Abstract
Herein we report a highly efficient and reliable membrane-assisted capillary isoelectric focusing (MA-CIEF) system being coupled with MALDI-FTMS for the analysis of complex neuropeptide mixtures. The new interface consists of two membrane-coated joints made near each end of the capillary for applying high voltage, while the capillary ends were placed in the two reservoirs which were filled with anolyte (acid) and catholyte (base) to provide pH difference. Optimizations of CIEF conditions and comparison with conventional CIEF were carried out by using bovine serum albumin (BSA) tryptic peptides. It was shown that the MA-CIEF could provide more efficient, reliable and faster separation with improved sequence coverage when coupled to MALDI-FTMS. Analyses of orcokinin family neuropeptides from crabs Cancer borealis and Callinectes sapidus brain extracts have been conducted using the established MA-CIEF/MALDI-FTMS platform. Increased number of neuropeptides was observed with significantly enhanced MS signal in comparison with direct analysis by MALDI FTMS. The results highlighted the potential of MA-CIEF as an efficient fractionation tool for coupling to MALDI MS for neuropeptide analysis.
Keywords: capillary isoelectric focusing, CIEF, MALDI-FTMS, mass spectrometry, neuropeptides, orcokinin
1. Introduction
The neuropeptides are a class of important signaling molecules expressed in neurons for intercellular communications. They are consisted of short chains of amino acids and involved in numerous physiological processes. The identification and characterization of these signaling peptides are critical first steps to deciphering their functions, however, the study of neuropeptides has long been challenging due to their low abundance (picomolar to nanomolar level), diverse dynamic range and chemical complexity. The current lack of DNA sequences for many organisms and the post-translational modifications (PTMs) commonly observed in the identified neuropeptides also make it difficult to predict the final products from the genome information. Extensive studies have been performed previously by using Edman degradation and immunocytochemistry [1–4]. These traditional methods are well established to provide accurate neuropeptide sequences, but they are limited by the need for large amounts of starting materials, extensive purification steps and/or specific antibodies [5]. In the past two decades, mass spectrometry (MS) has emerged as a central tool for peptide analysis. Taking advantage of matrix-assisted laser desorption/ ionization (MALDI) and electrospray ionization (ESI), it has been possible to detect and sequence neuropeptides with high mass accuracy and high sensitivity [6, 7]. Due to the high complexity of tissue extract samples and the need for more in-depth study at the cellular and molecular levels, a number of separation techniques, most commonly including liquid chromatography and capillary electrophoresis, have been coupled with mass spectrometry for the neuropeptide analysis for enhanced MS signals [8, 9]. Several MS-based strategies for neuropeptide analysis have been employed by our previous studies, where sample preparation methods and separation techniques are integrated for separation and purification of sample prior to MS analysis [8].
Among the separation techniques which have been coupled with mass spectrometry, capillary isoelectric focusing (CIEF) is an emerging tool for the separation and focusing of proteins and peptides. Both capillary zone electrophoresis (CZE) and CIEF are considered to have the greatest potential in MS-based proteomics [9]. In both cases, capillary is connected via buffer vials to the high voltage power supply. In classic CZE mode the separation is dependent on the analyte’s charge state and molecular size, while it is limited by the loading amount. For CIEF, separation and focusing of a larger amount of samples can be realized simultaneously based on the analyte’s pI value, and many applications has been found in proteomics and peptidomics [10–14], either directly coupled with ESI or MALDI MS [15–17], or as the first dimension in multi-dimensional separations [18, 19].
Despite the advantages in separation time and efficiency, the conventional CIEF setup suffers from several problems, including sample loss caused by carrier ampholyte band shift, MS signal suppression from carrier ampholyte, disruption of separation by gas bubbles and protein/peptide adsorption or precipitation resulted from unstable wall-coating or long time of focusing [20–22]. Optimizations have been made to the conventional CIEF, most commonly on capillary coating [23–29], carrier ampholytes [30–35], CIEF conditions [36–37] and sample collection/interface to mass spectrometers [38–40]. However, to date only a few papers reported the study of complex peptide mixtures by CIEF, coupled with limited types of mass spectrometers. To our knowledge, CIEF has not been previously coupled with MALDI-FTMS, or applied to the study of neuropeptide extracts.
In this paper we developed a modified CIEF system for neuropeptide analysis by adding two membrane-coated joints near both ends of the capillary to prevent carrier ampholytes band shift and interferences from gas bubbles. By coupling to MALDI-FTMS, systematic studies were conducted with BSA tryptic peptides. High separation efficiency and high sensitivity were observed, with good reproducibility. This new MA-CIEF/MALDI-FTMS based platform has also been applied to the analysis of orcokinin family neuropeptides extracted from crustacean model organisms.
2. Material and methods
2.1 Chemical and materials
Acetic acid, sodium hydroxide, ammonium hydroxide, acetone, acetonitrile, methanol, ammonium bicarbonate and urea were purchased from Fisher Scientific (Pittsburgh, PA). Trifluoroacetic acid, formic acid, cellulose acetate (39.7%, typical MW=50000), hydroxypropyl cellulose (HPC), Pharmalyte 3–10, iodoacetamide (IAA) and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO). 2,5-dihydroxybenzoic acid (DHB) was obtained from Alfa Aesar (Ward Hill, MA). Parafilm “M” was obtained from Pechiney Plastic Packaging (Menasha, WI). Sequencing grade modified trypsin and D/L-dithiothreitol (DTT) were from Promega (Madison, WI).
