Abstract
During interphase, the transport receptor importin-β carries cargoes into the nucleus, where RanGTP releases them. A similar mechanism operates in mitosis to generate a gradient of active spindle assembly factors around mitotic chromosomes. Importin-β and RanGTP have been implicated in additional cellular processes, but the precise roles of the Ran/importin-β pathway throughout the cell cycle remain poorly understood. We implemented a FRET-based, high-throughput small molecule screen for compounds that interfere with the interaction between RanGTP and importin-β and identified importazole, a 2,4-diaminoquinazoline. Importazole specifically blocks importin-β-mediated nuclear import both in Xenopus egg extracts and cultured cells, without disrupting transportin-mediated nuclear import or CRM1-mediated nuclear export. When added during mitosis, importazole impairs the release of an importin-β cargo FRET probe and causes both predicted and novel defects in spindle assembly. Together, these results indicate that importazole specifically inhibits the function of importin-β, likely by altering its interaction with RanGTP. Importazole is a valuable tool to evaluate the function of importin-β/RanGTP pathway at specific stages during the cell cycle.
Importin-β transport receptors, which comprise at least 22 members in vertebrates (1), bind to cargo molecules and mediate their import or export through nuclear pores (2). Directionality of transport depends on the nature of the receptor as well as the asymmetric distribution of nucleotide states of the small GTPase Ran, which is GTP-bound in the nucleus due to the chromatin interaction of its guanine exchange factor (GEF) RCC1, and GDP-bound in the cytoplasm where its GTPase activating protein, RanGAP, is localized. The founding member of this family, importin-β, together with its partner importin-α, recognize nuclear localization signal (NLS)-containing cargo molecules and transport them into the nucleus where RanGTP binds directly to importin-β, causing a conformational change that releases importin-α and NLS-cargoes. In addition to its vital interphase functions, importin-β and Ran are also important regulators during mitosis, contributing to chromatin-mediated spindle assembly (3–5). During mitosis, importin-β has an inhibitory function towards NLS-containing spindle assembly factors, binding them in the cytoplasm and impairing their microtubule-stabilizing or organizing activities. However, RanGTP remains enriched around condensed mitotic chromosomes in mitosis and generates a gradient of released cargoes that triggers spindle assembly (6, 7). The importin-β/RanGTP pathway has also been implicated in a variety of other cellular processes including postmitotic nuclear envelope assembly, nuclear pore complex assembly, protein ubiquitylation, and primary cilium formation (8–12).
Small-molecule inhibitors provide a promising approach to study the multifunctional importin-β/Ran pathway in living cells by acting like conditional mutations that allow disruption of a protein with temporal precision, at any phase of the cell cycle. Compounds targeting microtubules or microtubule-based motors have been successfully used to dissect their mitotic functions and also gain mechanistic insight into the complex events of mitosis. For example, the drug monastrol inhibits kinesin-5 (Eg5) (13) and causes a loss of spindle bipolarity, consistent with this motor’s microtubule cross-linking and sliding function, as well as the results of immunodepletion and antibody microinjection experiments (14, 15). However, monastrol has also provided novel insights through drug-washout experiments and in combination with other inhibitors, to assess how spindle bipolarity and microtubule attachment to chromosomes are established (16, 17) and how the cell division cleavage plane is positioned (18). Because of its fundamental role in many cellular functions including mitosis, nuclear transport is also an attractive target for small molecule inhibition. However, despite the importance of this process, surprisingly few inhibitors have been identified. With respect to nuclear export, leptomycin is a potent inhibitor, but binds covalently to its target, preventing washout experiments (19). Peptide inhibitors (20) and small molecule peptidomimetic inhibitors (21) of importin-α/β have been designed and used to study nuclear import in vivo. However, these inhibitors are not cell permeable. Recently, a new cell permeable small molecule inhibitor of the RanGTP/importin-β interaction named karyostatin 1A that binds specifically to importin-β and blocks importin-β-mediated nuclear import has been identified (22). However, the effects of karyostatin 1A on mitotic events have not yet been demonstrated.
