Abstract
Background
Since 2008 multiple studies have reported on copy number variations (CNVs) in schizophrenia. However, many regions are unique events with minimal overlap between studies. This makes it difficult to gain a comprehensive overview of all CNVs involved in the aetiology of schizophrenia. We performed a systematic CNV study based on a homogeneous genome-wide dataset aiming at all CNVs ≥50 kb. We complemented this analysis with a review of cytogenetic and chromosomal abnormalities for schizophrenia reported in the literature with the purpose to combine classical genetic findings and our current understanding of genomic variation.
Methods
We investigated 834 Dutch schizophrenia patients and 672 Dutch controls. CNVs were included if they were detected by QuantiSNP as well as PennCNV and contain known protein coding genes. The integrated identification of CNV regions and cytogenetic loci indicates regions of interest (CROIs).
Results
In total, 2,437 CNVs were identified with an average number of 2.1 CNVs per subject for both cases and controls. We observed significantly more deletions, but not duplications, in schizophrenia cases versus controls. The CNVs identified coincide with loci previously reported in the literature, confirming well-established schizophrenia CROIs 1q42 and 22q11.2, as well as indicating a potentially novel CROI on chromosome 5q35.1.
Conclusions
Chromosomal deletions are more prevalent in schizophrenia patients than in healthy subjects and therefore confer a risk factor for pathogenicity. The combination of our CNV data with previously reported cytogenetic abnormalities in schizophrenia provides an overview of potentially interesting regions for positional candidate genes.
Keywords: copy number variation, schizophrenia, cytogenetic abnormality, deletion, duplication, candidate gene
Introduction
Schizophrenia is a debilitating psychiatric disease showing a heterogeneous clinical phenotype and a life-time risk of 0.46–1% (1;2). It is characterized by psychotic symptoms, including delusions and hallucinations, reduced interest and drive, altered emotional reactivity, and disorganized behaviour (3). Its predisposition is influenced by a complex interaction of genetic and environmental factors (4;5). Family, twin and adoption studies have provided evidence for a substantial genetic contribution to the phenotypic variance of schizophrenia (6). It has been estimated that the heritability of developing schizophrenia is up to 80% (7), but the pattern of inheritance and related pathogenic pathways remain elusive.
Early genetic studies in schizophrenia were based on linkage studies in pedigrees and association testing of candidate genes (8). Linkage and association studies have been complemented by identification of chromosomal abnormalities in patients, as well as the more recently genome-wide association studies (GWAS) with single nucleotide polymorphisms (SNPs). These genomic microarrays also allowed for the systematic genome-wide analysis of submicroscopic cytogenetic variation, i.e. genomic copy number variation (CNV) which includes genomic deletions and duplications of more than 1 kb in size. Many recent studies support a polygenic basis of schizophrenia. Important in the understanding of the genetic architecture of schizophrenia is the common disease - common variant versus the common disease - rare variant hypothesis. The first refers to the possibility that common alleles with small to moderate disease risks may have an additive or multiplicative effect on schizophrenia. Recent large-scale association studies have supported this hypothesis for schizophrenia (9). However, common variants are unlikely to explain the total heritability of schizophrenia. The latter hypothesis suggests that multiple rare variants with relatively large effect play a role in the etiology of schizophrenia. It is likely that both common and rare alleles lead to the genetic heterogeneity of schizophrenia. Current CNV studies have mainly focussed on extremely rare and de novo variants. However, advances in CNV discovery techniques as well as the increasing number of data releases from SNP association studies may allow for the detection of smaller, more frequent variants (8).