Fused-silica capillary with 75 µm i.d. and 360 µm o.d. was purchased from Polymicro Technologies (Phoenix, AZ). Millipore C18 Ziptip column was used for sample cleaning, and all water used in this study was doubly distilled on a Millipore filtration system (Bedford, MA). The physiological saline consisted of 440 mM NaCl, 11mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, and 5 mM maleic acid in pH 7.45.
2.2 Protein digestion
The following solutions were used during tryptic digestion of BSA. 98 mg of ammonium bicarbonate was diluted in 50 ml water to make 25 mM ammonium bicarbonate solution. 8 M urea solution was made by diluting 0.96 g urea in 2.0 mL of 25 mM ammonium bicarbonate solution. 30 mg of DTT was dissolved in 0.20 mL of 25 mM ammonium bicarbonate solution to make 1 M DTT as reducing reagent. 36 mg of IAA was dissolved in 1 mL of 25 mM ammonium bicarbonate solution to make 200 mM IAA as alkylating reagent.
BSA was digested to make BSA tryptic peptides for CIEF separations. 30 µg of BSA was reconstituted in 20 µL of 8 M urea. 1 µL of reducing reagent was added to the tube followed by gentle vertex. After being kept in 37 °C for 1 h, 20 µL of alkylating reagent was added to the tube to alkylate for 1 h at room temperature in dark with shaking. 4 µL of reducing reagent was added to consume residual alkylating reagent, and 120 µL of 25 mM ammonium bicarbonate solution was added to dilute urea. Overnight digestion was performed at 37 °C after adding 1 µg of trypsin. In the next morning, 1 µL of formic acid was added to the sample and gently vortexed to quench the reaction. The BSA tryptic peptides sample was stored at −80 °C before usage.
2.3 Animal dissection and tissue extraction
The Jonah crabs (Cancer borealis) were purchased from the Fresh Lobster Company (Gloucester, MA) and the blue crabs (Callinectes sapidus) were purchased from local grocery store. They were kept in an artificial seawater tank at 10–12 °C without food. The detailed dissection procedure has been previously described [41, 42]. Briefly, the crabs were anesthetized for 15–30 min before dissection by packing in ice. Brains were dissected in physiological saline. The neuropeptides were extracted using acidified methanol as described previously [43]. The acidified methanol consisted of methanol, water and acetic acid in the ratio of 90:9:1 and was kept in ice during extraction. Brains were homogenized in 50 µL acidified methanol and centrifuged for the supernatants. The extraction was repeated for three times and the supernatants were combined and dried. The residue was reconstituted with 10 µL of 0.1% TFA and stored at −80 ° C before usage.
2.4 Sample preparations
In order to purify the extracted neuropeptides and BSA tryptic peptides to achieve better sensitivity on mass spectrometer, the samples were desalted by Ziptip C18 column before CIEF and mass spectrometry analysis. The samples were then eluted by 5 µL of ACN/water (50:50) containing 0.1% TFA. The eluted solution was dried and reconstituted in 0.5% Pharmalytes for analysis.
2.5 Fabrication of CIEF system
75 µm i.d. × 60 cm fused-silica capillary was flushed by 1 M NaOH for 10 min and dried with air before treatment. The capillary was further washed by water and dried by nitrogen. HPC was dissolved in water to the final concentration of 5% (w/w) and supersonically degassed. The capillary was (1) filled with 5% HPC; (2) flushed by hexagon at 100 psi; (3) filled with 5% HPC again; and (4) flushed by hexagon at 100 psi. An oven was used to heat the capillary from 60 °C to 140 °C in 16 min, and the temperature was maintained for 20 min. After that the capillary was removed from the oven, washed with water and dried before further process. Cellulose acetate membrane-coated porous joints were made 3 cm to both ends of the HPC-coated capillary as we previously described [44]. Briefly, the capillary was affixed near one end (about 3 cm) on a 1 cm × 0.3 cm plastic slide. QuickGrip glue (Beacon Adhesives Co., Mt. Vernon, NY) was loaded on each end of the plastic slide to attach the capillary and left about 0.5 cm capillary between the two glue spots. A small fracture section was made on the fixed capillary and was then covered by cellulose acetate solution (12% in acetone, w/v). Under a gentle stream of air a uniform cellulose acetate membrane could be formed over the fracture section. This porous joint was then placed into a 0.6 mL plastic vial (Fisher Scientific, Pittsburgh, PA) with about 2 cm capillary stretched out of the vial from a small hole on the bottom. 1% of acetic acid was filled in the vial and the electrode wire was inserted into the vial for electricity connection.
2.6 CIEF procedure and sample collection
CIEF was performed by using TriSep-2100 HV power supplier from Unimicro Technologies (Pleasanton, CA). 1% of acetic acid solution and 1% of sodium hydroxide solution were chosen as anolyte and catholyte, and were filled into the reservoirs, respectively. 1% of acetic acid was filled into the plastic vials containing membrane-coated joints, and electrodes were placed into these vials so that a circuit was formed via the porous joints, as shown in Figure 1. Prior to CIEF, the capillary was flushed with water and dried under air flow in sequence. 20 kV of constant voltage was then applied to the capillary and kept for 10–12 min depending on samples. After focusing, the fractions were mobilized by air pressure. MALDI plate pre-coated with Parafilm M was used for sample collection as described previously [45]. Briefly, the Parafilm M was cut into 2.5 cm × 0.4 cm pieces and stretched to approximately 3-fold long as its original length while the width was kept the same, and the film was directly placed and affixed onto the MALDI plate where the sample would be collected. 120 mg/ml of 2,5-dihydroxybenzoic acid (DHB) was pre-spotted on the MALDI plate as the matrix. For each of the spots, 250 nL of matrix solution was mixed with approximately 150 nL of collected fraction, and co-crystallized before MALDI-FTMS analysis.