To gain a better understanding of the functions of the importin-β/Ran pathway in mammalian cells without the limitations associated with microinjection of proteins or antibodies, or the time required for efficacy of RNA interference or peptide inhibitors (7), we aimed to identify a cell permeable, specific and reversible small molecule inhibitor that would provide high temporal precision, allowing dissection of the role of importin-β/RanGTP throughout the cell cycle. Here we report the discovery of importazole, which meets these criteria and suggests at least one previously uncharacterized role for this pathway in mitosis.
RESULTS AND DISCUSSION
Identification of importazole in a high throughput screen
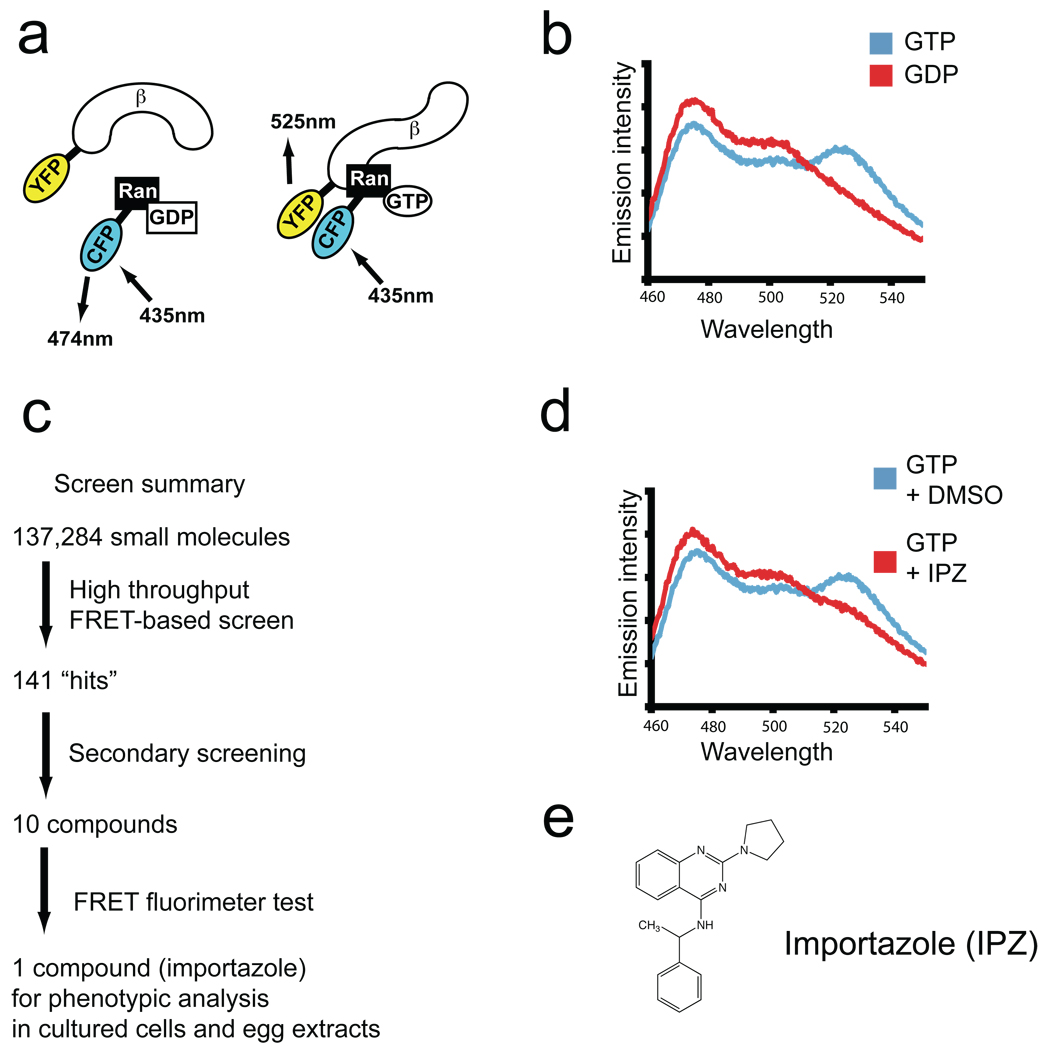
We applied a reverse chemical genetic high-throughput screen (HTS) to identify compounds that affect the interaction between RanGTP and importin-β using a fluorescence resonance energy transfer (FRET)-based assay with CFP-tagged Ran and YFP-tagged importin-β. These proteins bind one another only when CFP-Ran is GTP-bound, which can be detected by changes in FRET (Figure 1, panels a and b). When CFP-Ran is incubated with RCC1, GTP, and YFP- importin-β, and the mixture is excited with 435 nm fluorescence in a fluorometer, a strong FRET signal is generated, as indicated by a decrease in the fluorescence intensity at 475 nm (the emission wavelength of CFP) and an increase in the fluorescence intensity at 525 nm (the emission wavelength of YFP). No FRET signal is generated if GDP is substituted for GTP, and nucleotide-specific interaction could also be observed biochemically, as S-tagged YFP-importin-β pulls CFP-Ran out of solution only in the presence of GTP (Figure S1). These results demonstrate that the FRET signal generated by CFP-RanGTP and YFP-importin-β is due to a physical interaction dependent upon the nucleotide state of Ran, and that our approach could be used to identify compounds that interfere with the interaction between CFP-RanGTP and YFP-importin-β, resulting in a reduced FRET signal.
Figure 1.
A high-throughput screen identifies importazole as an inhibitor of FRET between CFP-Ran and YFP-importin-β. (a) Schematic of the fusion proteins that bind and undergo FRET in the presence of Ran-GTP but not Ran-GDP. (b) Fluorescence emission of the FRET pair detected between 460 nm and 550 nm following excitation at 435 nm, showing strong emission of CFP (475 nm) in the presence of GDP (red curve) that decreases in the presence of GTP (blue curve) concomitant with an increase at the emission wavelength of YFP (525 nm), indicative of FRET. (c) Summary of the screen. Of 137,284 small molecules screened in duplicate using the FRET-based assay, 141 putative hits were subjected to three secondary screens designed to eliminate false positives. Of the 10 compounds remaining after the secondary screens, only a single compound reproducibly diminished the FRET signal generated by CFP-Ran and YFP-importin-β in the original assay (d). (e) The structure of importazole, a 2,4-diaminoquinazoline.
The assay was tested for suitability for HTS using a 384-well format and a fluorescence plate reader. We calculated FRET ratios (IFRET/ICFP) for each well and determined two commonly used statistical parameters, the coefficient of variation (CV), which was 0.95% and 1.24% for reactions containing the GDP and GTP, respectively, and the Z’ value, which was 0.81, indicating that our assay was robust and appropriate for HTS (23). To facilitate rapid data analysis, we developed software to generate color-coded plate maps to identify compounds that reduced the FRET ratio by both an increase in CFP emission and a decrease in YFP emission, thereby eliminating compounds that altered the FRET ratio by contributing their own fluorescence at wavelengths in the range of our probes (Figure S2).
In total, we screened 137,284 compounds in duplicate (Figure 1, panel c), and selected 141 “hits” for further analysis. Compounds that showed activity upon retesting in the original assay were analyzed in a unimolecular CFP and YFP FRET-based assay using a YIC sensor (6) to confirm that the observed changes were not due to non-specific quenching or augmentation of either CFP or YFP emission. In a third assay, each compound was tested for its tendency to form aggregates that non-specifically inhibit β-lactamase (24). The 10 compounds that survived the secondary assays were obtained in larger quantities and tested again in the original CFP-Ran/YFP-importin-β FRET assay using a spectrofluorimeter. Only one of these compounds, a 2,4-diaminoquinazoline which we named “importazole”, reproducibly disrupted the FRET signal generated by CFP-Ran and YFP-importin-β and was analyzed further (Figure 1, panels d and e).
