Abstract
Phospholipase D (PLD), a superfamily of signaling enzymes that most commonly generate the lipid second messenger Phosphatidic Acid (PA), is found in diverse organisms from bacteria to man and functions in multiple cellular pathways. Since the early 1980’s when mammalian PLD activities were first described, most of the important insights concerning PLD function have been gained from studies on cellular models. Reports on physiological and pathophysiological roles for members of the mammalian PLD superfamily are now starting to emerge through from genetic models. In this review, we summarize recent findings on PLD functions in these model systems, highlighting newly appreciated connections of the superfamily to cancer, neuronal pathophysiology, cardiovascular topics, spermatogenesis, and infectious disease.
Keywords: Phospholipase D, mitochondria, mouse models, piRNA, influenza, cardiovascular, cancer
Introduction
Phospholipase D (PLD) is formally a transphosphatidylase that catalyzes head-group exchange on phosphodiester bonds linking a variety of substrates (Jenkins and Frohman, 2005). Most commonly, PLD employs water as a nucleophile to hydrolyze phospholipid substrates such as phosphatidylcholine to generate the membrane lipid phosphatidic acid (PA), releasing soluble choline into the cytosol. Outlying members of the superfamily however can use other phospholipid substrates, such as cardiolipin, to generate PA, or hydrolyze the phosphodiester bond found in the backbone of DNA or the protein-DNA linkage resulting from stalled Topoisomerase 1. Finally, some members of the superfamily can use nucleophiles other than water to create new lipids via exchange of a complex headgroup for a simple one on a phospholipid substrate.
PA is a negatively charged phospholipid with a small headgroup that promotes negative membrane curvature, which is thought to facilitate membrane vesicle fusion and fission. PA can also recruit proteins that have basic amino-acid rich regions that exhibit affinity for it, and in some cases, the proteins recruited are enzymes that are activated as a result of the recruitment. Finally, PA can be converted to other signaling lipids, specifically lyso-PA, by phospholipase A2, or diacylglycerol (DAG), by lipid phosphatases (Fig. 1A). PA, lyso-PA and DAG all have effects on multiple cellular pathways, including intracellular vesicle trafficking, endocytosis, exocytosis, actin cytoskeleton dynamics, cell proliferation, differentiation, migration and survival (Donaldson, 2009, Jenkins and Frohman, 2005, Rudge and Wakelam, 2009).
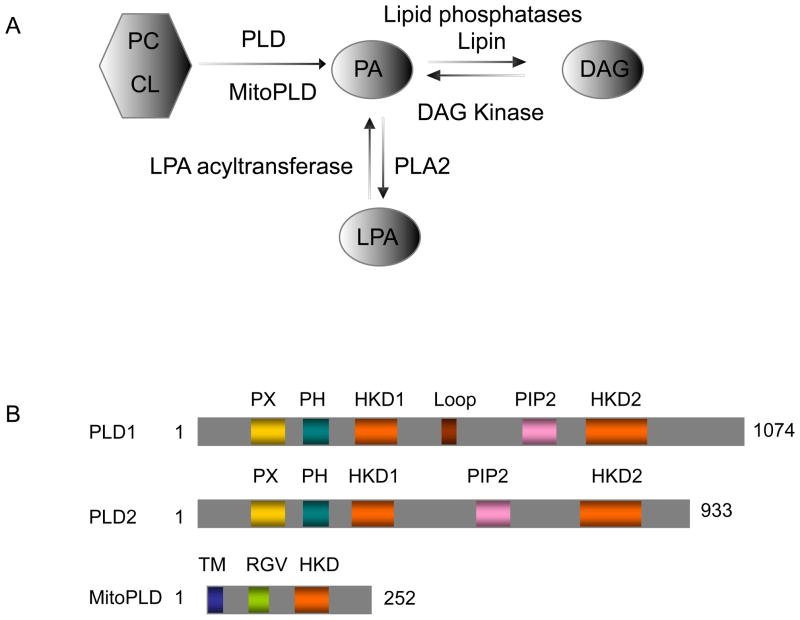
Fig. 1.
