Abstract
The inositol 1,4,5-trisphosphate receptors (IP3Rs) as ligand-gated Ca2+ channels are key modulators of cellular processes. Despite advances in understanding their critical role in regulating neuronal function and cell death, how this family of proteins impact cell metabolism is just emerging. Unexpectedly, a transgenic mouse line (D2D) exhibited progressive glucose intolerance as a result of transgene insertion. Inverse PCR was utilized to identify the gene disruption in the D2D mice. This led to the discovery that Itpr1 is among the 10 loci disrupted in chromosome 6. Itpr1 encodes for IP3R1, the most abundant IP3R isoform in mouse brain and also highly expressed in pancreatic β-cells. To study IP3R1 function in glucose metabolism, we utilized the Itpr1 heterozygous mutant mice, opt/+. Glucose homeostasis in male mice cohorts was examined by multiple approaches of metabolic phenotyping. Under regular diet, the opt/+ mice developed glucose intolerance but no insulin resistance. Decrease in second phase glucose-stimulated blood insulin level was observed in opt/+ mice, accompanied by reduced β-cell mass and insulin content. Strikingly, when fed with high-fat diet, the opt/+ mice were more susceptible to the development of hyperglycemia, glucose intolerance and insulin resistance. Collectively, our studies identify the gene Itpr1 being interrupted in the D2D mice, and uncover a novel role of IP3R1 in regulation of in vivo glucose homeostasis and development of diet-induced diabetes.
Keywords: IP3R1, mouse models, gene disruption, glucose intolerance, diet-induced diabetes
Introduction
The inositol 1,4,5-trisphosphate receptors (IP3Rs) are a family of ligand-gated Ca2+ channels. Located on the membrane of intracellular Ca2+ stores, such as endoplasmic reticulum (ER) and secretory vesicles, IP3Rs mediate Ca2+ release when bound with inositol 1,4,5-trisphosphate (IP3) (Foskett et al. 2007). As key modulators of cytosolic Ca2+ concentration, IP3Rs have been intensively studied in a variety of Ca2+-controlled cellular processes including muscle contraction, neuronal processing and cell death (Vanderheyden et al. 2009). In the central nervous system, IP3R-mediated Ca2+ signaling regulates synaptic transmission, secretion, excitability, learning and memory (Foskett 2010). Emerging evidence suggests that the IP3Rs may also play regulatory roles in pancreatic β-cell exocytosis and metabolism (Srivastava et al. 1999, Dyachok & Gylfe 2004). For example, mice heterozygous for the anx7 gene, which encodes for Ca2+-activated GTPase supporting Ca2+ channel activities, exhibit defects in IP3 receptor expression, Ca2+ signaling and insulin secretion in cells from pancreatic islets (Srivastava et al. 1999). It has been reported that protein kinase A-mediated promotion of Ca2+-induced Ca2+ release via IP3Rs is implicated as part of the mechanism by which cAMP amplifies insulin release (Dyachok & Gylfe 2004).
In mammals there are three isoforms of IP3R (IP3R1, 2 and 3) forming homo- or heterotetrameric channels (Foskett et al. 2007). Expression of mouse IP3R2 is most prominent in cardiac and skeletal muscle, as well as liver, kidney and other epithelial tissues, while mouse IP3R3 is expressed in both endocrine and exocrine pancreas, as well as testis, spleen, thymus and gastrointestinal tract (Taylor et al. 2009). Strikingly, double knockout of IP3R2 and IP3R3 in mice showed severe impairment of Ca2+ signaling and secretion in acinar cells of the exocrine tissues, and the mice are lean and hypoglycemic as a result of difficulty in nutrient digestion (Futatsugi et al. 2005). Furthermore, variations within IP3R3 have been identified as a risk factor for type 1 diabetes in humans (Roach et al. 2006).
IP3R1 is the most abundant isoform in mouse brain and pancreatic β-cells (De Smedt et al. 1997, Lee & Laychock 2001). In mice and humans, genetic mutation and functional impairment of IP3R1 have been linked to multiple neurological diseases, including spinocerebellar ataxias, Huntington’s and Alzheimer’s diseases (Foskett 2010). The IP3R1 gene (Itpr1), located on the distal portion of mouse chromosome 6, is critical for survival as its homozygous deletion in mice leads to ataxia and epileptic seizures resulting in death by 3 to 4 weeks of age (Matsumoto et al. 1996). Interestingly, these phenotypes are highly similar to that of the opisthotonous (opt) mouse (Street et al. 1997). The opt mutation arose spontaneously in a C57BL/Ks-db2J colony and classic genetic techniques localized opt to the same chromosomal localization as Itpr1. Subsequent genetic and molecular analysis revealed that the opt mutation results in a genomic deletion of 2 exons in Itpr1 and aberrant splicing of the Itpr1 mRNA transcripts (Street et al. 1997). Thus, the opt mutation leads to production of IP3R1 protein missing several potential kinase and ATP-binding regulatory sites, with decreased stability. In Western blot analysis, the IP3R1 protein level was about 10% and 67% of the wild-type level for opt homozygotes and heterozygotes respectively (Street et al. 1997), likely resulting from protein degradation mediated by cellular protein quality control mechanisms (Foskett 2010).
The GRP78 gene (Grp78) encodes for a stress-inducible ER chaperone which is a master regulator of the unfolded protein response (UPR) (Wang et al. 2009). Recent studies showed that Grp78 heterozygosity triggers adaptive UPR resulting in attenuation of diet-induced obesity and diabetes in C57BL/6 mice (Ye et al. 2010a). Transgenic mouse models are valuable tools for investigating gene expression patterns in vivo (Xian et al. 2001). While investigating the role of Grp78 promoter elements in regulating the expression of this gene in vivo, we created several transgenic mouse strains with Grp78 promoter deletions. Fortuitously, we observed progressive glucose intolerance in one of the transgenic mouse lines, referred to below as D2D, as a consequence of the genomic integration site of the transgene. With inverse PCR (IPCR), we determined that the transgene replaced 10 loci on chromosome 6, with the Itpr1 gene interrupted near the middle. As a first step towards elucidating the molecular mechanism leading to glucose intolerance in the D2D line, we investigated whether disruption of one Itpr1 allele is a contributing factor to the phenotype, assisted by the opt/+ mouse model. Our results revealed that opt/+ mice exhibited glucose intolerance but no insulin resistance. Decrease in second phase glucose-stimulated blood insulin level was observed in opt/+ mice, associated with reduction in β-cell mass and insulin content. The pathophysiological significance of IP3R1 heterozygosity was further revealed by the aggravation of high-fat diet (HFD)-induced diabetes in opt/+ mice. Collectively, our findings demonstrate a novel physiological role of IP3R1 in maintenance of glucose homeostasis.
Materials and Methods
Ethics statement
All protocols for animal use and euthanasia were reviewed and approved by the University of Southern California Institutional Animal Care and Use Committee. The animal assurance number is A3518-01. The protocol numbers are: 9964, 10621, 11307.
Plasmid construction
To construct the D2/LacZ plasmid, the BamHI fragment from −169/LacZ was first inserted into the BglII site at the internal deletion junction of D300/LacZ (Luo et al. 2003). The D2 promoter was then digested with HindIII and sub-cloned back to the SV40-β-gal plasmid at the HindIII site.