Importazole binds importin-β in vitro
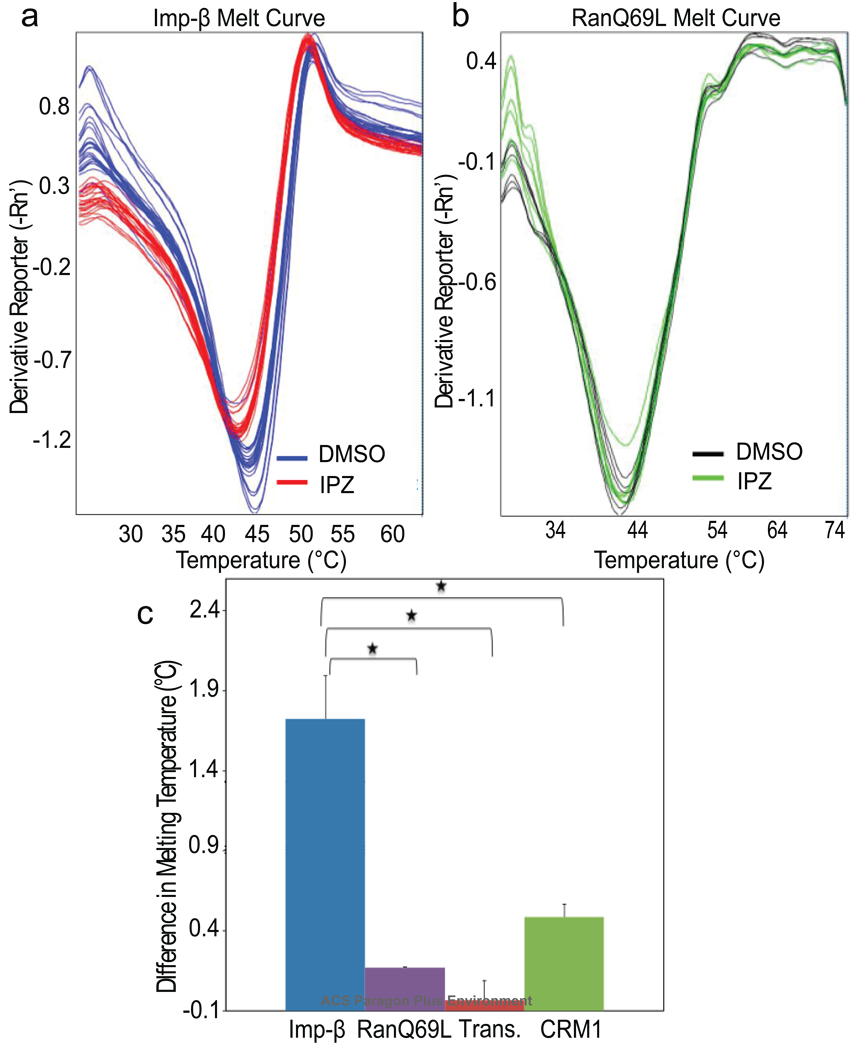
Although importazole blocked the FRET interaction between CFP-RanGTP and YFP-importin-β in vitro, it did not obviously affect the binding of the two proteins in pull-down assays (Figure S1). To begin elucidating the mechanism of importazole action, we tested whether importazole could alter the ability of importin-β to protect RanGTP from RanGAP-stimulated hydrolysis in vitro (25, 26). Binding curves calculated from these data do not indicate that importazole disrupts the RanGTP/importin-β interaction, and if anything, suggest that importazole may slightly stabilize the complex (Figure S3). The inability of importazole to disrupt the RanGTP/importin-β interaction is not entirely surprising considering the multiple large interaction surfaces between the two proteins (27). One possible explanation for the importazole-induced FRET change is that importazole binding causes a conformational change that disrupts the CFP-RanGTP/YFP-importin-β FRET interaction without preventing binding. To test whether importazole binds to importin-β in vitro, we used a fluorescent thermal shift assay with the dye Sypro® Orange, since small molecule binding is expected to affect the thermal stability of a protein (28). Importazole reduced the melting temperature of importin-β by 1.72+/− 0.27 °C (Figure 2, panels a and b), but was unaffected by a related compound of comparable hydrophobicity that did not interfere with CFP-RanGTP/YFP-importin-β FRET (compound 3016, Figure S4, panels c and d, and data not shown). In contrast, importazole did not significantly affect the melting curves of related importin-β family members transportin and CRM1, or that of RanGTP, suggesting that importazole binds preferentially to importin-β (Figure 2, panel c, Figure S5). Going forward, a crystal structure of the RanGTP/importin-β complex in the presence of importazole will likely be necessary to fully elucidate its biochemical mechanism of action.
Figure 2.
Importazole binds specifically to importin-β. (a) Negative first derivatives of melting curves of 2 µM importin-β in the presence of 50 µM importazole or DMSO where the minima indicate the melting temperature. Melting curves show the results from six experiments conducted in quadruplicate using the Applied Biosystems 7500 qPCR machine. (b) Negative first derivatives of melting curves of 2 µM RanQ69L in the presence of 50 µM importazole or DMSO as control where the minima indicate the melting temperature. (c) Mean changes in melting temperature of 2 µM importin-β, RanQ69L, transportin and CRM1 in the presence of 50 µM importazole. Error bars indicate standard error; asterisks denote statistical significance (p < 0.01).
Importazole disrupts importin-β/RanGTP-mediated nuclear import
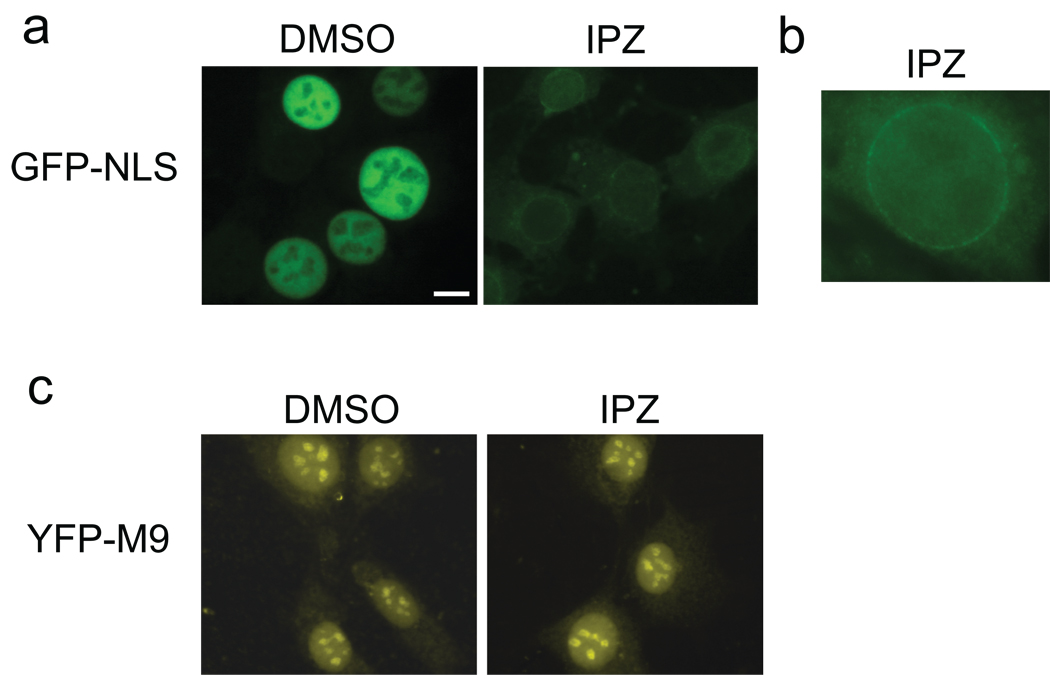
If importazole binds importin-β and affects the RanGTP/importin-β interaction, it should inhibit the nuclear import of any protein bearing a classical NLS. We first tested this prediction using permeabilized HeLa cells, in which nuclear import of a GFP-NLS reporter can be reconstituted in vitro (29). Digitonin-permeabilized cells were incubated with a GFP-NLS reporter plus Xenopus laevis egg extracts as a source of soluble transport factors including Ran, importin-α, and importin-β. Whereas rapid nuclear accumulation of GFP-NLS occurred in the presence of the solvent DMSO, importazole blocked import and the reporter became enriched at the nuclear envelope, where RanGTP functions to induce cargo release from importin-β (30, 31) (Figure 3, panels a and b). In contrast, importazole did not block nuclear import mediated by transportin, an importin-β family member that utilizes the M9 import signal together with RanGTP to import hnRNP proteins (Figure 3, panel c) (32).
Figure 3.
Importazole inhibits importin-β NLS-mediated nuclear import, but not transportin M9-mediated import. (a) NLS-GFP (importin-β import substrate) was added with Xenopus egg extract to permeabilized HeLa cells and assayed by fluorescence microscopy for nuclear import in the presence of DMSO or 100 µm importazole. (b) NLS-GFP accumulation at the nuclear rim in the presence of importazole. (c) M9-YFP (transportin import substrate) in the same assay. In all figures, scale bar = 10 µm.