A. signaling lipid pathway. PA can be generated via hydrolysis of PC or cardiolipin by PLD or MitoPLD, respectively. Once generated, the PA can be converted to DAG by Lipid Phosphatases (e.g. Lipin) or to LPA by PLA2 (phospholipase A2). B. Cartoon schematic of mammalian PLDs. PX domain (PHOX domain) and PH domain (Pleckstrin domain) mediate protein and lipid binding; HKD domain (HxKxxxxD, x is any amino acid)defines and mediates the catalytic reaction; PIP2 binding domain; Loop domain in PLD1 may function in autoinhibition of PLD1 activity; TM (transmembrane domain) is found in MitoPLD and functions to anchor the protein into the outer surface of mitochondria; RGV domain is enriched in basic amino acids that may interact with membrane surfaces or partner proteins.
There are two classic mammalian isoforms of phospholipase D: PLD1 (Hammond et al., 1995) and PLD2 (Colley et al., 1997) (Fig. 1B). Both PLD isoforms require phosphatidylinositol 4,5-bisphosphate (PIP2) as a co-factor for activity. Otherwise, however, PLD1 and PLD2 exhibit quite different regulatory properties and subcellular localization. PLD1 has a low basal activity in vitro and is activated by small G proteins (ARF, Rho and Rac) and protein kinase C, whereas PLD2 has high basal activity and is insensitive to the PLD1 activators (Du et al., 2000, Colley et al., 1997, Hammond et al., 1997). PLD1 and 2 cycle through a succession of subcellular localizations during signaling events (Du et al., 2003, Du et al., 2004). Most studies show that PLD1 is predominantly localized under steady-state conditions at the Golgi complex, endosomes, lysosomes, and secretory granules, whereas PLD2 is found at the plasma membrane, suggesting different cell biological roles for the PLDs (Brown et al., 1998, Du et al., 2004, Freyberg et al., 2001). However, PLD1 can also be found at the plasma membrane in some settings (Vitale et al., 2001), and PLD2 at the Golgi complex (Freyberg et al., 2002, Yang et al., 2008), illustrating their mobility. Finally, MitoPLD, a PLD family member more recently identified, localizes to the surface of mitochondria (Fig 1B), where it hydrolyzes cardiolipin to generate PA to promote mitochondrial fusion (Choi et al., 2006).
PLD activation has been implicated in many physiological and pathological actions via consideration of cell culture-based models, most of which have involved overexpression or downregulation of PLD genes or biochemical approaches. In this review, we will discuss the newest findings of functions of mammalian PLDs in physiological and pathological actions based on genetic approaches with knock-out and other in vivo models.
Functions of PLD in cancer
PLD function has been implicated in cancer progression in the context of many cellular pathways (Su et al., 2009a). Elevated phospholipase D activity and a driver mutation in PLD2 have been reported in malignant breast cancer biopsy samples (Noh et al., 2000, Uchida et al., 1997, Wood et al., 2007). PLD expression levels also correlate with tumor size and survival in progression of colorectal carcinoma, and increased expression of PLD2 is found in gastric carcinoma and renal cancer (Saito et al., 2007, Uchida et al., 1999, Zhao et al., 2000).
Nonetheless, the molecular mechanism through which PLD supports cancer progression remains unclear. A hypothesis that has been widely explored concerns potential roles for PLD and its product PA in activating mTOR (Fang et al., 2001, Toschi et al., 2009) to suppress cancer cell apoptosis (Sabatini, 2006). Understanding the signals that regulate PA levels and how PA impacts upon mTOR could be important for developing strategies to block the survival signals that suppress apoptosis.
PLD activity may also play an important role in cell motility and migration, a critical step in the spread of cancer. Increased PLD activity enhances the ability of MAD-MB-231 human breast cancer cells to migrate and invade matrigel (Zheng et al., 2006). PLD2 activation increases FAK (focal adhesion kinase) phosphorylation, Akt activation and cell invasion for EL4 lymphoma cells, whereas inactive PLD2 inhibits metastasis (Knoepp et al., 2008); PLD/PLD1 has been reported to be required for secretion of matrix metalloproteinase-9 by colorectal cancer cells (Kang et al., 2008) and MMP-2 by glioma cells (Park et al., 2009).