Figure 1. Schematic illustration of the membrane-assisted CIEF setup enhanced with cellulose acetate membrane-coated porous joints.
The sample was mixed with 0.5% of Pharmalyte and filled into the capillary, which was then inserted into the anolyte and catholyte reservoirs with acid and base, respectively. High voltage power supply was connected via the two membrane-coated joints near capillary ends to form a circuit, so that the shadowed sections were free from electrical field during focusing and formed “plugs” to prevent sample loss. After focusing, the reservoirs were removed, and the fractions were mobilized and collected on a MALDI target plate with pre-spotted matrix.
2.7 Mass spectrometry
The detection of CIEF fractions was performed on a Varian/IonSpec Fourier transform mass spectrometry (Lake Forest, CA), which was equipped with a 7.0 T actively shielded superconducting magnet and a 355nm Nd:YAG laser (Laser Science, Inc., Franklin, MA). Before transferred to the ion cyclotron resonance (ICR) cell through a quadrupole ion guide, the ions were accumulated in the external hexapole storage trap, and all of the mass spectra were collected in the positive ion mode. Prior to detection, the ions were excited by an rf sweep from 7050 to 7054 ms with an amplitude of 150 V base to peak. In order to baseline distortion of the peaks, the filament and quadrupole trapping were firstly raised to 15 V, and then ramped to 1 V from 6500 ms to 7000 ms. 50 laser shots were employed and all the mass spectra were recorded from m/z 108 to m/z 2500.
Autoflex III MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany) equipped with Smartbeam 2 laser was also employed and coupled with CIEF on the analysis of extracted neuropeptides. Mass spectra were acquired in a positive ion reflection mode with ion source 1 voltage 19.00 kV, ion source 2 voltage 16.35 kV, reflector 1 voltage 21.00 kV, reflector 2 voltage 9.9 kV and lens voltage 8.70 kV. 200 shots were acquired and all the mass spectra were recorded from m/z 500 to m/z 3000. External calibration was performed by using a standard peptide mixture provided by Bruker Daltonics.
Results and discussion
3.1 The membrane-coated joints on the MA-CIEF setup
The most important modification to the conventional CIEF setup in this work was to add two membrane-coated joints to both ends of the capillary. A similar interface was developed previously in our lab and used on capillary zone electrophoresis (CZE), but only one end was made into a porous joint. The two joints covered by porous cellulose acetate membrane allowed ions to pass through to form the current circuit, while samples were isolated from the buffers. Compared with other previously reported interfaces, our specially-designed membrane-coated joints were robust in the acid and neutral environment and easy to make. [40, 46–47]. 1% of acetic acid was added to the vials where the joints were immersed. During focusing, the electrodes were connected through these two joints, so that a circuit was built up and focusing was initiated. In order to form continuous pH gradient within the capillary, the anolyte reservoir with 1% of acetic acid and catholyte reservoir with 1% of sodium hydroxide were added to the ends of the capillary to provide pH difference. In this case, the two reservoirs were not connected to the electrode directly as what was usually seen in conventional CIEF setup, but due to the direct connection to the capillary, pH differences were still provided during focusing to form a continuous pH gradient. As a result, the two sections between anolyte (or catholyte) reservoir and membrane-coated joint (shadowed in Figure 1) were free from electrical field during focusing, and they formed “plugs” to prevent the CA bands shift which was thought to be the major reason of sample loss in previous setup [48]. In addition, the hydrolysis generated gas bubbles at electrode surface, mainly oxygen and hydrogen, significantly interfering with the separation in conventional CIEF [34]. This drawback was also eliminated by moving the electrodes to the membrane-coated joints. The gas bubbles were prevented from entering the capillary, which resulted in improved separation efficiency and better reproducibility.
3.2 Optimization of CIEF conditions
One major problem of coupling CIEF with MALDI MS is the suppression of MS signals caused by adding carrier ampholytes. As the concentration of carrier ampholytes increases, the CIEF resolution increases accordingly; however the MS ionization efficiency drops significantly due to ion suppression [15]. By using the modified CIEF setup, our experiments showed that Pharmalyte at concentration as low as 0.5% could be used. At this level satisfactory separation could be achieved with minimal MS signal suppression.
In addition to carrier ampholytes, the voltage and focusing time are critical for successful CIEF separation. If the voltage is too low or the focusing time is too short, there will not be enough time for the components and ampholytes to reach their isoelectric points. If the voltage is too high or the focusing time is too long, the carrier ampholytes band will migrate towards the reservoir much faster and break through the “plugs” and lead to sample loss. It was found that 20 kV of constant voltage worked best for both BSA tryptic peptides and extracted neuropeptide mixtures from crustaceans in our study. Depending on different sample conditions, the focusing was maintained for 10–12 min.
3.3 Reproducibility of MA-CIEF system
CIEF allows amphoteric molecules, such as peptides, to be separated by electrophoresis in a pH gradient generated between the cathode and anode. A solute will migrate with the carrier ampholytes to an isoelectric point (pI) where its net charge is zero. As the molecule reaches its pI during focusing, the migration stops and the net charge drops to zero, which consequently leads to electrophoretic current drop. Reproducible separations should exhibit identical current change in a same time course. Previous studies have suggested using current as an indicator for focusing process [49]. Here, the MA-CIEF system’s reproducibility was evaluated by monitoring the current change versus focusing time, which is independent to other possible variables in the whole analysis procedure, such as sample degradation or variations in MALDI-FTMS analysis.