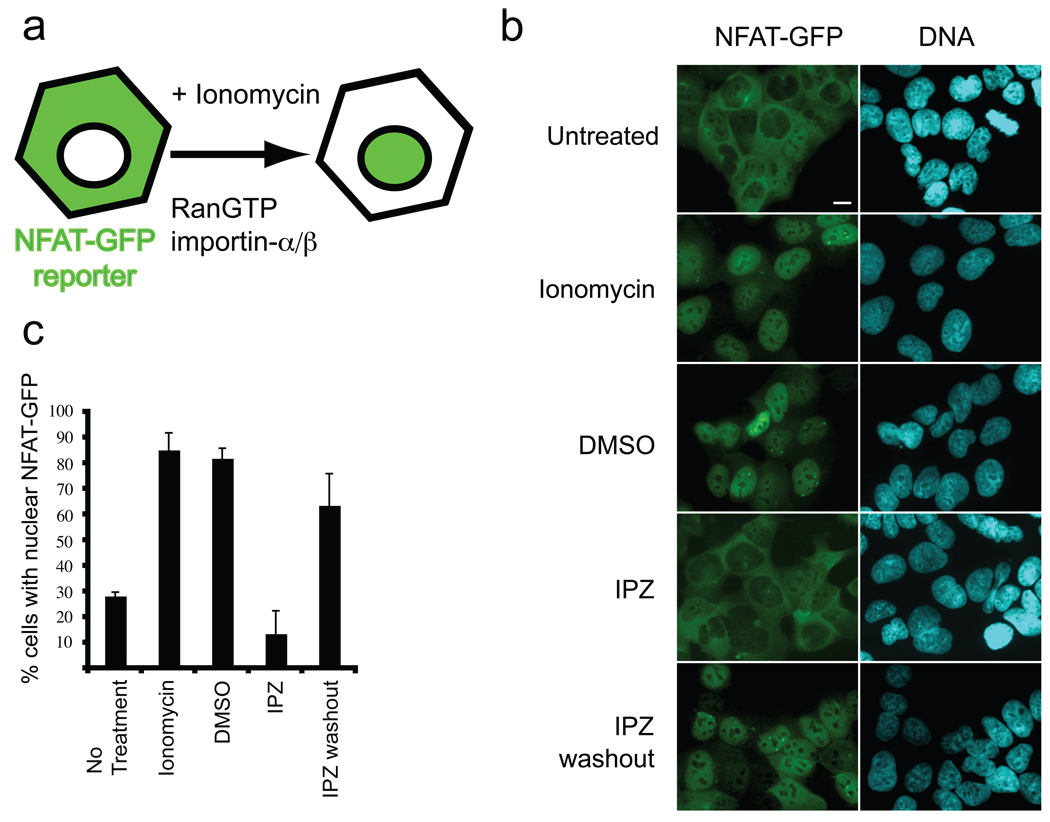
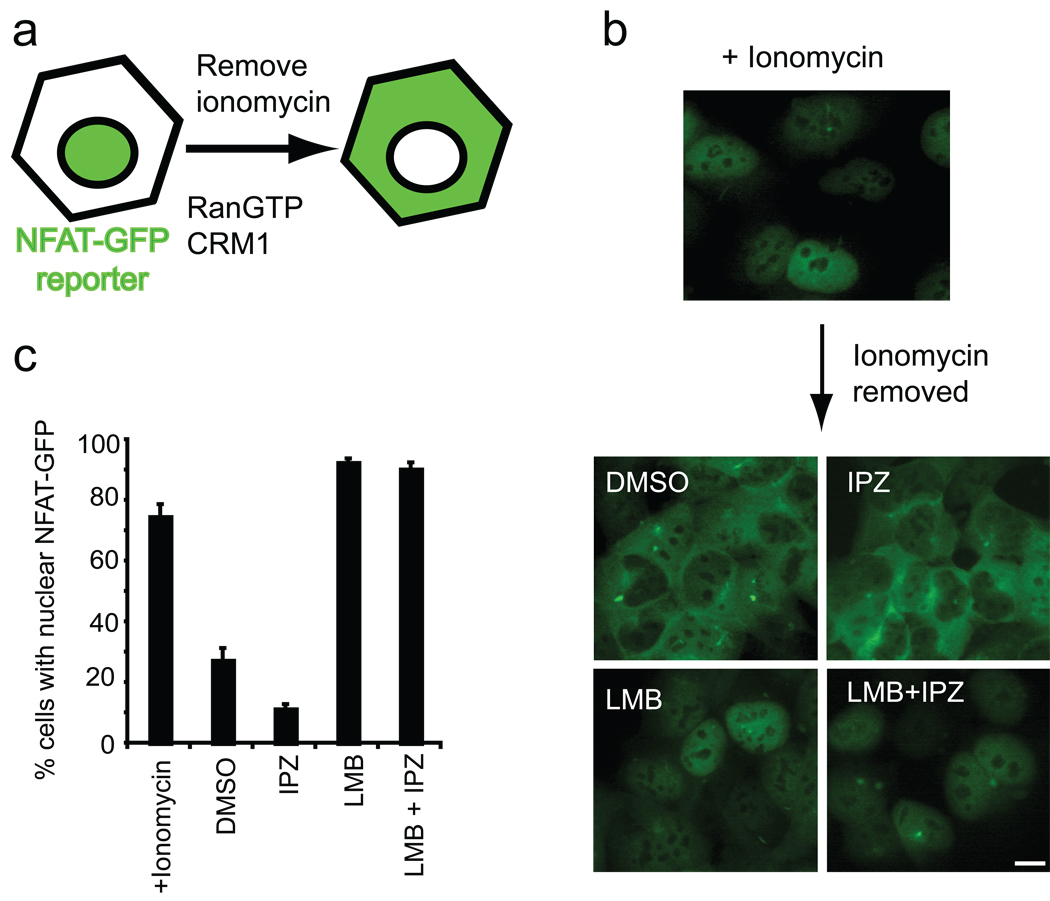
To investigate whether importazole is cell permeable and active in living human cells, we generated a cell line that stably expresses a GFP-tagged version of the transcription factor NFAT, which shuttles between the nucleus and the cytoplasm in a calcium-regulated manner (33, 34) and is imported by importin-α/β and exported by CRM1 (35, 36). At steady state NFAT is predominantly cytoplasmic. An increase in cytoplasmic calcium induced by the ionophore ionomycin leads to the accumulation of NFAT in the nucleus (Figure 4, panel a). NFAT import can be reverted upon ionophore withdrawal, (Figure 5, panel a) providing an inducible system ideal for testing the effects of importazole on importin-β-mediated nuclear import and CRM1-mediated nuclear export, both of which are dependent upon RanGTP.
Figure 4.