Animals
The D2/LacZ transgenic mouse lines (D2D, D2P and D2L) were generated by the same procedure as previously described (Dong et al. 2004). Itpr1 heterozygous mutant mice opt/+ were purchased from Jackson Laboratory, in the B6C3Fe genetic background. Both D2D and the opt/+ mice were maintained via sibling mating. Mice were fed on regular diet (RD, 11% fat by calories, Harlan Teklad) continuously after weaning at 3-week old, or changed to high-fat diet (HFD, 45% fat by calories, Research Diets) at 10-week old, with ad libitum food access. Only male mice were used in this study. Mouse body weight was measured after overnight fasting. Food intake was analyzed by daily food mass measurement for 4 or 5 successive days. Mouse stool was processed to Oil Red O staining for lipids as described (Ye et al. 2010a). After sacrifice, the epididymal fat pads and liver were removed and weighed, then calculated as percentage of whole body weight.
Blood glucose, insulin, triglyceride and free fatty acid
Mouse tail blood was measured for glucose by OneTouch Ultra System (Lifescan Inc., Milpitas, CA), or prepared for plasma and measured for insulin with ELISA kit (Linco Research). Retro-orbital blood was collected and prepared for serum, then assayed for triglyceride and free fatty acid by the Mouse Metabolic Phenotyping Core at University of Texas Southwestern Medical Center.
Glucose tolerance test
After overnight fasting, mice were subjected to intraperitoneal injection of glucose (1 mg/g body weight), followed by blood glucose or insulin measurement at multiple time points.
Insulin tolerance test
After 6 h fasting, mice were subjected to intraperitoneal injection of insulin (0.5 mU/g body weight), followed by blood glucose measurement at multiple time points.
Inverse PCR (IPCR)
The strategy of IPCR was described previously (Li et al. 1999). Genomic DNA was prepared from mouse tail biopsy with the previously described procedure (Laird et al. 1991). Five µg genomic DNA was digested with 40 U restriction endonuclease StuI (New England Biolabs, Ipswich, MA) in a total volume of 50 µl (37°C, overnight). Following inactivation of the restriction enzyme (65°C, 20 min) and precipitation in 70% ethanol, the restrictive DNA fragments were ligated with 3200 U of T4 DNA ligase (New England Biolabs, Ipswich, MA) in a total volume of 450 µl (16°C, overnight) to form circles. After precipitation in 70% ethanol, the DNA was dissolved in 40 µl storage buffer (10 mM Tris-Cl, 0.1 mM EDTA, pH 7.5) and served as the template of the subsequent nested PCR. The nested PCR was performed in a total volume of 50 µl, with 2.5 U Taq polymerase (New England Biolabs, Ipswich, MA) and 2.5 U Taq extender (Stratagene, La Jolla, CA). The primer sequences are listed in Table 1. PCR products were separated by agarose gel electrophoresis, purified with Gel Extraction Kit (QIAGEN, Valencia, CA), and sequenced.
Table 1.
Primers for inverse PCR (IPCR).
| For the 5’-end of the D2/LacZ transgene | |
| 1st pair | |
| G78P-1F | 5’ – GCAGCGTACTTCTTCCGAGTGAGA – 3’ |
| G78P-1R | 5’ – TCACAAAGATTGTCCTGCTTCTATCT – 3’ |
| 2nd pair | |
| G78P-2F | 5’ – GAAACGGTTTCCAGGTGAGAGGTCA – 3’ |
| G78P-2R2 | 5’ – CATGAGCCACCATATCTATCTCCT – 3’ |
| For the 3’-end of the D2/LacZ transgene | |
| 1st pair | |
| LacZ-1F | 5’ – CGCTACAGTCAACAGCAACTGATGGA – 3’ |
| LacZ-1R | 5’ – CTGCAAGGCGATTAAGTTGGGTAAC – 3’ |
| 2nd pair | |
| LacZ-2F | 5’ – CATGGCTGAATATCGACGGTTTCCA – 3’ |
| LacZ-2R | 5’ – CGGCTTACGGCAATAATGCCTTTCCA – 3’ |
Genotyping
The D2 transgenic mouse lines (D2D, D2P and D2L) were genotyped by PCR with the primer pair LacZ-901 and LacZ-1351, yielding a 451-bp product from the D2/LacZ transgene locus and no product from the wild-type allele. For direct determination of the junction sequences between genomic DNA and the transgene, the primers G78P-1R and ItprR were used for the 5’-end, and M106M-f6 and LacZ-3F for the 3’-end. To distinguish the homozygous D2D mice from the heterozygous ones, PCR primers Itpr-1f and ItprR-3 were used to amplify a 604-bp product exclusively from the wild-type allele, while LacZ-901 and LacZ-1351 were used to detect the D2D allele. The primer sequences are listed in Table 2. Opt mice were genotyped following the protocol from the Jackson Laboratory.
Table 2.
Primers for mouse genotyping.
| LacZ-901 | 5’ – CTGGCTGGAGTGCGATCTTCCTGAG – 3’ |
| LacZ-1351 | 5’ – GCGAGTGGCAACATGGAAATCGCTG – 3’ |
| G78P-1R | 5’ – TCACAAAGATTGTCCTGCTTCTATCT – 3’ |
| ItprR | 5’ – AGGAGGCCATTCCCAAGAGGCACGAT – 3’ |
| M106M-f6 | 5’ – CAGAGTATTGTTTCTGTTCTTGCCCTGATGG – 3’ |
| LacZ-3F | 5’ – GACAAACTCGGAACTTGTTTATTGCAGC – 3’ |
| Itpr-1f | 5’ – TTCATCTGGTTCGTAGTTGCGTGG – 3’ |
| Itpr-R3 | 5’ – CACAGATGGCCTGGCTAGAAACAGAGG – 3’ |
Immunoblotting
Whole cell lysates of mouse tissues were prepared as described (Ye et al. 2010a). To detect IP3R isoforms with high molecular weight (more than 240 kDa), protein lysates were separated on 6% SDS-PAGE. The IP3R antiserum and antibody were gifts from Dr. Richard Wojcikiewicz (State University of New York Upstate Medical University). These included rabbit polyclonal IP3R1 (CT1) antiserum against the conserved C terminus of rat IP3R1, rat monoclonal IP3R1 (4C11) antibody against the amino acids 679–727 of mouse IP3R1. β-actin primary antibody was from Sigma-Aldrich, St. Louis, MO.
Immunohistochemistry
Mouse pancreas was dissected and fixed in 10% PBS-buffered formalin, then processed for paraffin sections. Immunostaining was performed as previously described (Ye et al. 2010b). Primary antibodies used included insulin and glucagon (Signet Labs, Dedham, MA). Quantitation of insulin-positive β-cell area and insulin staining density was performed on multiple sections with the ImageJ 1.40 software (National Institutes of Health, Bethesda, MD).
Islet insulin content
After anesthesia and sacrifice, mouse islets of Langerhans were prepared as previously described (Ye et al. 2010b). Briefly, about 3.5 ml ice-cold digestion solution, which consisted of 0.233 mg/ml Liberase RI and 0.1 mg/ml DNase (Roche, Indianapolis, IN) in Hanks’ balanced salt solution (HBSS), was infused into the pancreas via the common bile duct and the pancreatic duct. The inflated pancreas was excised and transferred to a glass vial with 2.5 ml ice-cold digestion solution. After incubation in a 37°C water bath for 25 min, the vial was shaken by hand vigorously for 45 sec to disperse the digested content. After washing with ice-cold HBSS at least 5 times, the content was placed in a 10-cm dish, and the islets were hand-picked under a microscope. Individual islet was disrupted in 0.2 mL of 1 M acetic acid with 0.1% BSA and protease inhibitor cocktail (Roche, Indianapolis, IN) by sonication (30 sec, on ice). Islet lysates were 1:200 diluted and assayed for insulin concentration with ELISA kit (Linco Research).
Statistical analysis
Two-tailed student’s t-test was applied for all pairwise comparisons.