Since 2008 several large-scale whole-genome schizophrenia association studies have reported on the detection of CNVs (10–15). Over 41% of all CNVs identified overlap with known genes. This suggests that CNVs may play a substantial role in modulating gene expression (16). Two large-scale studies identified three recurrent but rare schizophrenia-associated microdeletions at chromosome 1q211, 15q13.3, and 15q11.2, each containing multiple genes (11;12). Many recurrent CNV loci are flanked by segmental duplications or low copy repeats (LCRs) which suggests that these deletions and duplications could be the result of nonallelic homologous recombination (NAHR) mediated by these LCRs. Large-scale CNV studies suggest that rare variants disrupting genes in neurodevelopmental pathways may significantly contribute to the risk for developing schizophrenia (OR ranging from 2.7 to 14.8) (8;12;14). However, many regions reported in those studies are unique events, with minimal overlap between studies with the exception of a few recurrent loci (8). Both phenotypic diversity associated with CNVs together with heterogeneity in diagnoses and ethnicities could explain the difficulties in replicating disease variants (17).
Few studies provided genome-wide evaluation of CNVs (11;12;18). However, most CNV studies thus far present a targeted and limited number of strong CNV candidates involved in disease. Relatively few investigators have yet reported their complete CNV data sets or released raw data to the scientific community, minimizing the opportunity for comparing and contrasting data or for meta-analyses (19). However, an overview of large-scale genomic variation will be a crucial resource in correlating genomic variations with experimental findings and clinical outcomes. A good example of a genome-wide overview of all CNVs found in schizophrenia is the ISC study (2008) (12) that reports all CNVs passing QC as an online datafile (http://pngu.mgh.harvard.edu/isc/). A systematic review of cytogenetic and CNV findings would provide researchers in the field of schizophrenia with an overview of potentially interesting regions for positional candidate genes. Good examples from other research fields are e.g. the Database of Genomic Variants and systematic information on chromosome rearrangements for autism spectrum disorders (20;21).
We performed a two-step study. First, we carried out a systematic genome-wide CNV analysis in a homogeneous group of Dutch schizophrenia cases versus unaffected controls, and provide data of all copy number variants of ≥50 kb, including common and rare variants. We further performed a systematic review of all cytogenetic and chromosomal abnormalities reported in the literature for schizophrenia with the purpose to present a comprehensive overview and combine classical cytogenetic findings and our current understanding of genomic variation (22;23). We hypothesize that (new) risk loci for schizophrenia may be revealed through overlap between genomic regions affected by previously reported cytogenetic abnormalities and those affected by CNVs. In addition, we also investigated CNVs in known candidate gene loci from literature. We hypothesize that those candidate gene loci will be affected by CNVs more often in cases compared to controls. Both steps are a first effort to understand the role of genomic chromosomal variation and schizophrenia susceptibility. We believe that these data, which we have made publicly available, are a necessary step toward that goal.
Methods and Materials
Systematic genome-wide CNV analysis
We studied CNVs within a cohort of 834 patients with schizophrenia and 672 unaffected control individuals. Inpatients and outpatients were recruited from a variety of psychiatric hospitals and institutions in The Netherlands, partly coordinated via academic hospitals in Amsterdam, Groningen, Maastricht, and Utrecht (The Genetic Risk and Outcome of Psychosis [GROUP] project). All patients had been diagnosed for subtypes of schizophrenia according to the DSM-IV-TR. Detailed medical and psychiatric histories were collected, including the Comprehensive Assessment of Symptoms and History (CASH), an instrument for assessing diagnosis and psychopathology. The controls were volunteers and were all screened for any psychiatric history, the majority via the CASH. Both cases and controls were of Dutch descent (with at least three out of four grandparents of Dutch ancestry) and they all gave informed consent. The study was approved by the Ethics Committee of the UMC Utrecht and by the appropriate local institutional review boards at all other participating hospitals.
Genomic DNA of all patients and controls was hybridized to HumanHap550v3 BeadArray (Illumina, San Diego, CA, USA) according to standard protocols. QuantiSNP (24) and PennCNV (25) were used to identify copy number deletions and duplications. Both QuantiSNP and PennCNV are based on a Hidden Markov Model (HMM) for kilobase-resolution detection of CNVs from Illumina high-density SNP genotyping data. The PennCNV program is probably the most frequently used program for CNV studies in recent publications. This may in part be due to the user-friendly design of the program and free access to users. Its low false positive rate is a promising aspect. On the other hand, QuantiSNP outperformed six other methods in a recent evaluation study of CNV calling algorithms (26).