Thus far, there have no studies of cancer susceptibility or resistance in mice lacking PLD isoforms. However, one hallmark of cancer cell metabolism has been examined and is of interest. Mice lacking PLD1, which are viable and grossly normal (Elvers et al., 2010), exhibit dramatically decreased levels of macroautophagy (Dall’Armi et al., 2010), a protein-and organelle-catabolizing process that generates energy and nutrients for cells under starvation conditions, and hence is vital for tumor cell spread (Xie and Klionsky, 2007). Autophagy plays a normal role in homeostasis, helping to maintain a balance between the synthesis, degradation, and subsequent recycling of cellular products, and decreases in autophagy have been linked to major diseases such as cancer, diabetes and neurodegeneration (Kundu and Thompson, 2008, Mizushima et al., 2008, Rubinsztein et al., 2007, Tooze and Schiavo, 2008). In cell lines, PLD1 was shown to co-localize with autophagosomes upon nutrient starvation, and FIPI, a PLD small molecular inhibitor (Su et al., 2009b), decreased starvation-induced autophagosome expansion and impaired autophage-mediated clearance of protein aggregation (Dall’Armi et al., 2010). Finally, these findings were validated using a genetic model. In wild-type mice restrained from food for 24 hours, significant autophagosome expansion was observed in the liver; however, food starvation failed to enlarge the autophagosome compartment in the livers of PLD1−/− mice. These studies suggest that PLD or PLD1-selective inhibitors might be of utility in approaches targeting the metabolism and survival of metastasizing cancer cells.
Functions of PLD in neurodegenerative diseases
PLD may also play physiological and pathological roles in the brain through affecting basic cellular functions such as signal transduction, vesicular trafficking and actin cytoskeletal reorganization. PLD1 promotes neurite outgrowth in the hippocampus in an in vivo model of learning and memory (Zhang et al., 2004, Zhang et al., 2005).
α-Synuclein has been implicated in the pathogenesis of Parkinson’s disease and Alzheimer’s diseases (Duda et al., 2000). A common pathological hallmark of Parkinson’s disease is the Lewy body, round eosinophilic inclusions found in the substantia nigra. Filamentous α-Synuclein is an important component of the Lewy bodies. Mutations in the PARK1 gene, which encodes α-Synuclein, lead to autosomal-dominant Parkinson’s disease. α-Synuclein normally exists in a naturally unfolded state but can form oligomers, called “protofibrils”, which are cytotoxic and readily aggregate into filaments that create the Lewy inclusion bodies (Lotharius and Brundin, 2002). PLD2 interacts and is inhibited by α-Synuclein (Jenco et al., 1998). PLD2 overexpression in the rat SNc (substantia nigra pars compacta) by viral-mediated gene transfer (Gorbatyuk et al.) was recently shown to lead to the loss of dopamine (DA) neurons resulting in a severe behavior rotational asymmetry. In this model system, α-Synuclein functioned to inhibit PLD2 in vivo and reduced its toxicity. These findings suggest that part of the consequence of α-Synuclein dysregulation may be unopposed PLD2 activity, with deleterious consequences.
Alzheimer’s disease (AD) has also been associated with PLD function. Brains in AD patients display amyloid plaques composed of aggregated Aβ (amyloid-beta peptides) on the outside of cells. Aβ is generated by the sequential cleavage of its precursor protein APP by α-, β- and γ-secretase (Tanzi and Bertram, 2005). PLD1 has been reported to interact with Presenilin1 (PS1), the major component of γ-secretase complex, in a manner that affects APP intracellular trafficking (Cai et al., 2006a, Cai et al., 2006b). In contrast, PLD2 has been linked to a subsequent step. After processing of APP to generate Aβ, the peptides have been observed to stimulate PLD activity via FPRL1 (formyl-peptide-receptor-like 1) (Brandenburg et al., 2008), which plays an essential role in Alzheimer’s disease (AD). Intriguingly, this step appears affected in PLD2−/− mice, which are otherwise viable and grossly normal (Oliveira et al., 2010). Di Paolo and colleagues found that PLD2 ablation rescued memory deficits and conferred synaptic protection in mice overexpressing APP that constitute a genetic model for Alzheimer’s Disease, despite a significant Aβ load. Mechanistically, Aβ42 peptides increased PLD activity in wild-type neurons approximately 2-fold, but no increase was observed in PLD2−/− neurons, indicating that PLD2 is a transducer for the Aβ signaling cascade. Consistent with this finding, ablation of PLD2 blocked the suppressive effect of Aβ oligomers on LTP (long-term potentiation), suggesting that the ability of Aβ42 oligomers to exhibit synaptotoxicity had been suppressed and raising the possibility that PLD2 inhibitors could pose a viable approach for therapeutics in this and similar types of neurodegenerative diseases.