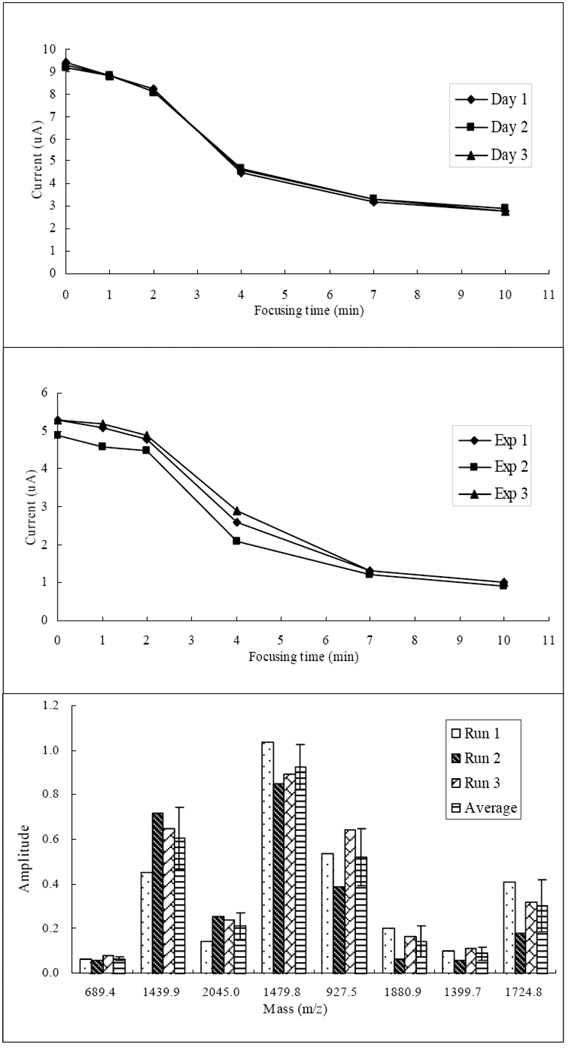
Both intra-day and inter-day evaluations have been performed with either BSA tryptic peptides or extracts from blue crab brains. For intra-day evaluation, BSA tryptic peptides were focused by CIEF under 20 kV for 10 min. After sample mobilization, the capillary was flushed by water for 10 min and dried with air for 5 min before the next sample loading. Three consecutive injections were made in the same day. For inter-day evaluation, extracted sample from blue crab brains were focused by CIEF also under 20 kV for 10 min. After sample mobilization, the capillary was flushed by water for 30 min and purged overnight with air flow. Three injections were made in three consecutive days. For each evaluation, six time points were recorded at 0, 1, 2, 4, 7 and 10 min. In Figure 2, the upper and middle panels show the results of reproducibility evaluation of the MA-CIEF setup for inter-day and intra-day, respectively. As we can see, very similar current behaviors were shown in both intra-day and inter-day experiments. It is notable that, the current plots were almost the same for the inter-day evaluation with the complex brain extract, which is even better than the intra-day result. The possible reason is that, for inter-day experiment the MA-CIEF system was fully conditioned with 30 min wash plus overnight air flow, while the 10-min-wash conditioning between intra-day runs might not completely eliminate the variation at the initial stage of CIEF. But the current of the three runs dropped to the same level after 10 min of focusing, showing a same level of separation and focusing. In addition, the current vs. time curves dropped significantly during 2–7 min in all CIEF experiments, showing efficient separations of the MA-CIEF platform within a short time. For both intra-day and inter-day evaluations similar mass spectra were obtained within each replicate. By comparing peptides with representative pI values in corresponding fractions from different runs, we observed similar peak intensities which were shown in the bottom panel in Figure 2. These results indicate excellent reproducibility and high efficiency of the newly developed MA-CIEF system, which is suitable for the analysis of complex peptide mixtures.
Figure 2. Reproducibility tests of the MA-CIEF system.
Three evaluations were made using the same condition in three consecutive days. Upper: inter-day evaluation using brain extract from C. sapidus (n=3). Middle: intra-day evaluation using BSA tryptic peptides. Bottom: Amplitudes and standard deviations of BSA tryptic peptides with representative pI values in three runs (m/z 689.4: pI=11.0. m/z 1439.9: pI=9.8. m/z 2045.0: pI=9.3. m/z 1479.8: pI=6.9. m/z 927.5: pI=6.8. m/z 1880.9: pI=6.4. m/z 1399.7: pI=4.1. m/z 1724.8: pI=4.1.The average bars are shown as average ± SD).