For QuantiSNP, a quality control step for GC-content was performed. For PennCNV, a gc-model for GC-correction was used and the following quality control steps were performed: (1) CNVs containing less than 10 consecutive SNP markers were excluded, (2) CNVs with a value below 0.5 for the confidence score (value for quality of the CNV call) divided by the number of SNP markers were excluded, and (3) CNVs with a SNP density below 10 kb were excluded.
CNVs were included only if they were detected by both PennCNV and QuantiSNP and meeting the quality control criteria described above. Several CNV detection methods are available, but differences in characteristics exist and every method has its own weaknesses (26). By including only overlapping CNVs we made an effort to limit the false positive rate of CNV detection, as suggested by Winchester et al. (2009) (27). CNVs detected by both algorithms were defined by overlapping start and stop positions with at least one position (start or stop SNP marker) similar or when a smaller CNV completely overlapped a larger CNV. Moreover, we only included CNVs containing known protein coding genes, using fuzzy border criteria of up to 50 kb surrounding the CNV boundaries. Compared with CNV size, gene content might be a more reliable indicator for clinical significance such that small, gene-rich CNVs are more likely to be pathogenic than larger, gene-poor CNVs (28). The gene content of each CNV was defined using the UCSC genome browser. We investigated the reliability of our CNV dataset by (1) calculating the concordance rate between calls from PennCNV and QuantiSNP, (2) calculating the percentage of overlap between calls from the two datasets when increasing filtering stringency, and (3) validating individual CNVs by MLPA and qPCR. Taken together, by focussing our analysis on those CNVs that were consistently called by the two algorithms and contain known protein coding (Refseq) genes, we generated a reliable data set for further testing.
We provide all data of CNVs ≥50 kb found in both cases and controls. Raw data is available for future research and meta-analysis. Additionally to the total overview of CNVs we compared the number of deletions and duplications for different size categories in cases and controls.
Reviewing literature
Studies were identified by searching the electronic NLM MEDLINE and Pubmed databases using different combinations of the search terms “schizophren*”, “genetic*”, “cytogenetic*”, “copy number”, “CNV”, and “chromosom*” for reports published until January 2010. All studies were examined with a special emphasis on the quality of the schizophrenia diagnosis and the definition of the chromosomal aberration. The quality of the phenotype was rated for each case by two investigators (JWM and JEB) (Cohen s kappa 0,68; p<0,001). Ratings were based on the clinical description, the presence of a psychiatric diagnosis or a classification system (Table S4 in the Supplement). This permitted a ranking of regions based on the most valid clinical diagnosis of schizophrenia.
Because we focussed on gene-containing CNVs most probable to have pathogenic effects in our CNV analysis, we compared the whole-genome CNV data with cytogenetic abnormalities found in case reports. Cytogenetic visible deletions and duplications may constitute a less common, but potentially stronger influence on risk for schizophrenia and these are likely to have direct effects on gene expression. Because of their relatively large size, these chromosomal aberrations may encompass multiple genes which suggests a role in modulating gene expression (29). For example, it is suggested that the combined haploinsufficient expression of multiple genes in the 22q11 microdeletion cause the high rates of schizophrenia observed in patients with this 3 Mb deletion syndrome (encompassing 30 genes) (22). From both the CNV and cytogenetic abnormality loci we were able to select cytogenetic regions of interest (CROIs): loci where at least one case report and two Dutch schizophrenia CNV cases overlapped. Together with a systematic review of CNV findings from the literature, we highlight (new) chromosomal variation loci for schizophrenia susceptibility.