Cardiovascular functions for PLD
Platelets aggregation is vital to terminate bleeding (Mauri, 1964). PLD1 is present in platelets (Chiang, 1994, Lee et al., 1994) and both isoforms rapidly translocate to the plasma membrane area upon platelet activation via agonists such as thrombin (Vorland and Holmsen, 2008a, Vorland and Holmsen, 2008b). Strikingly, platelets from PLD1−/− mice display impaired integrin αIIbβ3 activation and defective aggregate formation in vitro under high shear flow conditions, but normal function under low shear flow conditions (Elvers et al., 2010). In vivo, PLD1−/− mice are consequently resistant to pulmonary emboli, occlusive arterial thrombus formation, and neuronal damage following thrombotic-induced focal cerebral ischemia. Taken together, these findings suggest that a PLD inhibitor such as FIPI (Su et al., 2009b) or a PLD1-selective inhibitor (Lavieri et al., 2010, Scott et al., 2009) might be a promising candidate for an anti-platelet therapeutic.
Unexpectedly, PLD2 has been found to generate a second type of second messenger, cyclic PA (CPA), via hydrolysis of lysophosphatidylcholine (LPC) (Tsukahara et al., 2010). Moreover, CPA acts as an endogenous antagonist of PPARγ. PPARγ is a nuclear hormone receptor that is activated by fatty acids, thiazolidinedione drugs, and LPA and plays a role in insulin sensitivity and adipogenesis, leading to lipid accumulation, macrophage recruitment, and arterial wall thickening in settings of excessive activation. Activation of PLD2 in this setting inhibits PPARγ, blocking arterial wall thickening induced by agonists used to activate PPARγ as therapeutics for type 2 diabetes, raising the prospect of using CPA or CPA analogs as therapeutics for hypertensive disease caused through these mechanisms. Interestingly, mutations in PLD2 have been identified in large scale genetic screens for hypertensive risk factors (Hong et al., 2010). Replacement of Arg172 by Cysteine inversely correlated with risk of hypertension. Arg172 resides in the PX domain of PLD2 and could affect PLD2’s function in membrane trafficking, having one or more effects on PLD2 functions important in the long-term regulation of blood pressure.
Roles for MitoPLD in mitochondrial morphology and germ cell development
Mitochondrial dynamics are fundamentally important for mammalian cells, controlling not only the shape of mitochondria but also their function (Chen and Chan, 2009). Some of the key components that regulate mitochondrial dynamics have been identified, including the Mitofusins Mfn1 and Mfn2 (Chen et al., 2003), which are critical for mitochondrial fusion, and the dynamin-related GTPase Drp1 (Smirnova et al., 2001), which plays a key role in fission. Mitochondrial dynamics are essential for mammalian development, and partial defects in the fusion process can lead to neurodegenerative disease (Chen and Chan, 2009) or promote apoptosis (Wasilewski and Scorrano, 2009).
Our lab previously identified a Phospholipase D family member, denoted MitoPLD, which is anchored on the outer membrane surface of mitochondria (Choi et al., 2006). MitoPLD is an evolutionarily very divergent PLD superfamily member, exhibiting more similarity to the bacterial homolog called Nuc, which exhibits endonuclease activity (Stuckey and Dixon, 1999), than it does to mammalian PLD1 and PLD2. However, MitoPLD does not appear to be an endonuclease, but rather, acts as a phospholipase D to hydrolyze cardiolipin to generate Phosphatidic Acid (PA) and affect mitochondrial dynamics (Choi et al., 2006). Loss of MitoPLD activity decreases fusion, whereas increased MitoPLD activity causes mitochondria to aggregate, separated by a thin layer of electron-dense material.