Importazole reversibly blocks importin-β-mediated nuclear import in living cells. (a) Schematic showing that the GFP-tagged, NLS-containing transcription factor NFAT enters the nucleus upon treatment with the ionophore ionomycin in a RanGTP- and importin-β-dependent manner. (b) HEK 293 cells stably expressing GFP-NFAT were treated with DMSO or 40 µm importazole for 1 hour prior to a 30 min treatment with ionomycin to induce nuclear import. Importazole was washed out and after 1 hour prior to ionomycin re-treatment. (c) Results were quantified as the percentage of cells with nuclear NFAT-GFP. N=3, 100 or more cells counted under each condition. Bars represent standard error.
Figure 5.
Importazole does not inhibit Crm1-mediated nuclear export. (a) Schematic illustrating that upon removal of the ionophore ionomycin, GFP-NFAT exits the nucleus in a RanGTP- and Crm1-dependent manner. (b) Cells were treated with ionomycin to induce nuclear import of NFAT-GFP, then washed and treated with DMSO, importazole, leptomycin B, or importazole + leptomycin B for 1 hour. (c) Results were quantified as the percentage of cells with nuclear NFAT-GFP. N=3, 100 or more cells counted under each condition. Bars represent standard error.
Cells were pretreated with 40 µm importazole for 1 hour followed by 30 minutes of ionomycin treatment in the continued presence of importazole. Whereas control cells treated with DMSO or the control compound 3016 displayed a robust nuclear accumulation of the NFAT-GFP reporter after ionomycin addition, there was virtually no import of NFAT-GFP in importazole treated cells (Figure 4, panel b, quantified in panel c and Figure S6). Importazole displayed an IC50 of approximately 15 µm for inhibition of NFAT-GFP import (data not shown). This effect was reversible upon importazole washout, which restored ionomycin-induced import of NFAT-GFP to near control levels (Figure 4, panels b and c). Thus, it should be possible to use importazole in drug-washout experiments to study the Ran/importin-β pathway in cells. The reversibility of importazole required 1 hour of recovery time between washing out the drug and adding ionomycin, and did not require new protein synthesis (data not shown).
To further assess the specificity of importazole, we tested its effects on CRM1-mediated export of NFAT-GFP. Export of NFAT-GFP occurred efficiently in the presence or absence of importazole, but was blocked by leptomycin B, a specific CRM1 inhibitor (37) (Figure 5 panel b, quantified in panel c). Importantly, when cells were treated with both leptomycin B and importazole upon ionomycin washout, NFAT-GFP was still restricted to the nucleus (Figure 5, panels b and c), confirming that importazole treatment does not non-specifically damage the nuclear envelope allowing proteins to leak out into the cytoplasm. Consistent with the concentration of importazole sufficient to impair nuclear import, we found that importazole has an IC50 of approximately 22.5 µm in HeLa cells following treatment over a 24-hour period (Figure S7).
Taken together our nuclear import experiments indicate that importazole is likely specific for importin-β-mediated protein import. Although we have not tested importazole’s effect on all importin-β family members, no effect on transportin-mediated import or CRM1-mediated export was detected. Furthermore, these results also suggest that importazole does not impair RCC1-dependent loading of Ran with GTP or the function of RanGTP itself since the export function of CRM1 critically depends on the formation and function of RanGTP.
Importazole blocks spindle assembly in Xenopus egg extracts, but does not affect pure microtubules
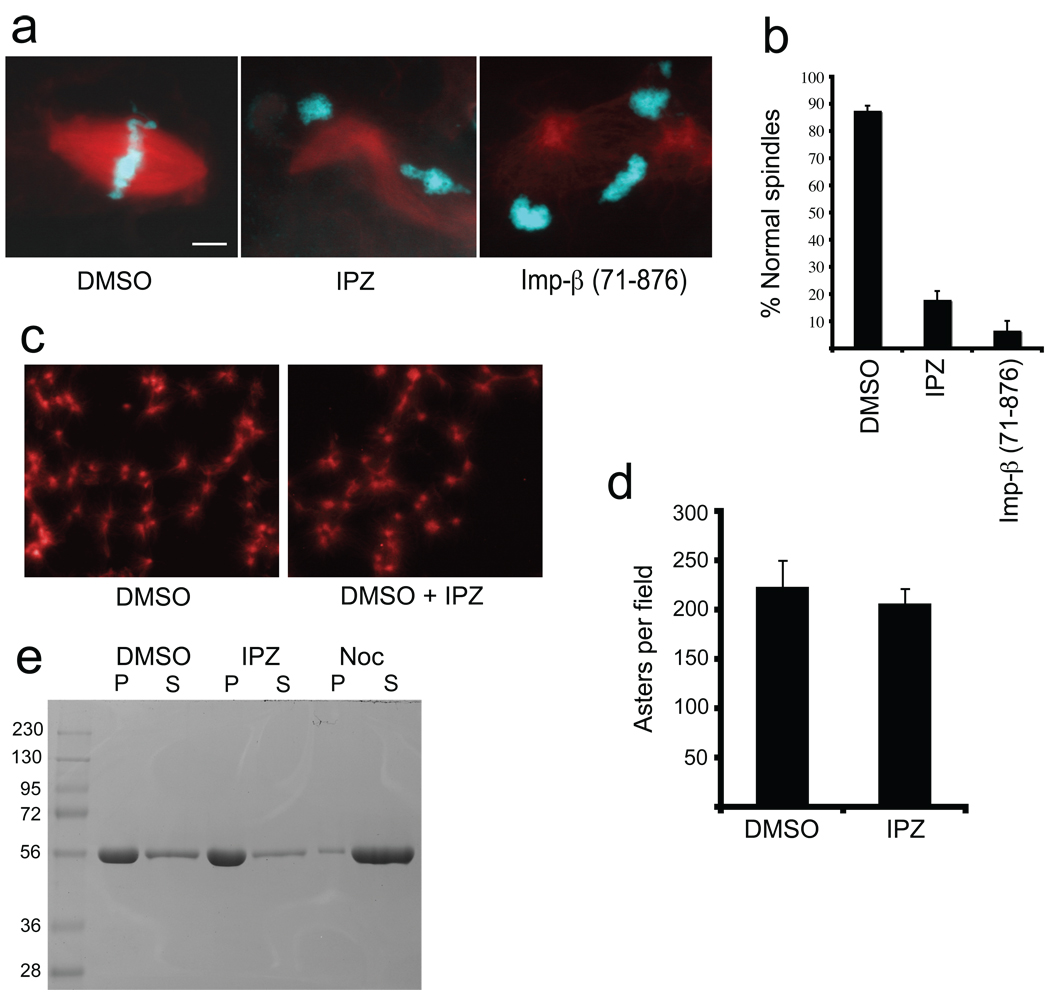
A specific inhibitor of importin-β/RanGTP should also disrupt mitosis. We first tested importazole in metaphase-arrested Xenopus egg extracts, which rely heavily on a RanGTP gradient for spindle assembly around sperm chromosomes. Addition of 100 µm importazole, but not the solvent DMSO, strongly inhibited spindle assembly, preventing normal bipolar microtubule structures from forming around 80% of sperm nuclei (Figure 6, panels a and b). The effect was similar to that of adding a truncated importin-β (amino acids 71–876), a version that no longer binds to RanGTP and therefore sequesters its cargoes (38). Although importazole significantly weakened spindle microtubule density, it was not a general microtubule inhibitor, since it did not impair the formation of microtubule asters in the extract induced by the microtubule stabilizing agent DMSO (Figure 6, panels c and d) or affect the polymerization of pure microtubules, in contrast to nocodazole (Figure 6, panel e). Thus, importazole caused dramatic effects on spindle assembly consistent with the known role of the importin-β/RanGTP pathway in the Xenopus egg extract system, and is not a general microtubule inhibitor.