3.4 Evaluation of MA-CIEF with BSA tryptic peptides
Before CIEF, the BSA tryptic peptides were dried in vacuum for 30 min and distributed into plastic vials. Each vial contained tryptic peptides originated from 5 µg of BSA, and was dissolved in 14.4 µL of 0.5% Pharmalyte. For MA-CIEF system testing, the BSA tryptic peptides were gently vortexed and filled into the capillary. CIEF was conducted for 12 min under 20 kV, and the collected fractions were further analyzed by MALDI-FTMS. Three representative fractions from basic, neutral and acid pH bands, together with the control of which the same sample was directly analyzed by MALDI-FTMS without CIEF, were shown in Figure 3. In fraction 8, all of the peptides detected were basic with high pI values, including AWSVAR (m/z 689.4, pI 11.0), ALKAWSVAR (m/z 1001.6, pI 11.5), HPEYAVSVLLR (m/z 1283.7, pI 9.6), RHPEYAVSVLLR (m/z 1439.8, pI 9.8), KVPQVSTPTLVEVSR (m/z 1639.9, pI 10.1) and RHPYFYAPELLYYANK (m/z 2045.0, pI 9.3). Depending on their concentration levels and slight pI value differences, some of them were only observed in fraction 8, while some were also found in fraction 9 or 10. In fraction 11, two neutral peptides YLYEIAR (m/z 927.5, pI 6.8) and LGEYGFQNALIVR (m/z 1479.8, pI 6.9) were the predominant peaks, together with a modified peptide RPCFSALTPDETYVPK (m/z 1880.9, original pI 6.4, modified by carbamidomethyl group on cysteine residue after alkylation by 2-iodoacetamide during BSA digestion). In fraction 14, a number of acidic peptides and modified peptides were detected, including TVMENFVAFVDK (m/z 1399.7, pI 4.1), DAFLGSFLYEYSR (m/z 1567.7, pI 4.1), MPCTEDYLSLILNR (m/z 1724.8, original pI 4.1, modified by carbamidomethyl group on cysteine residue) and NECFLSHKDDSPDLPK (m/z 1901.8, original pI 4.4, modified by carbamidomethyl group on cysteine residue). There were separations within the basic, neutral or acidic peptides among the nearby fractions, which were not shown in Figure 3. The only exception was that LGEYGFQNALIVR (m/z 1479.8, pI 6.9) was also observed in fraction 14 with basic peptides. Although diffusion was not significant during fraction mobilization, the most abundant peptides could be observed in several fractions. The overlapping of LGEYGFQNALIVR in neutral and basic fractions might be due to its highest abundance among the peptide mixture. However, the diffusion would not interfere with the separations of other peptides. Within 12 min of separation, majority of the tryptic peptides were focused into 2–3 fractions and it is worth mentioning that by using the same sample collection method, peak overlapping is much reduced in MA-CIEF system compared with conventional CIEF, mostly due to improved separation efficiencies. A table showing all of the 24 separated tryptic peptides and their distributions can be found as Table S1 in Supplementary Data. These results demonstrate that the MA-CIEF system can efficiently separate the complex peptide mixture according to their pI value in a short time. In addition, the signal has been significantly enhanced after CIEF separation with MALDI-FTMS detection.
Figure 3. Analysis of BSA tryptic peptides using the MA-CIEF/MALDI-FTMS platform.
Spectra of control sample without separation and three representative fractions were shown. #8, #11 and #14 show spectra of fractions from basic, neutral and acidic bands, respectively. Tryptic peptides originate from 3 µM BSA, voltage = 20 kV, focusing time = 12 min.
In order to compare the performance between the conventional CIEF setup and the MA-CIEF, we analyzed the same BSA tryptic peptide mixture with conventional and our CIEF systems. For conventional CIEF setup, 1% of acetic acid and 1% of sodium hydroxide were also used as anolyte and catholyte, respectively, which were directly connected to the anode and cathode. 12 min of focusing under 20 kV was performed, which was identical with the MA-CIEF setup. After all the fractions were analyzed with MALDI-FTMS, the sequence coverage, number of peptides found and ion intensity were compared with those from using the MA-CIEF/MALDI-FTMS platform. Table 1 shows the results of this comparison. In 12 min, MA-CIEF was able to efficiently separate the peptide mixture to achieve 43% of sequence coverage with 24 tryptic peptides detected, whereas for conventional CIEF, 12 min of separation time was not sufficient as reflected from the mass spectra. Lower sequence coverage and less number of peptides were obtained. An extension of separation time to 18 min was made with the conventional CIEF setup, and similar separation efficiency with MA-CIEF in 12 min was observed. However, as we prolonged the separation time on conventional CIEF, the peak intensity dropped, which could result from the loss of sample and peptide adsorption due to longer duration and interaction with capillary wall. As expected, by adding two membrane-coated joints, electrical field and driving force between the fractures and the capillary ends were eliminated, and the two sections (shadowed in Figure 1) played as two “plugs” to prevent the sample from flowing into the reservoir. This contributes to higher intensity observed in resulting mass spectra.
Table 1.
Comparison among current MA-CIEF system, conventional CIEF system and the control sample (no CIEF separation) in sequence coverage, number of peptides found and ion amplitude using BSA tryptic peptides
| CIEF with 0.5% pharmalyte |
MA-CIEF 12 min |
Conventional 12 min |
Conventional 18 min |
Control Average (n=3) |
|---|---|---|---|---|
| Sequence coverage | 43% | 38% | 43% | 20% |
| Number of peptide found | 24 | 21 | 23 | 12 |
| Ion amplitude* | 16.4 | 13.3 | 12.7 | 1.6 |
Amplitude is defined as intensity/5000 in Varian MALDI FTMS.
3.5 Profiling of orcokinin family neuropeptides from Cancer borealis brains
MA-CIEF system has been applied to the study of orcokinin family neuropeptides from Cancer borealis and Callinectes sapidus brains by coupling with MALDI-FTMS. Orcokinin is a conserved neuropeptide family found originally in crayfish, whose function is to enhance the activity on the hindgut contraction [50].