More recently, we have shown that PA recruits a novel PA phosphatase called Lipin 1 (Reue and Zhang, 2008) that converts the PA to DAG on the surface of mitochondria (Huang et al., 2011). Lipin1 mutations cause a form of lipodystrophy with similarities to type 2 diabetes (Peterfy et al., 2001). We have observed that Lipin 1 overexpression causes mitochondrial fragmentation, implying that Lipin 1 promotes mitochondrial fission. These data suggest that generation of the PA and DAG lipid signals on the surface of mitochondria play reciprocal roles in regulation of mitochondrial dynamics. Unexpectedly, male mice lacking MitoPLD are infertile, exhibiting meiotic arrest during spermatogenesis (Huang et al., 2011, Watanabe et al., 2011). Spermatogenesis is known to require assembly of piRNA, a specialized RNAi-generating protein complex on the nuage, an electron-dense “intermitochondrial cement” that adheres to mitochondria at this developmental stage (Morroni et al., 2008, Russell and Frank, 1978, Chuma et al., 2009). Loss of components of the piRNA machinery similarly blocks spermatogenesis during meiosis (O’Donnell et al., 2008, Soper et al., 2008). MitoPLD-deficient testes noticeably lack intermitochondrial cement, do not aggregate, and do not produce piRNA (Huang et al., 2011, Watanabe et al., 2011). The mitochondria also exhibit abnormal distribution, suggesting functional interaction of the MitoPLD-generated PA with mitochondrial transport on microtubules (Watanabe et al., 2011). MitoPLD is the first mitochondrial protein to be identified in this process, which reveals this mitochondrial-surface lipid signaling pathway as a novel molecular mechanism and mitochondrial disease underlying male germ cell differentiation. A similar phenotype has been found for the Drosophila homolog of MitoPLD known as Zucchini, although its biochemical activity was not known (Pane et al., 2007).
Other potential pathophysiological roles for PLD
Influenza virus pandemics are a major global public health concern. Current approaches rely largely on vaccines and modestly effective antiviral therapeutics such as neuraminidase inhibitors (e.g. Tamiflu). Given the yearly antigenic shift and rapid development of resistance when viral proteins are targeted, other approaches are crucial to develop therapeutics capable of stemming the severity of future pandemics. A promising approach is to identify host cellular factors crucial for viral replication that are temporarily dispensable for humans. Using a high throughput siRNA screening approach, 72 host genes were recently identified that when downregulated, reduced viral replication for avian H5N1, endemic H1N1, and the 2009 pandemic swine-origin influenza A strains (Karlas et al., 2010). Included in the list of host genes was PLD2, which has long been linked to membrane vesicle trafficking (Du et al., 2004) in part in connection with the secretory pathway (Huang and Frohman, 2007, Yang et al., 2008, Riebeling et al., 2009). PLD enzymatic activity more generally has been reported to regulate glycosylated viral protein transport, including that of influenza virus hemagglutinin (HA), from the endoplasmic reticulum to the Golgi complex (Bi et al., 1997) and from there on to the plasma membrane. In the presence of crude inhibitors of PLD activity, HA transport to the plasma membrane is decreased by 75–93% (Bi et al., 1997). The neuraminidase (NA) glycoprotein undergoes similar trafficking prior to becoming displayed with HA on the surface of mature influenza particles through virion budding from the plasma membrane. With the recent development of thus-far highly specific small molecule inhibitors of PLD (Lavieri et al., 2010, Scott et al., 2009, Su et al., 2009b), PLD2 represents a highly attractive target for therapeutic intervention.
Conclusion
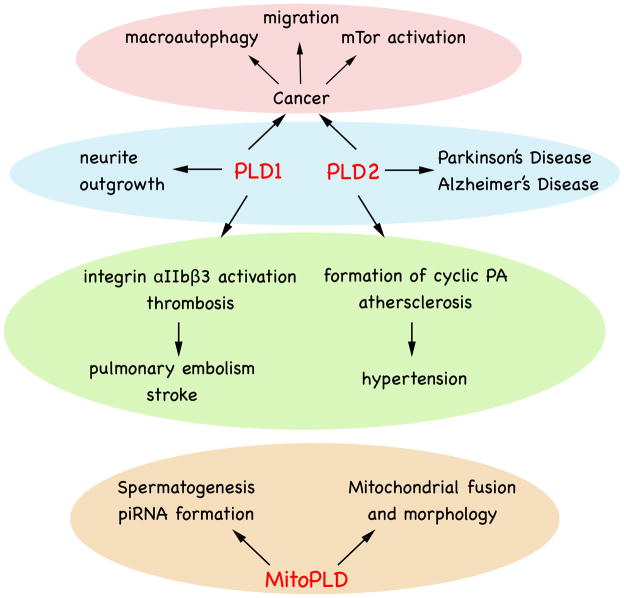
In summary, mammalian PLD genes have been studied for many years, and their structure and cellular regulation pointing to roles in cellular signaling, vesicle transport, membrane fusion, endocytosis, exocytosis and cytoskeletal reorganization have been defined largely using biochemical and cellular approaches. This body of knowledge is now undergoing refinement using genetic PLD knockouts, which will provide important insights into the functions that can be compensated for and those that are create phenotypic consequences. These genetic models, in combination with studying acute inhibition of PLD isoforms using small molecule inhibitors, will help establish appropriate settings in which to use PLD inhibitors as therapeutics, such as in the control of cancer metastasis, hypertension, thrombosis, neurodegenerative disease, and infectious viral disease (Fig. 2).