Results
Impaired glucose homeostasis in D2D transgenic mice
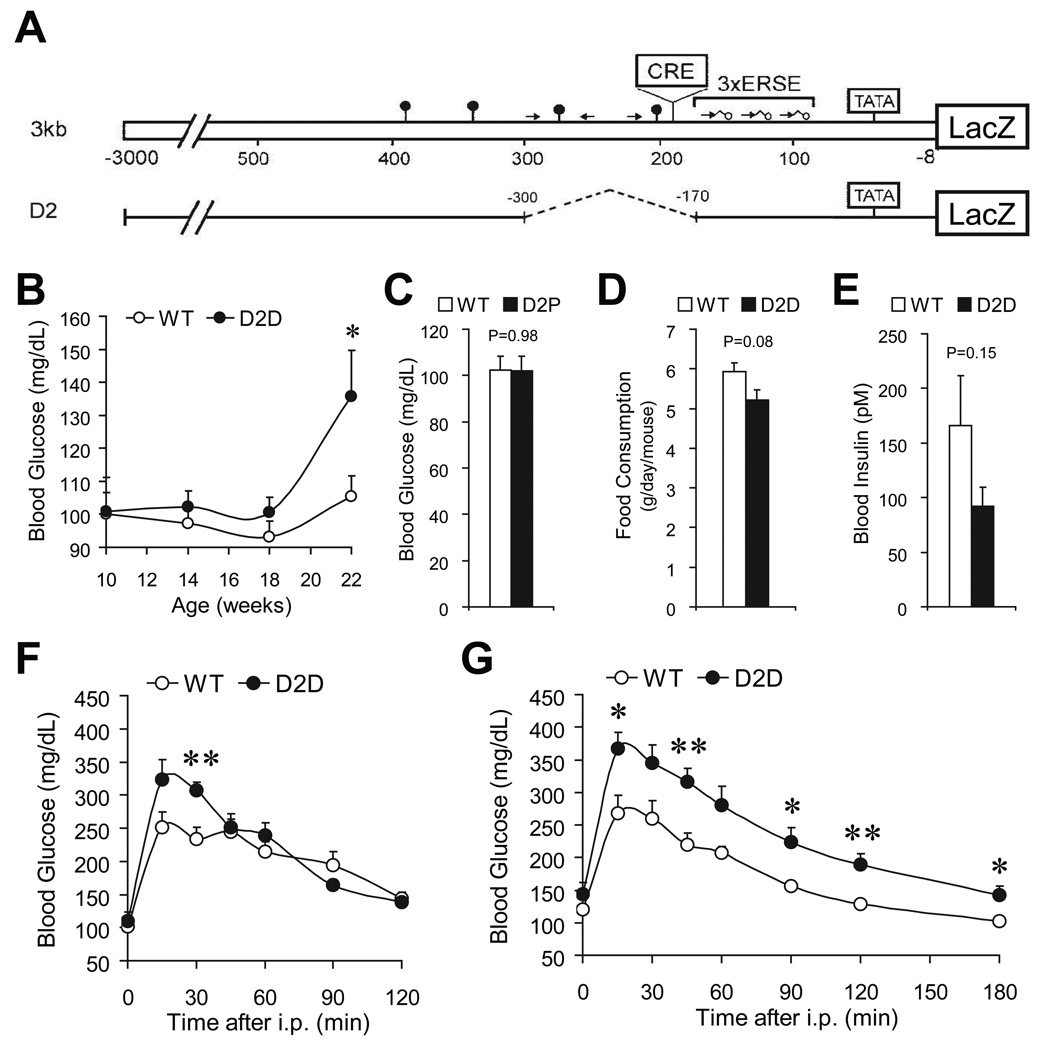
Towards understanding how Grp78 transcription is regulated in vivo, D2, an internal deletion mutant of the 3 kilobase rat Grp78 promoter, was constructed to drive a LacZ reporter gene (Fig. 1A). Within the D2 promoter, the sequence spanning −300 to −170 was deleted, eliminating the ATF/CRE-like site and adjacent upstream sequence. Three lines of the D2 transgenic mice were independently generated, denominated as D2D, D2P and D2L, respectively. Serendipitously, we observed elevated fasting blood glucose in male D2D mice (136±14 mg/dl versus 105±6 mg/dl in their sex-matched wild-type littermates, P=0.048) at the age of 22 weeks (Fig. 1B), but not in the D2P (Fig. 1C) or D2L (data not shown) lines. This moderate hyperglycemia was not due to excessive food intake (Fig. 1D). Rather, it associated with a 45% decrease in blood insulin (Fig. 1E). Strikingly, D2D mice developed progressive glucose intolerance, from 14-week old (Fig. 1F) to 23-week old (Fig. 1G), as revealed by intraperitoneal glucose tolerance test.
Figure 1.
D2D transgenic mice exhibited progressive impairment of glucose homeostasis. Schematic drawing of the 3kb/LacZ and D2/LacZ reporter genes. The locations of the TATA box, ERSE, and the CRE element are indicated. Additional CCAAT sequences and their orientation with respect to the TATA element are represented by arrows. The lollipop symbols indicate the occurrence of GC-rich sequences similar to Sp1 binding sites (A). Fasting blood glucose of D2D mice and their wild-type (WT) siblings at the indicated ages (n≥7 mice for each genotype) (B). Fasting blood glucose of 7-month old WT (n=5) and D2P mice (n=7) (C). Food intake measurement on 7-month old WT and D2D mice (n=3 per genotype) (D). Fasting blood insulin of 6-month old WT (n=14) and D2D mice (n=12) (E). Glucose tolerance test on 14-week old WT (n=6) and D2D (n=5) mice (F). Glucose tolerance test on 23-week old WT (n=6) and D2D (n=7) mice (G). Data are presented as the mean±SEM. *P<0.05, **P<0.01.