The crustacean brain extract is challenging to analyze due to the low concentrations of peptides and interferences from high salts and high abundance lipids in this complex sample [51]. Previously, study of orcokinin family neuropeptides from Cancer borealis brains has been performed in our lab. A total of 10 orcokinin family neuropeptides have been identified by combining multiple sample preparation methods, including direct tissue analysis, crude extraction, capillary electrophoresis and offline HPLC separation, with MALDI-FTMS [5] (Table 2). Here, by using the MA-CIEF/MALDI- FTMS based platform, the same experiment was performed twice with two biological replicates. It was found that after CIEF separation, the MS signals were significantly improved, and more neuropeptides were detected compared with those by direct MALDI-FTMS profiling. Since orcokinins have lower pI values compared with other neuropeptide families, they are focused into similar fractions with no overlapping with other neuropeptides. Figure 4 shows the analysis of orcokinin family neuropeptides from Cancer borealis brains using MA-CIEF/MALDI-FTMS. For sample one, five orcokinins were found using MALDI-FTMS, including EIDRSGFGFA (m/z 1098.52), NFDEIDRSGFA (m/z 1270.57), NFDEIDRSGFGFA (m/z 1474.66), NFDEIDRSGFGFV (m/z 1502.69) and NFDEIDRSSFGFV (m/z 1532.70). After CIEF, four additional orcokinins were observed, including NFDEIDRSGFa (m/z 1198.5487), NFDEIDRSGFG (m/z 1256.5542), NFDEIDRSGFGF (m/z 1403.6226) and NFDEIDRSSFGFN (m/z 1547.6761). Another neuropeptide NFDEIDRTGFGFH (m/z 1554.70) was detected in the replicate analysis. This result demonstrates that all orcokinins found previously from Cancer borealis brain by using a combination of multiple sample preparation methods and instruments (as shown in Table 2) can be comprehensively characterized by a single MA-CIEF/MALDI-FTMS platform reported here.
Table 2.
List of orcokinin family neuropeptides detected with MA-CIEF/MALDI-FTMS based platform from Cancer borealis and Callinectes sapidus brains
| m/z | Sequence | Estimated pI value |
Found in |
|---|---|---|---|
| 1098.52 | EIDRSGFGFA | 4.1 | J, B, P |
| 1198.55 | NFDEIDRSGFa | 4.3 | J, P |
| 1256.55 | NFDEIDRSGFG | 3.7 | J, P |
| 1270.57 | NFDEIDRSGFA | 3.7 | J, B, P |
| 1271.55 | DFDEIDRSGFA | 3.5 | B |
| 1403.62 | NFDEIDRSGFGF | 3.7 | J, B, P |
| 1474.66 | NFDEIDRSGFGFA | 3.7 | J, B, P |
| 1502.69 | NFDEIDRSGFGFV | 3.7 | J, B, P |
| 1532.70 | NFDEIDRSSFGFV | 3.7 | J, B, P |
| 1547.68 | NFDEIDRSSFGFN | 3.7 | J, B, P |
| 1554.70 | NFDEIDRTGFGFH | 4.3 | J, P |
J: Jonah crab (Cancer borealis), B: Blue crab (Callinectes sapidus), P: previously found by Ma et al. (Ref 5) from Cancer borealis brains by coupling four sample preparation methods, including direct tissue analysis, crude extraction, capillary electrophoresis and offline HPLC separation, with MALDI-FTMS.
Figure 4. Analysis of orcokinin family neuropeptides from brain of Cancer borealis using MA-CIEF/MALDI-FTMS.
A: Control sample without CIEF separation. Orcokinin family neuropeptides are marked with stars. B: Fraction 11, rich in orcokinin family peptides after CIEF. Peaks with closed circles indicate additional peptides detected with CIEF not observed in direct MALDI FTMS.
3.6 Profiling of orcokinin family neuropeptides from Callinectes sapidus brains
To date, there was no report on the orcokinin family neuropeptide profiling from the brain of Callinectes sapidus. We employed the MA-CIEF/MALDI-FTMS platform to analyze the Callinectes sapidus brain extracts for the orcokinin family neuropeptides. In direct analysis with MALDI-FTMS (Figure 5A), only four most abundant orcokinins were detected, including NFDEIDRSGFGFA (m/z 1474.7), NFDEIDRSSFGFA (m/z 1502.7), NFDEIDRSSFGFV (m/z 1532.7) and NFDEIDRSSFGFN (m/z 1547.7) in a relatively low abundance. The same sample was then analyzed by the newly developed MA-CIEF/MALDI-FTMS platform. After CIEF, orcokinins with similar pI values were separated from other neuropeptides and focused to a narrow band corresponding to their similar pI values. As compared with direct MALDI-FTMS analysis, four additional putative orcokinins were observed, including EIDRSGFGFA (m/z 1098.5), NFDEIDRSGFA (m/z 1270.6), DFDEIDRSGFA (m/z 1271.6) and NFDEIDRSGFGF (m/z 1403.6) (Figure 5B). Compared with Cancer borealis brain extract, one orcokinin family neuropeptide DFDEIDRSGFA (m/z 1271.6) starting with aspartic acid at the N-terminus rather than asparagine at the N-terminus was found in C. sapidus brains, suggesting a possible deamidation modification occurring in this peptide in the blue crab.
Figure 5. Analysis of orcokinin family neuropeptides from brain of Callinectes sapidus using MA-CIEF/MALDI-FTMS.
A: Control sample without CIEF separation. Orcokinin family neuropeptides are marked with stars. B: Fraction 16, rich in orcokinin family peptides after CIEF. Peaks with closed circles indicate additional peptides detected with CIEF not seen in the control sample.