Figure 6.
Importazole impairs spindle assembly in Xenopus egg extracts but does not affect pure microtubule polymerization. (a) Spindle assembly reactions containing X-rhodamine labeled tubulin in the presence of DMSO, 100 µM importazole, or a truncated form of importin-β that is unable to bind to RanGTP. Microtubules are red and DNA is blue. (b) Quantification of the percentage of normal spindle structures. N=3, 100 structures counted under each condition. Bars represent standard error. (c) Aster assembly induced by addition of 5% DMSO to extracts containing X-rhodamine labeled tubulin in the presence of DMSO or importazole. (d) Quantification of the number of asters per field. 10 fields were counted under each condition. (e) DMSO induced pure tubulin polymerization assay. Reactions were supplemented with additional DMSO, importazole, or nocodazole, and microtubules pelleted through a sucrose cushion and samples from the pellet (P) and supernatant (S) analyzed by SDS PAGE.
Importazole impairs mitotic cargo release and reveals novel functions for the importin-β/RanGTP pathway in human cells
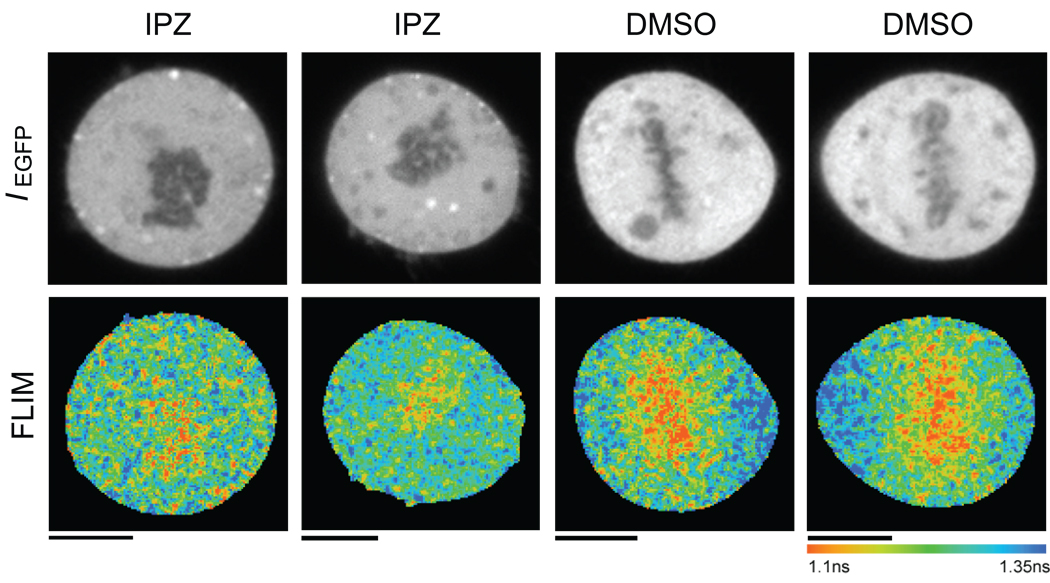
A major advantage of a cell-permeable importin-β/RanGTP inhibitor is its potential for dissecting novel roles of this pathway in dividing human cells, which also provide a system to analyze mitotic gradients of released cargos using FRET probes (7). If importazole disrupts the interaction of importin-β with RanGTP, then the chromatin-localized FRET gradient of the cargo probe Rango should be reduced, since it undergoes FRET when released from importin-β in HeLa cells (7). As predicted, the difference in fluorescence lifetime of the donor GFP of the Rango-3 probe between the chromosomes and distal cytoplasm was significantly reduced in the presence of importazole compared to controls, from an average of 0.12 +/− 0.4 ns to 0.07+/−0.03 ns due to reduced FRET (Figure 7, p-value: 1.3 ×10−7). Thus, importazole impairs mitotic importin-β cargo release in HeLa cells.
Figure 7.
Importazole disrupts mitotic cargo release monitored by the FRET probe Rango. Donor fluorescence (top panels) and pseudo-colored FLIM images (bottom panels) of mitotic HeLa cells expressing the Rango-3 FRET sensor. Rango-3 displays a greater fluorescence lifetime around the chromosomes of cells treated with importazole compared to that of cells treated with DMSO, resulting from importazole’s disruption of sensor release from importin-β. N=3, 30 cells counted for each condition.