Disruption of the inositol 1,4,5-trisphosphate receptor 1-encoding gene (Itpr1) in D2D mice
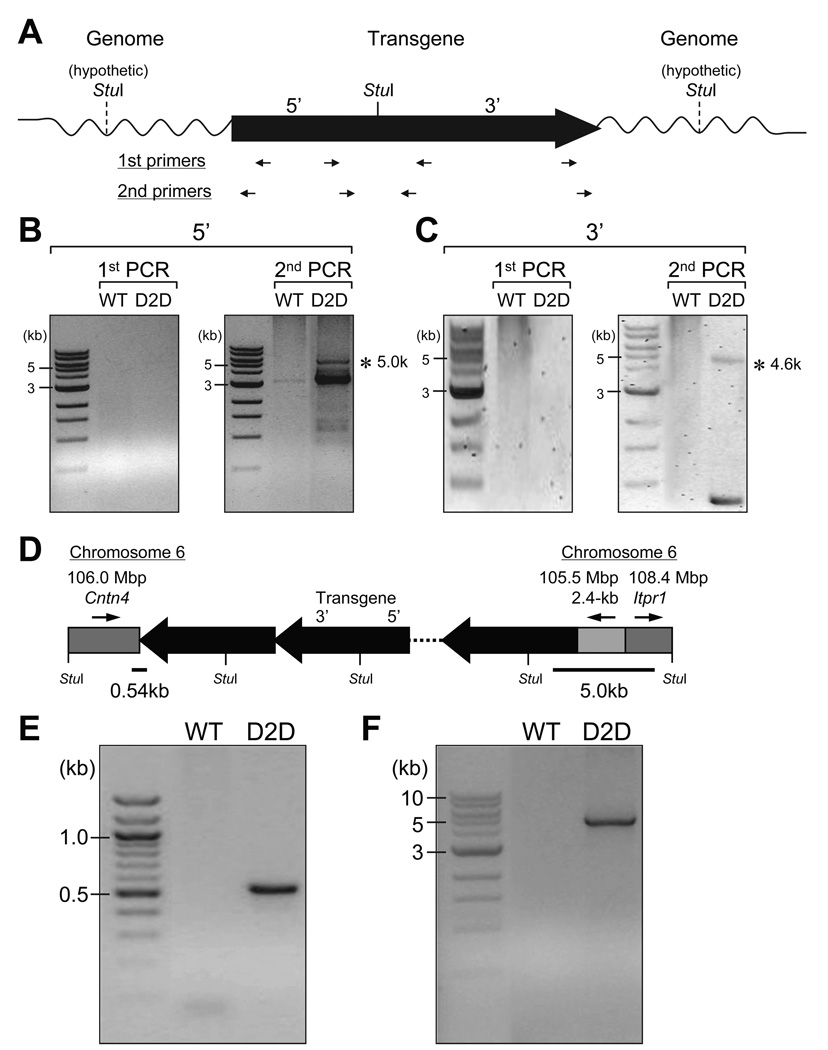
Our observation that perturbed glucose homeostasis was only observed in the D2D line but not in the other lines of the D2 transgenic mice suggests that it likely resulted from the genomic integration site of the D2 transgene in the D2D line rather than an inherent property of the D2 transgene. To identify the genomic DNA sequences flanking the transgene in the D2D mice, we employed the inverse PCR (IPCR) strategy described previously (Li et al. 1999). As depicted in Figure 2A, the genomic DNA from the D2D mice was digested with the restriction endonuclease StuI, which cut the D2/LacZ transgene once in the middle. Following the circular ligation of the restricted DNA fragments, the genomic DNA flanking the 5’-end (Fig. 2B) and 3’-end (Fig. 2C) of the transgene was amplified by nested PCR. Sequencing of the nested PCR products revealed that the two loci flanking the D2 transgene were located on mouse chromosome 6 and identified as the Itpr1 and Cntn4 (contactin 4) (Cottrell et al. 2011) genes (Fig. 2D). Additionally, a 2.4-kb upstream fragment on chromosome 6 was incorporated between the 5’-end of the transgene and the 3’-half of the Itpr1 locus, in a reverse-complementary manner. The nested PCR also amplified sequences from the self-ligated transgene, suggesting the presence of tandem repeats of the transgene in the genome. The junctions between the transgene and the mouse genome were independently determined by PCR amplification from the D2D genomic DNA and sequencing of the PCR products (Fig. 2E and F), providing direct evidence that the Cntn4 and Itpr1 genes were interrupted in the D2D mice.
Figure 2.
Identification of the genomic DNA sequences flanking the transgene in D2D mice. Strategy of the inverse PCR (IPCR). The D2/LacZ transgene is presented with 5’ to 3’ orientation. The single StuI restriction site on the transgene is indicated by a solid line, with the assumed ones on the flanking genomic DNA represented by dash lines. The restrictive DNA fragments were ligated to form circles. The position and orientation of the nested PCR primers are indicated by arrows underneath the transgene, for detection of the genomic sequences flanking either the 5’-end or the 3’-end of the transgene (A). Agarose gel electrophoresis of the products from the nested PCR. Asterisks indicate the PCR products containing mouse genomic DNA, flanking the 5’-end (B) or the 3’-end (C) of the transgene, as revealed by sequencing and BLAST with assembled mouse genome. Schematic drawing of the transgene tandem repeats with flanking genomic sequences on mouse chromosome 6, as suggested by IPCR. The gray boxes represent the genomic DNA, with arrows above indicating their 5’ to 3’ orientation on the chromosome 6. Their positions on the chromosome and the involved loci are also noted on top. The StuI sites on the transgene repeats and the flanking DNA are labeled (D). To independently determine the junction sequences, the primer pair M106M-f6 and LacZ-3F was used for the 3’-end of the transgene, yielding a 0.54-kb PCR product (E) and the G78P-1R and ItprR pair was used for the 5’-end, yielding a 5.0-kb PCR product (F).
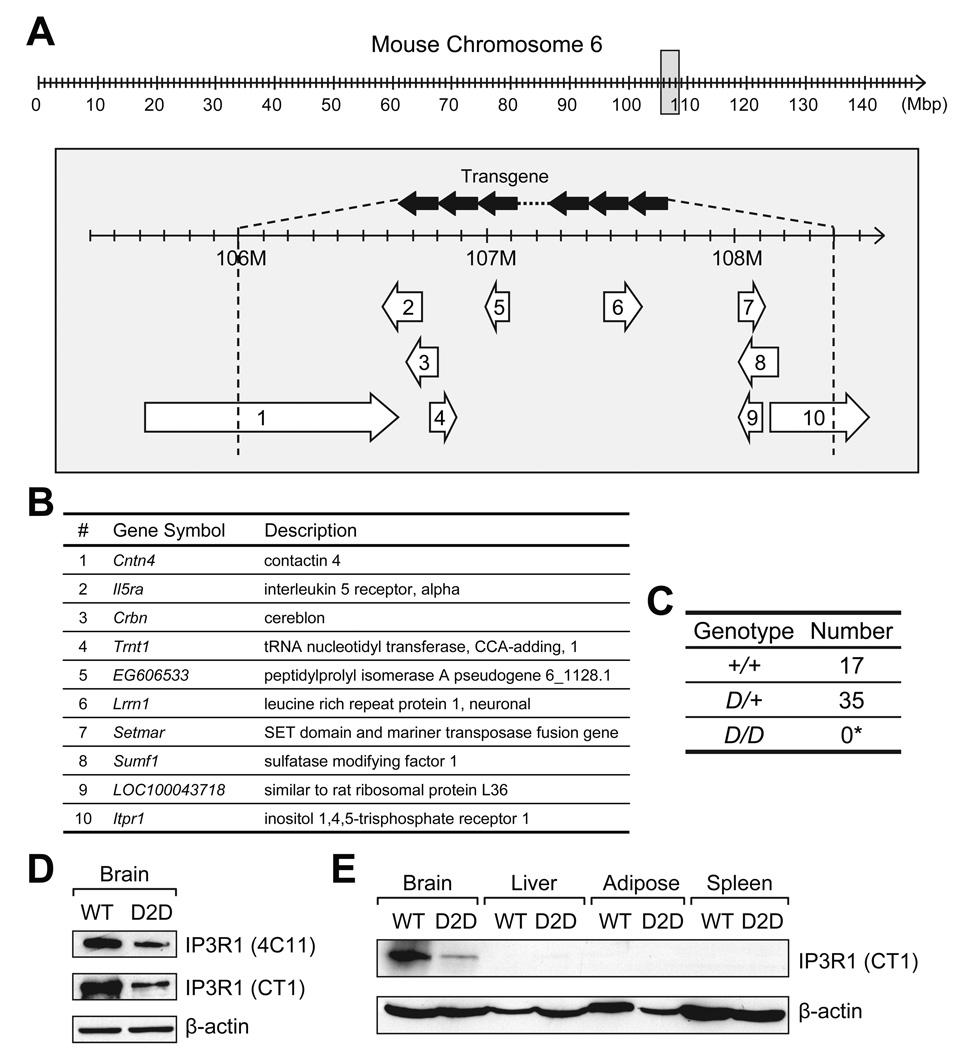
Genomic mapping by IPCR implied that on chromosome 6 of the D2D mice, the 2.4-Mbp genomic DNA between the two junctions was replaced by the tandem repeats of D2/LacZ transgene, with its 5’ to 3’ orientation in inverse to that of chromosome 6 (Fig. 3A). The 2.4-Mbp sequences contain 8 intact loci, as well as the 3’-half of Cntn4 and the 5’-half of Itpr1 (Fig. 3B). The identification of the junction sequences enabled us to distinguish the homozygous D2D (D/D) from the heterozygous mice (D/+) by determining the presence of the wild-type allele (+) and the D2D allele (D) with genotyping via PCR. In 3-week old offspring (n=52) from D/+ × D/+ mating pairs, there was no D/D, and the +/+:D/+ ratio was approximate to 1:2 (Fig. 3C). The lethality for the homozygous D2D mice was consistent with that reported for the homozygous Itpr1 knockout and opt mutant mice (Matsumoto et al. 1996, Street et al. 1997). However, the contribution by the other 9 disrupted loci remains to be determined. In agreement with the genetic and molecular data that the D2D mice were heterozygous for the transgene insertion (D/+), the level of the Itpr1-encoded IP3R1 protein in D2D mouse brain was reduced by about 63% compared to wild-type siblings, as revealed independently in Western blots using antibodies against the amino acids 679–727 (4C11) or the carboxyl end (CT1) of IP3R1 (Fig. 3D). In contrast to its prominent expression in the brain, IP3R1 protein was barely detectable in the liver and below detection limit in white adipose and spleen of both wild-type and D2D mice (Fig. 3E).