Fig. 2.
Physiological and Pathophysiological Roles for PLD family members. See text for details.
Acknowledgments
Supported by NIH GM071520 and GM084251 to MAF.
Footnotes
Conflicts of Interest
The authors do not declare any conflicts of interest.
References
- Bi K, Roth MG, Ktistakis NT. Phosphatidic acid formation by phospholipase D is required for transport from the endoplasmic reticulum to the Golgi complex. Curr Biol. 1997;7:301–307. doi: 10.1016/s0960-9822(06)00153-9. [DOI] [PubMed] [Google Scholar]
- Brandenburg LO, Konrad M, Wruck C, Koch T, Pufe T, Lucius R. Involvement of formyl-peptide-receptor-like-1 and phospholipase D in the internalization and signal transduction of amyloid beta 1–42 in glial cells. Neurosci. 2008;156:266–276. doi: 10.1016/j.neuroscience.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT, Solari R, Wakelam MJ. Phospholipase D1 localises to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr Biol. 1998;8:835–838. doi: 10.1016/s0960-9822(98)70326-4. [DOI] [PubMed] [Google Scholar]
- Cai D, Netzer WJ, Zhong M, Lin Y, Du G, Frohman M, Foster DA, Sisodia SS, Xu H, Gorelick FS, Greengard P. Presenilin-1 uses phospholipase D1 as a negative regulator of beta-amyloid formation. Proc Natl Acad Sci U S A. 2006a;103:1941–1946. doi: 10.1073/pnas.0510708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Zhong M, Wang R, Netzer WJ, Shields D, Zheng H, Sisodia SS, Foster DA, Gorelick FS, Xu H, Greengard P. Phospholipase D1 corrects impaired betaAPP trafficking and neurite outgrowth in familial Alzheimer’s disease-linked presenilin-1 mutant neurons. Proc Natl Acad Sci U S A. 2006b;103:1936–1940. doi: 10.1073/pnas.0510710103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang TM. Activation of phospholipase D in human platelets by collagen and thrombin and its relationship to platelet aggregation. Biochim Biophys Acta. 1994;1224:147–155. doi: 10.1016/0167-4889(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Dall’Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, Yu WH, Robinson KS, Yeku O, Small SA, Duff K, Frohman MA, Wenk MR, Yamamoto A, Di Paolo G. The Phospholipase D1 Pathway Modulates Macroautophagy. Nat Commun. 2010;1:142–152. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Phospholipase D in endocytosis and endosomal recycling pathways. Biochim Biophys Acta. 2009;1791:845–849. doi: 10.1016/j.bbalip.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Altshuller YM, Kim Y, Han JM, Ryu SH, Morris AJ, Frohman MA. Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Mol Biol Cell. 2000;11:4359–4368. doi: 10.1091/mbc.11.12.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ, Bader MF, Frohman MA. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J Cell Biol. 2003;162:305–315. doi: 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Huang P, Liang BT, Frohman MA. Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol Biol Cell. 2004;15:1024–1030. doi: 10.1091/mbc.E03-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda JE, Lee VM, Trojanowski JQ. Neuropathology of synuclein aggregates. J Neurosci Res. 2000;61:121–127. doi: 10.1002/1097-4547(20000715)61:2<121::AID-JNR1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Elvers M, Stegner D, Hagedorn I, Kleinschnitz C, Braun A, Kuijpers ME, Boesl M, Chen Q, Heemskerk JW, Stoll G, Frohman MA, Nieswandt B. Impaired alpha(IIb)beta(3) integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1. Sci Signal. 2010;3:ra1. doi: 10.1126/scisignal.2000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Freyberg Z, Bourgoin S, Shields D. Phospholipase D2 is localized to the rims of the Golgi apparatus in mammalian cells. Mol Biol Cell. 2002;13:3930–3942. doi: 10.1091/mbc.02-04-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyberg Z, Sweeney D, Siddhanta A, Bourgoin S, Frohman M, Shields D. Intracellular localization of phospholipase D1 in mammalian cells. Mol Biol Cell. 2001;12:943–955. doi: 10.1091/mbc.12.