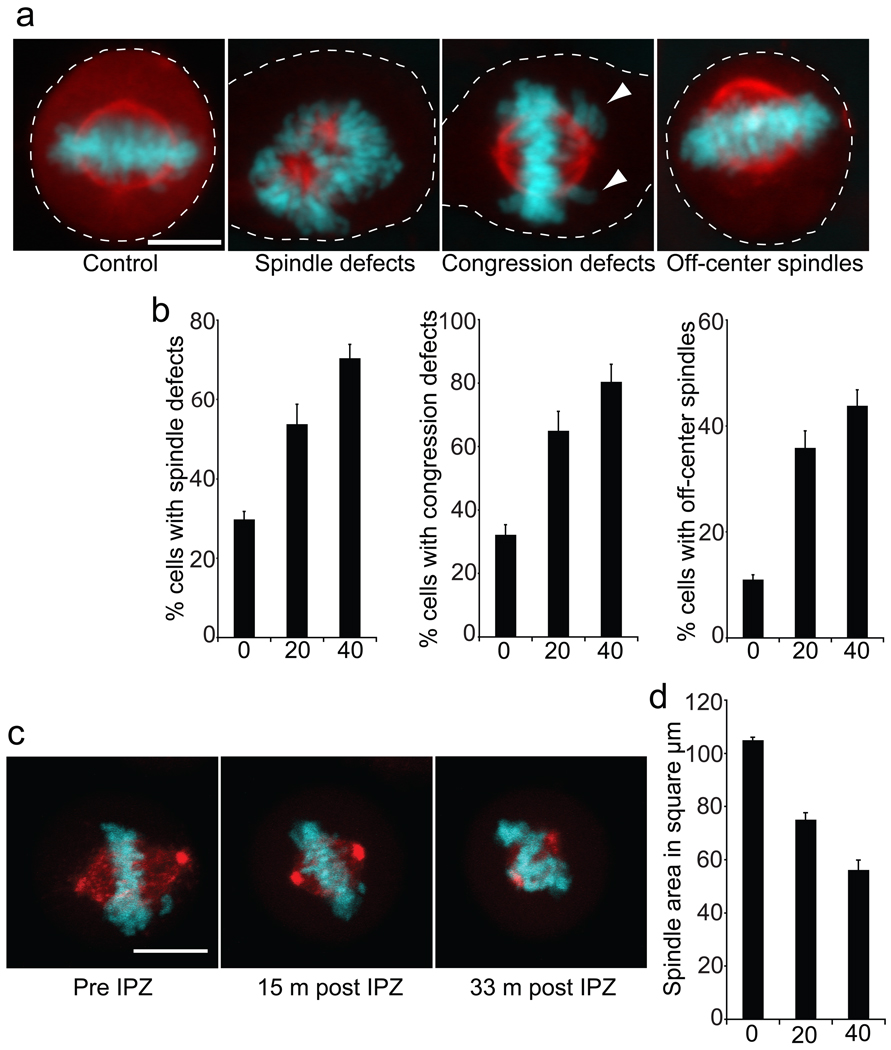
To examine the consequences of importazole on mitosis, HeLa cells treated for one hour were fixed and stained for tubulin and chromosomes. Control metaphase figures displayed robust spindles with a mean area of 105 µm2, and were centrally located within the cell with chromosomes aligned on the metaphase plate (Figure 7, panels a and d). Importazole treatment caused dose-dependent defects in spindle assembly, chromosome alignment, and spindle size (Figure 8, Supplemental Movie 1). Interestingly, importazole also led to spindle positioning defects, with more than 40% of the cells displaying off-center spindles (Figure 8, panels a and b). Spindle positioning was not previously attributed to the Ran pathway and this phenotype may be a consequence of astral microtubule disruption by importazole (data not shown). Previous studies have most likely not revealed this role of the Ran pathway in mitosis because they were performed in cell free systems such as Xenopus extract where spindle positioning could not be assessed. The discovery of this spindle misalignment phenotype demonstrates the importance of importazole as a tool to study the Ran pathway in mitosis.
Figure 8.
Importazole disrupts mitotic spindles in living HeLa cells. (a) Asynchronously growing cultures were treated with DMSO or importazole for 1 hour prior to fixation and staining for DNA (blue) and tubulin (red). Note defects including chromosome congression (white arrowheads point to misaligned chromosomes) and spindle positioning upon importazole treatment. Dashed white lines indicate cell boundaries. (b) Quantification of spindle defects in cells treated with DMSO, 20 µm importazole, or 40 µm importazole. N=5. In each case, 100 metaphase cells were counted and the fraction of those displaying defects were scored. (c) Time-lapse fluorescence microscopy of a metaphase HeLa cells treated with 50 µm importazole. Frames were captured every 3 minutes. See Supplemental Movie 1. (d) Asynchronous HeLa cells were treated with 0 to 40 µm importazole for 1 hour prior to fixation, and the size of the spindle in mitotic cells was measured. N=4, 100 metaphase spindles were measured per condition. Bars represent standard error.
The Ran pathway members Ran and importin-β are highly conserved, and an inhibitor of the RanGTP/importin-β interaction may have considerable value as a research tool across multiple species. Additionally, as the Ran pathway has been shown to be upregulated in some forms of cancer (39), importazole may have some potential as a therapeutic compound. Development of more potent, related compounds should allow a more complete disruption of the Ran/importin-β interaction as well as limit any possible non-specific effects of compound treatment, further increasing the value of these inhibitors in both the academic and medical fields. Overall, we have shown importazole to be an effective inhibitor of the Ran/importin-β interaction in vitro and in cells with great potential for future use as a tool to study the Ran pathway in mitosis.
METHODS
Protein expression and purification
pET30a-derived constructs encoding importin-β with an N-terminal YFP fusion (pKW1532), a CFP-Ran fusion (pKW1543), and importin-β (pKW485) were transformed into BL21 cells (Invitrogen). Additionally, pQE32-derived Ran constructs (pKW356 [WT Ran], pKW 590 [RanQ69L]), a pQE9-derived Crm1 construct (pKW812), and a pQE60-derived transportin construct (pKW738) were transformed in to SG13 cells. All constructs were induced with IPTG at room temperature. Harvested cells were lysed using a French press. Fusion proteins were purified with Ni NTA resin using a standard protocol followed by gel filtration. RCC1 was purified as described previously (40).
FRET assay
The following reaction buffer was used for all FRET assays, including the high-throughput screen: 1X PBS, 5 % glycerol, 2 mM MgCl2, 1 mM DTT, 0.01 % NP-40. For standard CFP-Ran/YFP-importin-β FRET assays, 50 – 100 nM CFP-Ran mixed with 20 nM RCC1 and 200 µm GDP or 200 µm GTP was immediately followed by addition of 50 – 100 nM YFP-importin-β. The reaction was excited using a Fluorolog 3 spectrofluorometer with 435 nm fluorescence and the emission was read between 460 nm and 550 nm. For the high-throughput screen, the concentrations of reaction components were as follows: CFP-Ran: 62.5 nM; YFP-importin-β: 62.5 nM; GTP or GDP: 200 µm; RCC1: 20 nM
The high-throughput screen
The screen was carried out in collaboration with the Small Molecule Discovery Center (SMDC) at the University of California, San Francisco. Compounds were from ChemBridge, ChemDiv, SPECS, ChemRX, and Microsource. The complete content of this library can be found through the Small Molecule Discover Center website (http://smdc.ucsf.edu/). A detailed screening protocol can be found in the supplemental methods section. The software used to analyze the screening data is available upon request.
Secondary screening
141 hit compounds from the primary screen were tested for non-specific effects on fluorescence with FRET probe YIC that contains the importin-β-binding domain of importin-α flanked by CFP and YFP. When unbound in solution, this probe undergoes intramolecular FRET (6). In the second assay, we tested our 141 hits for nonspecific inhibition due to aggregation using a β-lactamase-based assay as described (24). Importazole was found to be soluble up to approximately 100 µm in water. Additionally, importazole was characterized by mass spectrometry and NMR to confirm its identity and purity.
Fluoresecent thermal shift assay
Experiments were performed using Applied Biosystems StepOnePlus™ Real-Time PCR (RT-PCR) System as previously described (28, 41). Protein stocks were diluted in PBS and added 70% v/v to a Microamp® Fast 96-well Reaction Plate and maintained on ice. Compounds (importazole and control compound 3016) were then added at 30% v/v in 3% DMSO. Freshly prepared 100X water based-dilution of Sypro® Orange Protein Gel Stain was then added at 1% v/v to reach a final reaction volume of 20 µl. Samples were mixed by gentle pipetting. After sealing the plates with Microamp™ Optical Adhesive Film, the plate was subjected to a heating cycle composed of a 10 sec prewarming step at 25°C and a gradient between 25°C and 95°C with a 0.3°C ramp. Data was analyzed using the StepOnePlus™Software v2.1.