Figure 3.
D2D mice are Itpr1-heterozygous. Schematic drawing of D2D mouse chromosome 6 with the transgene insert. In the 148-Mbp of mouse chromosome 6 (upper panel), the area surrounding the transgene insertion site is represented by the gray box expanded in the lower panel. Schematic drawing of the insertion site was modified from the MGI Mouse Genome Browser on build 37, NCBI transcripts. The transgene in reverse-complementary tandem repeats (solid arrows) displaces a 2.4-Mbp (106.0–108.4 Mbp) segment of genomic DNA (A). The 10 open reading frames in this segment are represented by the numbered open arrows underneath, and annotated in (B). The arrows indicate the orientation of the genes. Genotypes of 3-week old offspring from mating pairs of heterozygous D2D mice (D/+ × D/+) with observed numbers. (*Probability of finding no homozygous D2D (D/D) progeny among 52 individuals by chance; P=3.2×10−7) (C). Western blotting for IP3R1 in the brain of D2D mice and their wild-type (WT) littermates (D). Primary antibodies against amino acids 679–727 (4C11) or the conserved C terminal (CT1) of IP3R1 were used. β-actin served as the loading control. Western blotting for IP3R1 in the brain, liver, white adipose and spleen of WT and D2D mice, with β-actin as the loading control (E).
Perturbed glucose homeostasis in the Itpr1 heterozygous mutant (opt/+) mice
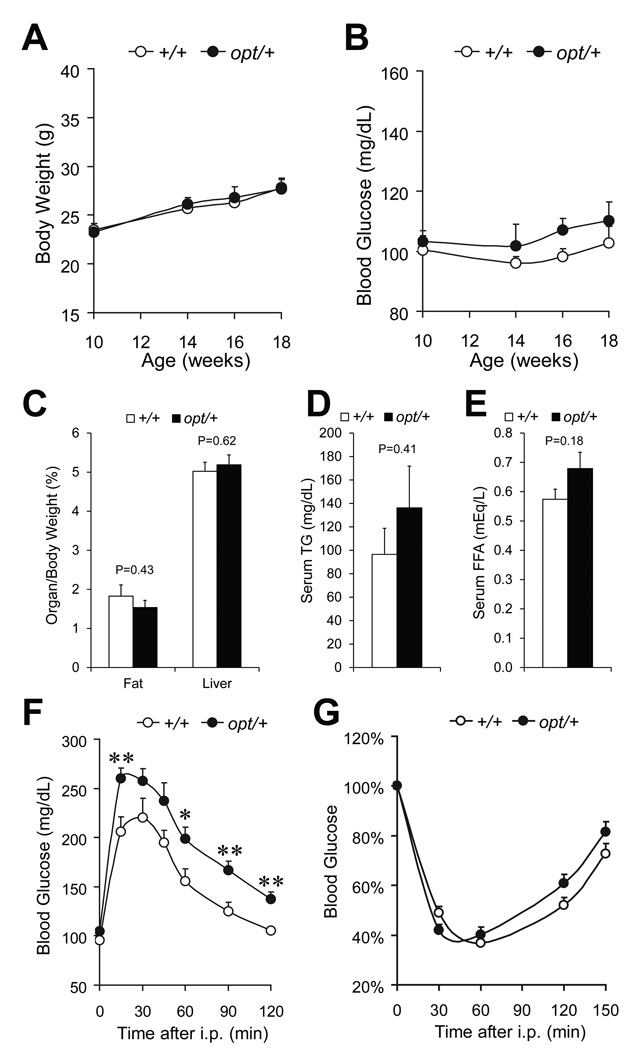
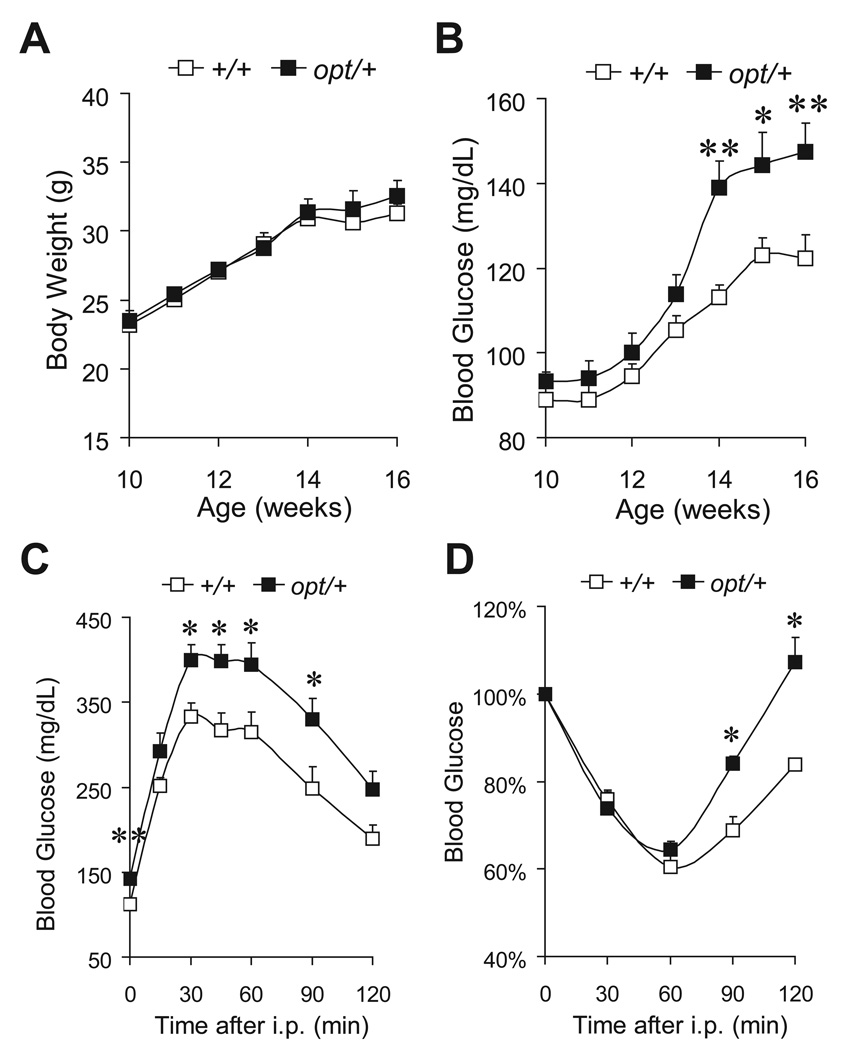
IP3R1 is the most abundant isoform in mouse neurons, serving critical functional roles (Foskett 2010). It is also highly expressed in pancreatic β-cells together with IP3R3 (De Smedt et al. 1997, Lee & Laychock 2001), with implied function in Ca2+ signaling and exocytosis (Dyachok & Gylfe 2004, Dyachok et al. 2004). Since both central nervous system and endocrine pancreas are essential for glucose homeostasis and energy metabolism (Prentki & Nolan 2006, Cota et al. 2007), we directly examined the role of IP3R1 in glucose metabolism utilizing the opt/+ mouse model, which had been established to express reduced level of IP3R1 (Street et al. 1997). Only male mice were used for examination, since the hormonal cycle in females might interfere with metabolism and complicate the results. During the age of 10–18 weeks, the fasting body weight (Fig. 4A) and blood glucose (Fig. 4B) of opt/+ mice were similar to those of their wild-type littermates. The weight of epididymal fat pads and liver were similar between the two genotypes (Fig. 4C). Analysis of serum levels of triglycerides (Fig. 4D) and free fatty acids (Fig. 4E) showed trends towards elevation in opt/+ mice, although neither of them reached statistical significance at the sample size being examined. However, at the age of 10 weeks, opt/+ mice exhibited overt glucose intolerance (Fig. 4F). During the intraperitoneal glucose tolerance test, opt/+ mice reached a higher level of blood glucose than wild-type (260±10 mg/dL versus 206±10 mg/dL, P=0.009) 15 min after an injection of glucose (1 mg/g body weight). Two h after the glucose bolus, while the blood glucose level in wild-type (105±3 mg/dL) was restored to the fasting level, the blood glucose level in the opt/+ mice (138±7 mg/dL) remained significantly higher (P=0.003). The defective glucose clearance in opt/+ mice was not a likely outcome of insulin resistance, since the whole body insulin sensitivity was similar between the two genotypes, as revealed by the intraperitoneal insulin tolerance test (Fig. 4G).