Additionally to the selection of CROIs, we were interested to see if we were able to combine our CNV findings and literature loci in a different way. For this sub analysis we included all articles describing CNVs in schizophrenia patients (Table S3 in the Supplement) with special emphasis on the description of candidate genes residing in these loci. We counted all deletions and duplications from our CNV analysis showing overlap with the candidate gene loci from the literature. Cases and controls were compared on the number of deletions and duplications within these regions.
See Tables S2, S3 and S4 in the Supplement for further details on the methods.
Results
Systematic genome-wide CNV analysis
In total, 7,211 CNVs passed the quality control for QuantiSNP and 21,182 CNVs for PennCNV. More than 50% of all CNVs was gene containing: 3,767 for QuantiSNP and 13,849 for PennCNV. In total, 2,437 gene-containing CNVs were called by both algorithms and were included in the study. These CNVs were found in 659 unique cases and 508 unique controls.
The poor overlap between QuantiSNP and PennCNV is striking, although not unusual (26;27;30). A recent study by Dellinger et al. (2010) compared among others the PennCNV and QuantiSNP algorithms and found that the average number of detected CNVs differed depending on the algorithm. The call numbers were correlated to sensitivity, specificity and kappa (26). Tsuang et al. (2011) (31) also demonstrated that the number of CNVs identified depends on the algorithm(s) used. In accordance to our own findings, they showed that PennCNV called far more CNVs compared to QuantiSNP.
It is known that modifying parameters affects CNV detection. Not only the number of calls is affected, but also the size of predicted CNVs (26;27). When relaxing some of the parameter settings, we saw differences in the number of CNV calls also influencing the number of overlapping CNVs of both algorithms (data not shown). When increasing the filtering stringency from 10 to 30 consecutive SNP markers we observed more overlapping CNVs between PennCNV and QuantiSNP, which affect especially CNVs of larger size. The total overlap percentage between the two datasets increased from 27.4% to 46.8%. The use of overlapping calls from both algorithms should increase the confidence of our dataset and gives clearer indications of the CNV boundaries (27;31).
In order to investigate the reliability of our CNV dataset, we visually inspected a set of common and rare CNVs. Rare CNVs were detected in one patient only. Inspection of their intensity plots revealed that they were likely to be true CNV loci. Based on their gene content we selected and validated four CNVs by either multiplex ligation-dependent probe amplification (MLPA; duplication 2p25.3, deletion 2p16.3, and deletion 9q33.1) or genomic quantitative PCR (qPCR; duplication 5p15.2), as described previously (13). All four CNVs were confirmed as true positive finding. We also investigated nine duplicate samples in order to calculate a concordance rate between calls from PennCNV and QuantiSNP. The calls were highly correlated with a correlation coefficient of 0.81.
The average number of gene containing CNVs per subject was 2.1 for both cases and controls. One subject can have multiple deletions or duplications of different lengths. Therefore, groups of deletions or duplications categorized by size across subjects are not mutually exclusive. When we limited our analysis to a subject just having one or more CNVs per type and size, we indicated a total of 796 deletions (Table 1) and 827 duplications (Table 2). Cases appeared to have significantly more deletions compared to controls for all different size categories (Table 1). However, we did not find any significant difference between cases and controls for duplications (Table 2). This is in accordance with the literature, stating that deletions might be more pathogenic than duplications (28).
Table 1.
Deletions in cases versus controls
| Size | Cases | Controls | Total | # CNVs per subject Cases/controls | Case/control ratio | P-value (1-sided) |
|---|---|---|---|---|---|---|
| All | 470 | 326 | 796 | 0.71/0.64 | 1.11 | 0.00559 |
| ≥ 50 kb | 375 | 226 | 601 | 0.57/0.44 | 1.28 | 1.37e-5 |
| ≥ 500 kb | 18 | 4 | 22 | 0.027/0.008 | 3.47 | 0.00817 |
| ≥ 1 Mb | 11 | 2 | 13 | 0.017/0.004 | 4.24 | 0.02422 |
Table 2.