Finally, the MA-CIEF system was coupled with a MALDI-TOF/TOF mass spectrometer and compared with conventional CIEF-MALDI MS in analysis of C. sapidus brain extracts. Same procedure has been adopted as described above. Similar to the results from BSA tryptic peptides, it was found that both MA-CIEF and conventional CIEF separated the peptides in the extract according to their pI values, but MA-CIEF resulted in higher peak intensities. Representative mass spectra have been shown in Figure S1 along with the mass spectrum via direct analysis with MALDI TOF/TOF. In addition to the orcokinin family neuropeptides found by MA-CIEF/MALDI-FTMS platform, we were able to observe two additional orcokinins, including NFDEIDRSGFG (m/z 1256.6) and NFDEIDRSSFA (m/z 1300.6) by employing MA-CIEF/MALDI-TOF/TOF, which confirmed the high separation efficiency of the MA-CIEF system.
4. Conclusions
In summary, a membrane-assisted capillary isoelectric focusing (MA-CIEF) technique has been developed, coupled to MALDI-FTMS for the analysis of complex neuropeptides. By applying the membrane-coated joints, the new platform exhibits better performance than the conventional CIEF in terms of separation efficiency, reduced sample loss and improved reproducibility. Evaluation of protein tryptic digests demonstrated that the MA-CIEF/MALDI-FTMS platform can efficiently separate and focus the peptide mixture in shorter time with higher sequence coverage and enhanced MS signals in comparison with conventional CIEF. This newly built system has been applied to the analysis of orcokinin family neuropeptides in complex crustacean brain extracts. Increased number of putative neuropeptides has been detected with higher sensitivity, showing great potential for the analysis of specific neuropeptide families according to their similar pI values. Collectively, the MA-CIEF/MALDI-FTMS platform offers an attractive new tool for proteomics and peptidomics applications.
Supplementary Material
Acknowledgement
The authors thank the Analytical Instrument Center at UW School of Pharmacy for the access of MALDI-FTMS. This work was supported in part by the National Science Foundation grant (CHE-0957784), and National Institutes of Health through grant 1R01DK071801. L. Li acknowledges an Alfred P. Sloan Research Fellowship and a Vilas Associate Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keller R. Experientia. 1992;48:439. doi: 10.1007/BF01928162. [DOI] [PubMed] [Google Scholar]
- 2.Kobierski LA, Beltz BS, Trimmer BA, Kravitz EA. J. Comp. Neurol. 1987;266:1. doi: 10.1002/cne.902660102. [DOI] [PubMed] [Google Scholar]
- 3.Spittaels K, Schoofs L, Grauwels L, Smet H, Van Damme J, Proost P, de Loof A. Peptides. 1991;12:31. doi: 10.1016/0196-9781(91)90162-i. [DOI] [PubMed] [Google Scholar]
- 4.Christie AE, Lundquist CT, Nassel DR, Nusbaum MP. J. Exp. Biol. 1997;200:2279. doi: 10.1242/jeb.200.17.2279. [DOI] [PubMed] [Google Scholar]
- 5.Ma M, Wang J, Chen R, Li L. J. Proteome Res. 2009;8:2426. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Q, Li L. Anal. Chem. 2005;77:7783. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. J. Neurochem. 2003;87:642. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Sweedler JV. Ann. Rev. Anal. Chem. 2008;1:451. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 9.Simpson D, Smith RD. Electrophoresis. 2005;26:1291. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- 10.Wu W, Li Y, Deng C, Zhang X. Electrophoresis. 2006;27:2100. doi: 10.1002/elps.200500820. [DOI] [PubMed] [Google Scholar]
- 11.Storms HF, van der Heijden R, Tjaden UR, van der Greef J. J. Chromatogr. B. 2005;824:189. doi: 10.1016/j.jchromb.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Storms HF, Heijden R, Tjaden U, van der Greef J. Electrophoresis. 2004;25:3461. doi: 10.1002/elps.200406087. [DOI] [PubMed] [Google Scholar]
- 13.Silvertnd LLH, Torano J, Bennekom W, de Jong G. J. Chromatogr. A. 2008;1204:157. doi: 10.1016/j.chroma.2008.05.057. [DOI] [PubMed] [Google Scholar]
- 14.Crowley T, Hayes M. Proteomics. 2005;5:3798. doi: 10.1002/pmic.200401212. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda Y, Yukinaga H, Kitano M, Noguchi T, Nemati M, Shibukawa A, Nakagawa T, Matsuzaki K. J. Pharm. Biomed. Anal. 2005;37:423. doi: 10.1016/j.jpba.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Silvertand LLH, Torano J, de Jong G, Bennekom W. Electrophoresis. 2008;29:1985. doi: 10.1002/elps.200700434. [DOI] [PubMed] [Google Scholar]