Figure 4.
Glucose intolerance in opt/+ mice. Fasting body weight (A) and fasting blood glucose (B) of opt/+ mice and their wild-type littermates (+/+), n≥6 mice per genotype. Epididymal fat pad and liver weight normalized against whole body weight, n=7 (+/+) or 6 (opt/+) (C). Serum concentration of triglyceride (D) and free fatty acid (E), n=4 (+/+) or 5 (opt/+). Glucose tolerance test on 10-week old mice, n=6 (+/+) or 8 (opt/+) (F). Insulin tolerance test on 11-week old mice, n=12 (+/+) or 15 (opt/+). For an individual mouse, the blood glucose level is calculated as percentage of that at 0 min (G). Data are presented as the mean±SEM. *P<0.05, **P<0.01.
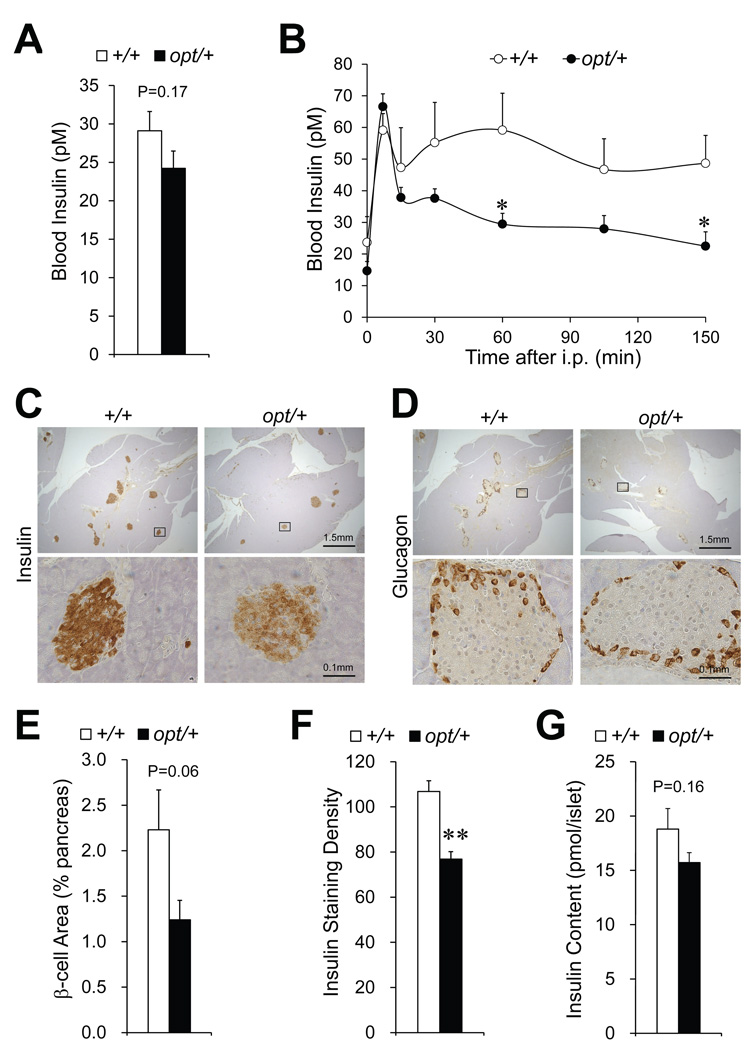
To investigate whether defective insulin production contributes to the glucose intolerance, we first measured the fasting insulin (Fig. 5A), and observed a 17% decrease in opt/+ mice (29±3 pM versus 24±2 pM in wild-type, P=0.17). After a bolus of glucose stimulation via intraperitoneal injection, while the wild-types exhibited a biphasic insulin response in blood, opt/+ mice showed an intact first phase followed by a significantly diminished second phase (Fig. 5B). Immunostaining on pancreas sections revealed decreased β-cell mass and insulin content in opt/+ mice (Fig. 5C). In contrast, the α-cell distribution was comparable to wild type (Fig. 5D). Quantitative analysis confirmed lower β-cell area (Fig. 5E) and insulin staining density (Fig. 5F) in opt/+ mice. To directly examine the islet insulin content, we freshly isolated pancreatic islets from +/+ and opt/+ mice and selected for size-matched ones via microcopy. A 16% decrease in the average insulin content was observed in opt/+ islets compared to wild-type (Fig. 5G), which is consistent with the immunohistochemistry quantitation (Fig. 5F). Collectively, these data suggest defective insulin production in opt/+ mice, which could contribute in part to the glucose intolerance.
Figure 5.
Reduced pancreatic β-cell mass and insulin content in opt/+ mice. For 10-week old opt/+ mice and their wild-type littermates (+/+): Fasting blood insulin, n=9 mice per genotype (A). Blood insulin during glucose tolerance test, n=6 mice per genotype (B). Immunostaining of insulin (C) and glucagon (D) on pancreas, n=5 (+/+) or 6 (opt/+). Lower panels demonstrate enlarged view of the boxed area in the corresponding upper panels. Quantitation of the β-cell area (E) and brown staining density (F) of the insulin-stained cells. Brown areas are normalized against the corresponding whole pancreas section areas. Insulin content in size-matched isolated islets, n=13 per genotype (G). Data are presented as the mean±SEM. *P<0.05, **P<0.01.
