phosphopeptides during collision-induced dissociation by malondialdehyde modification of arginine residues. J Mass Spectrom. 2007;42:950–959. doi: 10.1002/jms.1233. [DOI] [PubMed] [Google Scholar]
- 16.Palumbo AM, Tepe JJ, Reid GE. Mechanistic insights into the multistage gas-phase fragmentation behavior of phosphoserine- and phosphothreonine-containing peptides. J Proteome Res. 2008;7:771–779. doi: 10.1021/pr0705136. [DOI] [PubMed] [Google Scholar]
- 17.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, Christensen JG, Settleman J, Haber DA. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 22.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 23.Bakalarski CE, Elias JE, Villen J, Haas W, Gerber SA, Everley PA, Gygi SP. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7:4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Bassermann F, von Klitzing C, Illert AL, Munch S, Morris SW, Pagano M, Peschel C, Duyster J. Multisite phosphorylation of nuclear interaction partner of ALK (NIPA) at G2/M involves cyclin B1/Cdk1. J Biol Chem. 2007;282:15965–15972. doi: 10.1074/jbc.M610819200. [DOI] [PubMed] [Google Scholar]
- 27.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 28.Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, Marette A. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 30.Buchanan G, Ricciardelli C, Harris JM, Prescott J, Yu ZC, Jia L, Butler LM, Marshall VR, Scher HI, Gerald WL, Coetzee GA, Tilley WD. Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Res. 2007;67:10087–10096. doi: 10.1158/0008-5472.CAN-07-1646. [DOI] [PubMed] [Google Scholar]
- 31.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 32.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phosphorylation of SGTA at S305.
Akt-RSK-S6-kinase networks downstream of RTKs
Summary of all peptide assignments from all mass spectrometric experiments in this study
Proteins with RxRxxS*/T* or RxxS*/T* sites phosphorylation changed by inhibitors
Proteins with RxRxxS*/T* or RxxS*/T* sites and pY sites both phosphorylation changed by RTK inhibitors (overlap).
33 proteins tested in siRNA inhibition experiments.