- 17.Yu W, Li Y, Deng C, Zhang X. Electrophoresis. 2006;27:2100. doi: 10.1002/elps.200500820. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Berger S, Anderson G, Smith RD. Anal. Chem. 2000;72:2154. doi: 10.1021/ac991367t. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, Zhang L, Liu H, Zhang W, Zhang Y. J. Chromatogr. A. 2003;1018:97. doi: 10.1016/j.chroma.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 20.Righetti PG, Gelfi C, Bossi A, Olivier E, Castelletti L, Verzola B, Stoyanov AV. Electrophoresis. 2000;21:4046. doi: 10.1002/1522-2683(200012)21:18<4046::AID-ELPS4046>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Simpson DC, Smith RD. Electrophoresis. 2005;26:1291. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- 22.Righetti PG, Simo C, Sebastiano R, Citterio A. Electrophoresis. 2007;28:3799. doi: 10.1002/elps.200700232. [DOI] [PubMed] [Google Scholar]
- 23.Busch MHA, Kraak JC, Poppe H. J. Chromatogr. A. 1995;695:287. [Google Scholar]
- 24.Belder D, Deege AA, Husmann H, Kohler F, Ludwig M. Electrophoresis. 2001;22:3813. doi: 10.1002/1522-2683(200109)22:17<3813::AID-ELPS3813>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Palm A, Zaragoza-Sundqvist M, Marko-Varga G. J. Sep. Sci. 2004;27:124. doi: 10.1002/jssc.200301596. [DOI] [PubMed] [Google Scholar]
- 26.Gao L, Liu SR. Anal. Chem. 2004;76:7179. doi: 10.1021/ac049353x. [DOI] [PubMed] [Google Scholar]
- 27.Tsai SW, Loughran M, Hiratsuka A, Yano K, Karube I. Analyst. 2003;128:237. doi: 10.1039/b207871f. [DOI] [PubMed] [Google Scholar]
- 28.Horka M, Planeta J, Ruzicka F, Slais K. Electrophoresis. 2003;24:1383. doi: 10.1002/elps.200390177. [DOI] [PubMed] [Google Scholar]
- 29.Knittle JE, Roach D, Horn PB, Voss KO. Anal. Chem. 2007;79:4978. doi: 10.1021/ac071537z. [DOI] [PubMed] [Google Scholar]
- 30.Poitevin M, Morin A, Busnel JM, Descroix S, Hennion MC, Peltre G. J. Chromatogr. A. 2007;1155:230. doi: 10.1016/j.chroma.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Lalwani S, Vigh G. Electrophoresis. 2005;26:3. doi: 10.1002/elps.200406174. [DOI] [PubMed] [Google Scholar]
- 32.Mohan D, Lee CS. J. Chromatogr. A. 2002;979:271. doi: 10.1016/s0021-9673(02)01442-5. [DOI] [PubMed] [Google Scholar]
- 33.Huang T, Wu X, Pawliszyn J. Anal. Chem. 2000;72:4758. doi: 10.1021/ac000599l. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Y, Lemma T, Musteata MF, awliszyn JP. J. Chromatogr. A. 2009;1216:2928. doi: 10.1016/j.chroma.2008.07.093. [DOI] [PubMed] [Google Scholar]
- 35.Yang C, Wang S, Chang C, Wang Y, Hu X. Anal. Chem. 2010;82:1580. doi: 10.1021/ac902223y. [DOI] [PubMed] [Google Scholar]
- 36.Balgley BM, Wang W, Song T, Fang X, Yang L, Lee CS. Electrophoresis. 2008;29:3047. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 37.Silvertand LHH, Torano JS, de Jong GJ, van Bennekom WP. Electrophoresis. 2009;30:1828. doi: 10.1002/elps.200800740. [DOI] [PubMed] [Google Scholar]
- 38.Minarik M, Foret F, Karger BL. Electrophoresis. 2000;21:247. doi: 10.1002/(SICI)1522-2683(20000101)21:1<247::AID-ELPS247>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C, Xiang F, Pasa-Tolic L, Anderson GA, Veenstra TD, Smith RD. Anal. Chem. 2000;72:1462. doi: 10.1021/ac9912653. [DOI] [PubMed] [Google Scholar]
- 40.Lechner M, Seifner A, Rizzi AM. Electrophoresis. 2008;29:1974. doi: 10.1002/elps.200700836. [DOI] [PubMed] [Google Scholar]
- 41.Kutz KK, Schmidt JJ, Li L. Anal. Chem. 2004;76:5630. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 42.Fu Q, Kutz KK, Schmidt JJ, Hsu Y-WA, Messinger DI, Cain SD, de la Iglesia HO, Christie AE, Li L. J. Comp. Neurol. 2005;493:607. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- 43.DeKeyser SS, Li L. Analyst. 2006;131:281. doi: 10.1039/b510831d. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Ma M, Chen R, Li L. Anal. Chem. 2008;80:6168. doi: 10.1021/ac800382t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Chen R, Ma M, Li L. Anal. Chem. 2008;80:491. doi: 10.1021/ac701614f. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z, Pawliszyn J. Anal. Chem. 2003;75:4887. doi: 10.1021/ac034587m. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Zhang L, Onoda K. Electrophoresis. 2005;26:563. doi: 10.1002/elps.200410028. [DOI] [PubMed] [Google Scholar]
- 48.Mack S, Cruzado-Park I, Chapman J, Ratnayake C, Vigh G. Electrophoresis. 2009;30:4049. doi: 10.1002/elps.200800690. [DOI] [PubMed] [Google Scholar]
- 49.Wehr T. Handbook of Isoelectric Focusing and Proteomics. San Diego: Elsevier; 2005. p. 181. [Google Scholar]
- 50.Yasuda-Kamatani Y, Yasuda A. Gen. Comp. Endocrinol. 2000;118:161. doi: 10.1006/gcen.1999.7453. [DOI] [PubMed] [Google Scholar]
- 51.Redeker V, Toullec JY, Vinh J, Rossier J, Soyez D. Anal. Chem. 1998;70:1805. doi: 10.1021/ac971309c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.