Susceptibility to diet-induced diabetes in opt/+ mice
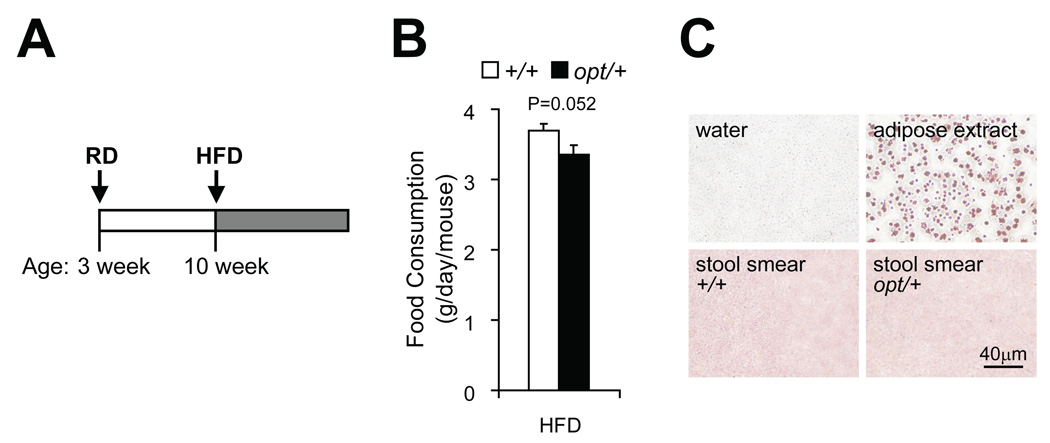
To test whether Itpr1 heterozygosity exposes the mice to diet-induced diabetes, we fed cohorts of opt/+ mice and their wild-type littermates with high-fat diet (HFD) from 10-week old (Fig. 6A). The opt/+ mice showed about 90% of food intake as their wild-type siblings (Fig. 6B), and no fat mal-absorption as revealed by Oil Red O staining of stool smear (Fig. 6C).
Figure 6.
HFD-fed opt/+ mice showed normal food intake and fat absorption. Scheme of high-fat diet (HFD) feeding. Cohorts of opt/+ and +/+ male littermates were fed regular diet (RD) after being weaned at 3-week old, and switched to HFD feeding from 10-week old (A). For mice during the fifth week of HFD. Food intake measurement. n=6 for each genotype. Data are presented as the mean±SEM (B). Oil Red O staining of stool smear. Negative control: dH2O; positive control: white adipose extract. n=4 for each genotype (C).
During the first 6 weeks of HFD regimen, the body weight gain was similar between the two genotypes (Fig. 7A). However, opt/+ mice showed a rapid increase in fasting blood glucose from the fourth week of HFD (139±6 mg/dL versus 113±3 mg/dL in wild-type, P=0.0005) (Fig. 7B). Hyperglycemia remained evident in opt/+ mice after 6 weeks of HFD (148±7 mg/dL versus 122±6 mg/dL in wild-type, P=0.008) (Fig. 7B). Correspondingly, exacerbated glucose intolerance was observed in opt/+ mice after 4 weeks of HFD (Fig. 7C). As further evidence of perturbed glucose metabolism, opt/+ mice were more insulin resistant than wild-type, as revealed by the intraperitoneal insulin tolerance test after 6 weeks of HFD (Fig. 7D). Taken together, our results suggest that the glucose homeostasis in opt/+ mice was more vulnerable upon HFD challenge. With reduced IP3R1 protein expression, opt/+ mice were predisposed to HFD-induced diabetes.
Figure 7.
opt/+ mice were predisposed to HFD-induced diabetes. Fasting body weight (A) and fasting blood glucose (B) of mice fed high-fat diet (HFD) from 10-week old, n≥8 mice per genotype. Glucose tolerance test on mice after 4-week HFD, n=9 (+/+) or 7 (opt/+) (C). Insulin tolerance test on mice after 6-week HFD, n=10 (+/+) or 4 (opt/+) (D). Data are presented as the mean±SEM. *P<0.05, **P<0.01.
Discussion
The D2D transgenic mouse line was originally constructed for the study of Grp78 promoter analysis. Unexpectedly, D2D mice exhibit progressive glucose intolerance not seen in other independently derived lines from the same transgene. This led to the discovery that the D2/LacZ transgene has serendipitously inserted into mouse chromosome 6 and along with displacement of several other loci, split Itpr1 into half. Considering that IP3R1 is both a receptor for IP3 and a calcium channel, generation of viable mouse models with mutated IP3R1 function provides a valuable tool for the understanding of its function in vivo. However, since the D2D harbors other chromosomal aberrations in addition to Itpr1 disruption, it is not possible to dissect the contribution of Itpr1 from the other perturbed loci.
Mouse models targeting Itpr1 function have been previously reported including the Itpr1 knockout model (Matsumoto et al. 1996) and the opt mutant mouse model (Street et al. 1997). While the former is not available, the opt/+ mutant mice with an altered Itpr1 gene are commercially available and aptly serve as a surrogate for the D2D transgenic line with a single disruption of the Itpr1 locus. Previous studies on IP3R1 have focused on its regulation of calcium channels in relation to neurological function. For example, both the opt (Street et al. 1997) and more recently, the Δ18 mice (van de Leemput et al. 2007) bearing spontaneous mutation of the Itpr1 gene led to neuronal dysfunction. The fact that homozygous deletion of the Itpr1 alleles in mice results in ataxia and seizure followed by death by a few weeks after birth did not lend itself to studies of its function in metabolic diseases which require the mice to reach adulthood in relatively stable health. Since the opt/+ mice are phenotypically normal, their potential defects in metabolism have not been apparent in past studies.
Upon matching the D2D mice with the opt/+ mice, our studies extend characterization of the opt/+ mice and reveal several new findings on IP3R1 function in regulation of glucose homeostasis. First, in mouse peripheral tissues including liver and white adipose, IP3R1 protein is hardly detectable by immunoblotting, in agreement with the previous report on its tissue-specific mRNA expression (De Smedt et al. 1997). Consistent with this notion, insulin sensitivity is not altered in regular diet-fed opt/+ mice compared to wild-type, rendering it unlikely that reduced IP3R1 suppresses insulin-stimulated glucose metabolism in peripheral tissues. Importantly, reduction of IP3R1 in opt/+ mice leads to impaired glucose tolerance, correlating with the observations in D2D mice. Although the underlying mechanisms could be complex involving multiple pathways in different organs, our in vivo and in vitro data support the notion that Itpr1 heterozygosity reduces both the mass and insulin production of pancreatic β-cells. These could synergistically contribute to the attenuation of second phase glucose-stimulated insulin response in opt/+ mice. Considering IP3R1 is the dominant isoform expressed in the mouse central nervous system which regulates satiety and neuronal regulated insulin secretion (Foskett 2010), it is also tempting to speculate that the glucose intolerance in opt/+ mice could be the functional defect in neurons responsible for nutrient sensing and glucose regulation (Cota et al. 2007). Another possibility could relate to the emerging role of IP3Rs in regulation of cytosolic Ca2+ and thus insulin secretion in pancreatic β-cells (Srivastava et al. 1999, Dyachok & Gylfe 2004), where IP3R1 is also highly expressed (Lee & Laychock 2001). The resolution of these issues awaits further investigation such as comprehensive characterization of insulin sensitivity in individual organs (Kim 2009) and may be best addressed in future studies utilizing mouse models with tissue specific ablation of IP3R1.
Our studies demonstrate that upon high-fat diet the requirement of IP3R1 to maintain glucose homeostasis is more acute. Unlike mice on regular diet, these mice develop hyperglycemia, severe glucose intolerance and insulin resistance, suggesting predisposition to diabetes. In support of our findings, it was recently reported that parasympathetic stimulation of pancreatic islets augments glucose-stimulated insulin secretion by inducing IP3R-mediated Ca2+ release, such that defective glycemic regulation through loss of ankyrin-B-dependent stabilization of IP3R is a potential risk factor for type 2 diabetes (Healy et al. 2010). Future studies on the molecular pathways leading to development of diabetes due to IP3R deficiency may provide novel strategies to combat diet and age onset diabetes.