Duplications in cases versus controls
| Size | Cases | Controls | Total | # CNVs per subject Cases/controls | Case/control ratio | P-value (1-sided) |
|---|---|---|---|---|---|---|
| All | 455 | 372 | 827 | 0.69/0.73 | 0.94 | 0.0675 |
| ≥ 50 kb | 417 | 344 | 761 | 0.63/0.68 | 0.93 | 0.0607 |
| ≥ 500 kb | 39 | 27 | 66 | 0.06/0.05 | 1.11 | 0.35135 |
| ≥ 1 Mb | 14 | 9 | 23 | 0.021/0.018 | 1.2 | 0.4162 |
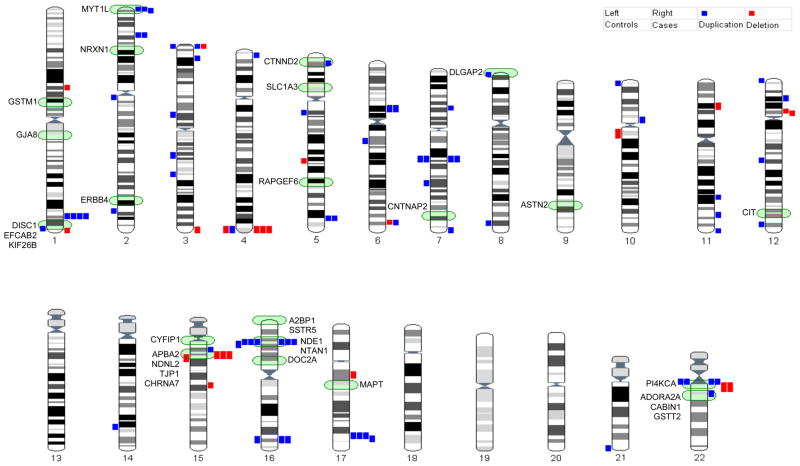
We identified a total of 1,896 CNVs of ≥50 kb observed in 433 schizophrenia patients and 355 controls. For this effort we only included overlapping results of the two CNV detection methods with the purpose to increase our confidence of the CNV calls. QuantiSNP yielded 3,195 CNVs while PennCNV analysis resulted in detection of 10,641 CNVs; overlap between the two methods was 27.4%. These findings (including exact basepair boundaries) are presented in Tables S1 and S2 (in the Supplement). Some of our CNVs overlap regions of low copy repeats (LCRs). Previous analyses showed significant associations between LCRs and CNV regions (32–34). However, the HumanHap550v3 BeadArray we used for SNP genotyping primarily targets SNP loci at non-repetitive genomic regions. Furthermore, the rate of LCR flanking CNVs is not different between cases and controls (data not shown). LCR mediated CNVs should therefore have limited effect in our analyses. We found no evidence for increased double hit rate (i.e. multiple CNVs per subject) in schizophrenia patients compared to controls. In Figure 1, all CNVs ≥500 kb (n=60) are represented to scale on each chromosome. Genes reported to be associated with schizophrenia are also indicated.
Figure 1. Copy Number Variations in 834 Dutch schizophrenia patients and 672 Dutch controls.
Representation of a systematic genome-wide survey for CNVs in 834 Dutch schizophrenia cases and 672 Dutch controls. On the left side of each chromosome, CNVs of controls are indicated. On the right side of each chromosome, CNVs of schizophrenia cases are indicated. Blue bars represent duplications, red bars represent deletions. In green, schizophrenia associated genes from CNV literature are indicated.