Cell lines and tissue culture
A GFP-NFAT expression plasmid (pKW520) was generated by inserting a BamH I/Hind III-cleaved NFATC1 cDNA fragment into pEGFPC1 (Clontech) digested with Bgl II and Hind III. The plasmid was a gift of K. Reif. The construct was stably transfected into HEK 293 cells and a single clone expressing moderate levels of NFAT-GFP was selected and maintained in Opti-mem media (Gibco) plus 4 % fetal bovine serum, 1 % penicillin/streptomycin, and 200 µg/ml G418. HeLa cells were grown and maintained according to standard protocols.
Nuclear import with permeabilized HeLa cells
HeLa cells were permeabilized and treated with an import reporter and cytosol from Xenopus laevis oocytes as described previously (29).
NFAT-GFP nuclear import and export
For all import and export experiments, HEK 293 cells stably expressing NFAT-GFP were grown on glass coverslips to approximately 50% confluency prior to drug treatment. In all cases, importazole was used at 40 µm and leptomycin B was used at 10 ng/ml. For controls, DMSO was used at a concentration of 0.4%. Ionomycin was added at 1.25 µm. Importazole and leptomycin B treatments were all for 1 hour. In all experiments cells were fixed with 4% formaldehyde prior to fluorescence microscopy. DNA was visualized with 1 µg/ml Hoechst dye. For quantification, 100 cells from each condition were analyzed and the percentage that showed nuclear accumulation of NFAT-GFP calculated.
In vitro microtubule polymerization and spindle assembly
Xenopus laevis egg extracts were prepared as described (42). For in vitro spindle assembly, Xenopus laevis sperm DNA was added to egg extracts supplemented with rhodamine-labeled tubulin. Asters were formed by addition of 5% DMSO. DNA was stained with Hoechst dye. The formation of microtubule-based structures was assessed using epifluorescence microscopy after a 30 minute room temperature incubation. In vitro microtubule polymerization and pelleting assays were performed by incubating 25 µM bovine tubulin, 1 mM GTP, and 5% DMSO in BRB80 buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8) at 37°C for 30 minutes. Polymerized microtubules were pelleted through a sucrose cushion, resuspended, and analyzed by SDS PAGE.
Fluorescence lifetime imaging microscopy (FLIM)
The Rango-3 FRET sensor is an improved version of Rango and was created by replacing Cerulean-EYFP donor-acceptor pair in Rango (7) with EGFP as a donor and non-fluorescent acceptor sREACh (43) which was modified by the introduction of mild dimerization mutations. Time-correlated single photon counting (TCSPC) datasets were acquired with a Plan-Apochromat 63×/1.40 NA oil immersion lens on an inverted Zeiss LSM710 NLO microscope equipped with a Becker & Hickl SPC-830 TCSPC controller. Samples were excited by one-photon 485nm pulses generated by a frequency doubling 970nm 80MHz Ti:Sapphire laser (Coherent MiraSHG). The emission was collected from a custom side port, filtered through a 525 nm bandpass filter (ET525/50 Chroma) and detected by a HPM-100-40 module (Becker & Hickl) containing a hybrid Hamamatsu R10467-40 GaAsP photomultiplier. Two to three days before the experiment, HeLa cells were transfected with a pSG8 plasmid containing the Rango-3 open reading frame (pK135) to induce sensor expression. Treatment with importazole or DMSO was started one hour before imaging and continued for up to one hour in an environmental chamber built on the microscope (37°C, 5% CO2). Recording conditions were chosen to limit emission to approx 1–2×106 counts per second, and images of 128 × 128 pixels (1024 time bins/pixel) were averaged over 60 seconds. Fluorescence lifetime images were produced and analyzed using SPCI software (Becker & Hickl).
Immunofluorescence microscopy
Cells were fixed in 4% formaldehyde and 0.1% glutaraldehyde in PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgSO4) at 37°C for 15 minutes followed by permeabilization with 0.1% triton X-100 for 2 minutes at room temperature. Cells were then washed and blocked (PHEM + 5% FBS + 0.2% saponin) and stained by standard techniques using the E7-A anti β tubulin antibody (Developmental Studies Hybridoma Bank) diluted 1:1000 and Hoechst dye.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Brian Wolff, Janice Williams, Brian Feng and James Wells at the Small Molecule Discovery Center at UCSF for help with the screen, Elisa Dultz for live cell imaging of imporatzole effects, Karin Reif for NFAT cDNA, David Halpin for biotin-labeled RCC1, and Lili Wang at the Broad Institute for developing the thermal shift assay to probe binding of importazole to importin-β. This work was supported by the NIH (R01 GM065232 and R21 NS53592, KW and RH), the Cancer Research Coordinating Committee (SLB), and the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (PK and KH).
ABBRIEVATIONS USED
- NLS
nuclear localization signal
- FRET
fluorescence resonance energy transfer
- IPZ
importazole
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 4.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 5.Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 6.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 7.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 8.Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 10.Ryan KJ, Zhou Y, Wente SR. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol Biol Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 14.Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- 15.Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- 19.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 20.Kosugi S, Hasebe M, Entani T, Takayama S, Tomita M, Yanagawa H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. doi: 10.1016/j.chembiol.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Ambrus G, Whitby LR, Singer EL, Trott O, Choi E, Olson A-J, Boger DL, Gerace L. Small molecule peptidomimetic inhibitors of importin alpha/beta mediated nuclear transport. Bioorg Med Chem. 2010;18:7611–7620. doi: 10.1016/j.bmc.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hintersteiner M, Ambrus G, Bednenko J, Schmied M, Knox AJ, Meisner NC, Gstach H, Seifert JM, Singer EL, Gerace L, Auer M. Identification of a small molecule inhibitor of importin beta mediated nuclear import by confocal on-bead screening of tagged one-bead one-compound libraries. ACS Chem Biol. 2010;5:967–979. doi: 10.1021/cb100094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floer M, Blobel G. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- 26.Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 27.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 28.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 29.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–603. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 33.Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 34.Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 35.Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT In vitro. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 37.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 38.Chi NC, Adam EJ, Adam SA. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- 39.Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 2008;68:1826–1833. doi: 10.1158/0008-5472.CAN-07-5279. [DOI] [PubMed] [Google Scholar]
- 40.Azuma Y, Seino H, Seki T, Uzawa S, Klebe C, Ohba T, Wittinghofer A, Hayashi N, Nishimoto T. Conserved histidine residues of RCC1 are essential for nucleotide exchange on Ran. J Biochem. 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- 41.Uniewicz KA, Ori A, Xu R, Ahmed Y, Wilkinson MC, Fernig DG, Yates EA. Differential scanning fluorimetry measurement of protein stability changes upon binding to glycosaminoglycans: a screening test for binding specificity. Anal Chem. 2010;82:3796–3802. doi: 10.1021/ac100188x. [DOI] [PubMed] [Google Scholar]
- 42.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 43.Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.