Acknowledgements
We thank Dr. Richard Wojcikiewicz for the generous gift of antibodies. We thank the Richard Bergman lab for expert assistance in insulin assays and helpful discussion.
We thank the USC Norris Comprehensive Cancer Center Transgenic/Knockout Mouse Core for generation of the transgenic mice, and the Molecular Genomics Core for DNA sequencing. The USC core facilities described were supported in part by P30CA014089 from the National Cancer Institute. We also thank Dr. Philipp Scherer and the Mouse Metabolic Phenotyping Core at University of Texas Southwestern Medical Center for assistance in triglyceride and free fatty acid assays.
Funding
This research was supported by grants from the National Institutes of Health R01CA027607, R01DK070582 and R01DK079999 awarded to ASL, and R01DK060623 and R01GM085791 to RHC.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Publisher's Disclaimer: Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in Journal of Endocrinology, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the Society for Endocrinology accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at http://dx.doi.org/10.1530/JOE-11-0012. Copyright 2011 Society for Endocrinology.
References
- Cota D, Proulx K, Seeley RJ. The role of CNS fuel sensing in energy and glucose regulation. Gastroenterology. 2007;132:2158–2168. doi: 10.1053/j.gastro.2007.03.049. [DOI] [PubMed] [Google Scholar]
- Cottrell CE, Bir N, Varga E, Alvarez CE, Bouyain S, Zernzach R, Thrush DL, Evans J, Trimarchi M, Butter EM, et al. Contactin 4 as an autism susceptibility locus. Autism Research. 2011 doi: 10.1002/aur.184. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smedt H, Missiaen L, Parys JB, Henning RH, Sienaert I, Vanlingen S, Gijsens A, Himpens B, Casteels R. Isoform diversity of the inositol trisphosphate receptor in cell types of mouse origin. Biochemical Journal. 1997;322(Pt 2):575–583. doi: 10.1042/bj3220575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Dubeau L, Bading J, Nguyen K, Luna M, Yu H, Gazit-Bornstein G, Gordon EM, Gomer C, Hall FL, et al. Spontaneous and controllable activation of suicide gene expression driven by the stress-inducible grp78 promoter resulting in eradication of sizable human tumors. Human Gene Therapy. 2004;15:553–561. doi: 10.1089/104303404323142006. [DOI] [PubMed] [Google Scholar]
- Dyachok O, Gylfe E. Ca(2+)-induced Ca(2+) release via inositol 1,4,5-trisphosphate receptors is amplified by protein kinase A and triggers exocytosis in pancreatic beta-cells. Journal of Biological Chemistry. 2004;279:45455–45461. doi: 10.1074/jbc.M407673200. [DOI] [PubMed] [Google Scholar]
- Dyachok O, Tufveson G, Gylfe E. Ca2+-induced Ca2+ release by activation of inositol 1,4,5-trisphosphate receptors in primary pancreatic beta-cells. Cell Calcium. 2004;36:1–9. doi: 10.1016/j.ceca.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Foskett JK. Inositol trisphosphate receptor Ca2+ release channels in neurological diseases. Pflügers Archiv European Journal of Physiology. 2010;460:481–494. doi: 10.1007/s00424-010-0826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiological Reviews. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futatsugi A, Nakamura T, Yamada MK, Ebisui E, Nakamura K, Uchida K, Kitaguchi T, Takahashi-Iwanaga H, Noda T, Aruga J, et al. IP3 receptor types 2 and 3 mediate exocrine secretion underlying energy metabolism. Science. 2005;309:2232–2234. doi: 10.1126/science.1114110. [DOI] [PubMed] [Google Scholar]
- Healy JA, Nilsson KR, Hohmeier HE, Berglund J, Davis J, Hoffman J, Kohler M, Li LS, Berggren PO, Newgard CB, et al. Cholinergic augmentation of insulin release requires ankyrin-B. Science Signaling. 2010;3:ra19. doi: 10.1126/scisignal.2000771. [DOI] [PubMed] [Google Scholar]
- Kim JK. Hyperinsulinemic-euglycemic clamp to assess insulin sensitivity in vivo. Methods in Molecular Biology. 2009;560:221–238. doi: 10.1007/978-1-59745-448-3_15. [DOI] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Research. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Laychock SG. Inositol 1,4,5-trisphosphate receptor isoform expression in mouse pancreatic islets: effects of carbachol. Biochemical Pharmacology. 2001;61:327–336. doi: 10.1016/s0006-2952(00)00559-1. [DOI] [PubMed] [Google Scholar]
- Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Leukaemia disease genes: large-scale cloning and pathway predictions. Nature Genetics. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- Luo S, Baumeister P, Yang S, Abcouwer SF, Lee AS. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through an upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. Journal of Biological Chemistry. 2003;278:37375–37385. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nakagawa T, Inoue T, Nagata E, Tanaka K, Takano H, Minowa O, Kuno J, Sakakibara S, Yamada M, et al. Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature. 1996;379:168–171. doi: 10.1038/379168a0. [DOI] [PubMed] [Google Scholar]
- Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. Journal of Clinical Investigation. 2006;116:1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, Sheridan CM, Glusman G, Hood L, Lernmark A, et al. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. American Journal of Human Genetics. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Atwater I, Glasman M, Leighton X, Goping G, Caohuy H, Miller G, Pichel J, Westphal H, Mears D, et al. Defects in inositol 1,4,5-trisphosphate receptor expression, Ca(2+) signaling, and insulin secretion in the anx7(+/−) knockout mouse. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13783–13788. doi: 10.1073/pnas.96.24.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street VA, Bosma MM, Demas VP, Regan MR, Lin DD, Robinson LC, Agnew WS, Tempel BL. The type 1 inositol 1,4,5-trisphosphate receptor gene is altered in the opisthotonos mouse. Journal of Neuroscience. 1997;17:635–645. doi: 10.1523/JNEUROSCI.17-02-00635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CW, Taufiq-Ur-Rahman, Pantazaka E. Targeting and clustering of IP3 receptors: key determinants of spatially organized Ca2+ signals. Chaos. 2009;19 doi: 10.1063/1.3127593. 037102. [DOI] [PubMed] [Google Scholar]
- van de Leemput J, Chandran J, Knight MA, Holtzclaw LA, Scholz S, Cookson MR, Houlden H, Gwinn-Hardy K, Fung HC, Lin X, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genetics. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheyden V, Devogelaere B, Missiaen L, De Smedt H, Bultynck G, Parys JB. Regulation of inositol 1,4,5-trisphosphate-induced Ca2+ release by reversible phosphorylation and dephosphorylation. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2009;1793:959–970. doi: 10.1016/j.bbamcr.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wey S, Zhang Y, Ye R, Lee AS. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxidants & Redox Signaling. 2009;11:2307–2316. doi: 10.1089/ars.2009.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian M, Zhang J, Lau YF. Sry promoters from domesticus (Tirano) and C57BL/6 mice function similarly in embryos and adult animals. Journal of Experimental Zoology. 2001;290:632–641. doi: 10.1002/jez.1114. [DOI] [PubMed] [Google Scholar]
- Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010a;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Mareninova OA, Barron E, Wang M, Hinton DR, Pandol SJ, Lee AS. Grp78 heterozygosity regulates chaperone balance in exocrine pancreas with differential response to cerulein-induced acute pancreatitis. American Journal of Pathology. 2010b;177:2827–2836. doi: 10.2353/ajpath.2010.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]