The graphical overview of this systematic genome-wide CNV analysis shows sites of increased deletions in cases versus controls for: 4q35.2, 15q13.2-q13.3, and 22q11.21. Interesting sites of increased duplications in cases versus controls are: 1q42.3–43, 2p25.3, and 17q25.1. When examining loci of candidate genes for schizophrenia indicated by previous CNV studies, we observe significantly more CNV deletions of ≥50 kb within these gene regions for cases compared to controls (p-value = 0.0004). However, there is no significant difference for CNV duplications of ≥50 kb within these candidate gene regions (p-value = 1; see Table 3). The regions on chromosome 1q, 2p, 15q, and 22q have previously been highlighted in cytogenetic and CNV studies (10–15;18;35–38). The 4q35.2 region (188,329,837 – 191,164,126 bp) is a possible novel CNV region-of-interest, indicated by three schizophrenia cases with the deletion versus only one control subject. Our duplications at chromosome 17q (69,345,596 – 71,746,800 bp) in 4 cases and no controls were also not previously described to be associated with schizophrenia.
Table 3.
Schizophrenia candidate genes residing in CNVs in Dutch case-control study
| Gene | Position | # CNVs overlapping | # Cases (%) | # Controls (%) | |||
|---|---|---|---|---|---|---|---|
| Del | Dup | Del | Dup | Del | Dup | ||
| A2BP1 (15) | 16p13.2 | 4 | 3 | 3 (0.69) | - | 1 (0.28) | 3 (0.85) |
| ADAMTSL3 (18) | 15q25.2 | 0 | 0 | - | - | - | - |
| ADORA2A (47) | 22q11.23 | 0 | 0 | - | - | - | - |
| APBA2 (18;48;49) | 15q13.1 | 0 | 0 | - | - | - | - |
| ASTN2 (13) | 9q33.1 | 0 | 1 | - | 1 (0.23) | - | - |
| CABIN1 (47) | 22q11.23 | 0 | 0 | - | - | - | - |
| CHRNA7 (11;12) | 15q13.3 | 3 | 2 | 3 (0.69) | - | - | 2 (0.56) |
| CIT (15) | 12q24.23 | 0 | 0 | - | - | - | - |
| CNTNAP2 (50) | 7q35-q36.1 | 3 | 0 | 3 (0.69) | - | - | - |
| COMT (37;38) | 22q11.21 | 1 | 0 | 1 (0.23) | - | - | - |
| CTNND2 (13) | 5p15.2 | 0 | 1 | - | 1 (0.23) | - | - |
| CYFIP1 (11) | 15q11.2 | 7 | 10 | 4 (0.92) | 6 (1.39) | 3 (0.85) | 4 (1.13) |
| DISC1 (51) | 1q42.2 | 0 | 5 | - | 3 (0.69) | - | 2 (0.56) |
| DLGAP2 (48) | 8p23.3 | 0 | 0 | - | - | - | - |
| DOC2A (48) | 16p11.2 | 0 | 0 | - | - | - | - |
| EFCAB2 (18) | 1q44 | 0 | 1 | - | 1 (0.23) | - | - |
| ERBB4 (14) | 2q34 | 1 | 0 | 1 (0.23) | - | - | - |
| GJA8 (11) | 1q21.1 | 0 | 0 | - | - | - | - |
| GSTM1 (47) | 1p13.3 | 0 | 0 | - | - | - | - |
| GSTT2 (47) | 22q11.23 | 0 | 1 | - | - | - | 1 (0.28) |
| KIF26B (18) | 1q44 | 1 | 1 | 1 (0.23) | 1 (0.23) | - | - |