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk OS, Li S, Nha Nguyen F, Manfredsson FP, Kondrikova G, Sullivan LF, Meyers C, Chen W, Mandel RJ, Muzyczka N. alpha-Synuclein expression in rat substantia nigra suppresses phospholipase D2 toxicity and nigral neurodegeneration. Mol Ther. 2010;18:1758–1768. doi: 10.1038/mt.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Prestwich GD, Frohman MA, Morris AJ. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-α. J Biol Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- Hong KW, Jin HS, Lim JE, Cho YS, Go MJ, Jung J, Lee JE, Choi J, Shin C, Hwang SY, Lee SH, Park HK, Oh B. Non-synonymous single-nucleotide polymorphisms associated with blood pressure and hypertension. J Hum Hypertens. 2010;24:763–774. doi: 10.1038/jhh.2010.9. [DOI] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD pro-fusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Frohman MA. The potential for phospholipase D as a new therapeutic target. Expert Opin Ther Targets. 2007;11:707–716. doi: 10.1517/14728222.11.5.707. [DOI] [PubMed] [Google Scholar]
- Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of phospholipase D2 - Selective inhibtion of mammalian phospholipase D isoforms by a- and b-synucleins. Biochem. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Park MH, Lee YJ, Kim HS, Kwon TK, Park WS, Min do S. Phorbol ester up-regulates phospholipase D1 but not phospholipase D2 expression through a PKC/Ras/ERK/NFkappaB-dependent pathway and enhances matrix metalloproteinase-9 secretion in colon cancer cells. J Biol Chem. 2008;283:4094–4104. doi: 10.1074/jbc.M707416200. [DOI] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Knoepp SM, Chahal MS, Xie Y, Zhang Z, Brauner DJ, Hallman MA, Robinson SA, Han S, Imai M, Tomlinson S, Meier KE. Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells. Mol Pharmacol. 2008;74:574–584. doi: 10.1124/mol.107.040105. [DOI] [PubMed] [Google Scholar]
- Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- Lavieri RR, Scott SA, Selvy PE, Kim K, Jadhav S, Morrison RD, Daniels JS, Brown HA, Lindsley CW. Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J Med Chem. 2010;53:6706–6719. doi: 10.1021/jm100814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kim HS, Pai JK, Ryu SH, Suh PG. Activation of phospholipase D induced by platelet-derived growth factor is dependent upon the level of phospholipase C-γ1. J Biol Chem. 1994;269:26842–26847. [PubMed] [Google Scholar]
- Lotharius J, Brundin P. Pathogenesis of Parkinson’s disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- Mauri S. the Phenomenon of Adhesiveness and Aggregation of Blood Platelets. Collective Review. G Gerontol. 1964;12:413–428. [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morroni M, Cangiotti AM, Marzioni D, D’Angelo A, Gesuita R, De Nictolis M. Intermitochondrial cement (nuage) in a spermatocytic seminoma: comparison with classical seminoma and normal testis. Virchows Arch. 2008;453:189–196. doi: 10.1007/s00428-008-0610-0. [DOI] [PubMed] [Google Scholar]
- Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, Ryu SH, Lee KH, Han JS. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Burns KH, Boeke JD. A descent into the nuage: the maelstrom of transposon control. Dev Cell. 2008;15:179–181. doi: 10.1016/j.devcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira TG, Chan RB, Tian H, Laredo M, Shui G, Staniszewski A, Zhang H, Wang L, Kim TW, Duff KE, Wenk MR, Arancio O, Di Paolo G. Phospholipase D2 ablation ameliorates Alzheimer’s disease-linked synaptic dysfunction and cognitive deficits. J Neurosci. 2010;30:16419–16428. doi: 10.1523/JNEUROSCI.3317-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Ahn BH, Hong YK, Min do S. Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/Sp1-mediated signaling pathways. Carcinogen. 2009;30:356–365. doi: 10.1093/carcin/bgn287. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Reue K, Zhang P. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 2008;582:90–96. doi: 10.1016/j.febslet.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebeling C, Morris AJ, Shields D. Phospholipase D in the Golgi apparatus. Biochim Biophys Acta. 2009;1791:876–880. doi: 10.1016/j.bbalip.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Wakelam MJ. Inter-regulatory dynamics of phospholipase D and the actin cytoskeleton. Biochim Biophys Acta. 2009;1791:856–861. doi: 10.1016/j.bbalip.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Russell L, Frank B. Ultrastructural characterization of nuage in spermatocytes of the rat testis. Anat Rec. 1978;190:79–97. doi: 10.1002/ar.1091900108. [DOI] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Saito M, Iwadate M, Higashimoto M, Ono K, Takebayashi Y, Takenoshita S. Expression of phospholipase D2 in human colorectal carcinoma. Oncol Rep. 2007;18:1329–1334. [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat Struct Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- Su W, Chen Q, Frohman MA. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 2009a;5:1477–1486. doi: 10.2217/fon.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009b;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Tooze SA, Schiavo G. Liaisons dangereuses: autophagy, neuronal survival and neurodegeneration. Curr Opin Neurobiol. 2008;18:504–515. doi: 10.1016/j.conb.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T, Tsukahara R, Fujiwara Y, Yue J, Cheng Y, Guo H, Bolen A, Zhang C, Balazs L, Re F, Du G, Frohman MA, Baker DL, Parrill AL, Uchiyama A, Kobayashi T, et al. Phospholipase D2-dependent inhibition of the nuclear hormone receptor PPARgamma by cyclic phosphatidic acid. Mol Cell. 2010;39:421–432. doi: 10.1016/j.molcel.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19:671–675. [PubMed] [Google Scholar]
- Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. J Cancer Res Clin Oncol. 1997;123:280–285. doi: 10.1007/BF01208639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N, Caumont AS, Chasserot-Golaz S, Du G, Wu S, Sciorra VA, Morris AJ, Frohman MA, Bader MF. Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 2001;20:2424–2434. doi: 10.1093/emboj/20.10.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorland M, Holmsen H. Phospholipase D activity in human platelets is inhibited by protein kinase A, involving inhibition of phospholipase D1 translocation. Platelets. 2008a;19:300–307. doi: 10.1080/09537100801910838. [DOI] [PubMed] [Google Scholar]
- Vorland M, Holmsen H. Phospholipase D in human platelets: presence of isoenzymes and participation of autocrine stimulation during thrombin activation. Platelets. 2008b;19:211–224. doi: 10.1080/09537100701777329. [DOI] [PubMed] [Google Scholar]
- Wasilewski M, Scorrano L. The changing shape of mitochondrial apoptosis. Trends Endocrinol Metab. 2009;20:287–294. doi: 10.1016/j.tem.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MitoPLD Is a Mitochondrial Protein Essential for Nuage Formation and piRNA Biogenesis in the Mouse Germline. Developmental Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yang JS, Gad H, Lee S, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, Baldanzi G, Graziani A, Bourgoin S, Frohman MA, Luini A, Hsu VW. COPI vesicle fission: a role for phosphatidic acid and insight into Golgi maintenance. Nat Cell Biol. 2008;10:1146–1153. doi: 10.1038/ncb1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang P, Du G, Kanaho Y, Frohman MA, Tsirka SE. Increased expression of two phospholipase D isoforms during experimentally induced hippocampal mossy fiber outgrowth. Glia. 2004;46:74–83. doi: 10.1002/glia.10322. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kanaho Y, Frohman MA, Tsirka SE. Phospholipase D1-promoted release of tissue plasminogen activator facilitates neurite outgrowth. J Neurosci. 2005;25:1797–1805. doi: 10.1523/JNEUROSCI.4850-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y, Deguchi T, Ohishi N, Yagi K, Nozawa Y. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem Biophys Res Commun. 2000;278:140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, Foster DA. Phospholipase D couples survival and migration signals in stress response of human cancer cells. J Biol Chem. 2006;281:15862–15868. doi: 10.1074/jbc.M600660200. [DOI] [PubMed] [Google Scholar]