| MAPT (48) | 17q21.31 | 0 | 0 | - | - | - | - |
| MYT1L (13) | 2p25.3 | 0 | 2 | - | 2 (0.46) | - | - |
| NDE1 (52) | 16p13.1 | 0 | 6 | - | 3 (0.69) | - | 3 (0.85) |
| NDNL2 (48) | 15q13.1 | 0 | 0 | - | - | - | - |
| NOTCH4 (41;53) | 6p21.32 | 0 | 0 | - | - | - | - |
| NRGN (41) | 11q24.2 | 0 | 0 | - | - | - | - |
| NRXN1 (13;14;18;48;49) | 2p16.3 | 3 | 2 | 3 (0.69) | 1 (0.23) | - | 1 (0.28) |
| NTAN1 (52) | 16p13.1 | 0 | 4 | - | 3 (0.69) | - | 1 (0.28) |
| PGBD1 (41) | 6p22.1 | 0 | 0 | - | - | - | - |
| PI4KCA (47) | 22q11.21 | 2 | 1 | 2 (0.46) | - | - | 1 (0.28) |
| PRODH (37;38) | 22q11.21 | 11 | 23 | 9 (2.08) | 13 (3.0) | 2 (0.56) | 10 (2.82) |
| RAPGEF6 (15) | 5q31.1 | 0 | 0 | - | - | - | - |
| SLC1A3 (14) | 5p13.2 | 0 | 0 | - | - | - | - |
| SSTR5 (47) | 16p13.3 | 0 | 0 | - | - | - | - |
| TJP1 (48) | 15q13.1 | 0 | 0 | - | - | - | - |
| Total | 36 | 63 | 30 (6.9) | 35 (8.1) | 6 (1.7) | 28 (7.9) | |
| Del p = 0.0004 | Dup p = 1 | ||||||
Comparison with literature
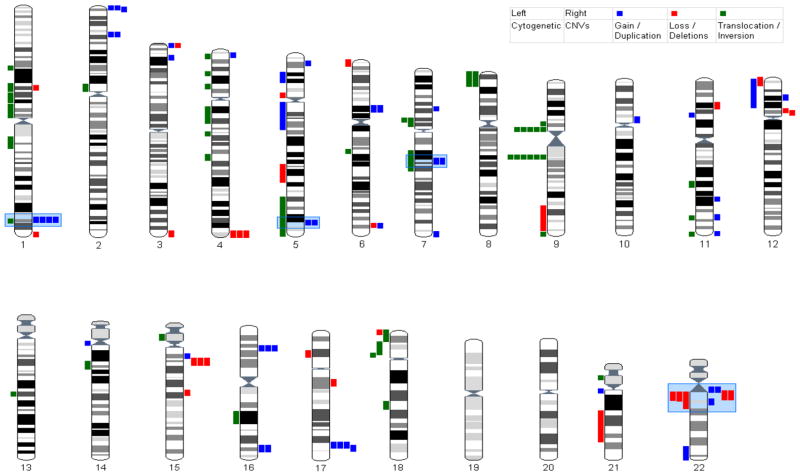
An overview of all cytogenetic abnormalities described in literature can be found in Table S4 and Figure S2 (in the Supplement). It appears that schizophrenia susceptibility loci are not equally distributed across the human genome, but rather cluster together in specific chromosomal regions of interest (CROIs). When we compare cytogenetic abnormalities to CNVs found in Dutch schizophrenia cases and controls, we observe the following CROIs: 1q42, 5q35.1, 7q21.12-q21.13, and 22q11 (Figure 2). These regions are reported by at least one cytogenetic study and have CNVs in at least 2 cases. The regions on chromosome 1q and 22q have been described before (10–12;14;15;18;35–38) and also appeared as interesting sites in our CNV analysis. Our CNV on 1q42 although overlapping the large region from the 1;11 translocation, it is not overlapping the exact boundaries of the DISC1 gene (approximately 3 Mb between CNV and gene region). For the 22q11.2 region we find both deletions and duplications (see Figure S3 in the Supplement). This region is known from the 22q11.2 Deletion Syndrome (MIM ID #188400). Most patients with this syndrome share a 3-Mb loss, although a nested 1.5-Mb deletion is also observed along with infrequent atypical deletions. Low copy repeats (LCRs) are flanking and mediating the deletion regions (39). We identified 12 deletions within the 22q11.2 interval. Of these, 1 (case) was consistent with the larger deletion, 0 were consistent with the shorter deletion, and 11 (9 cases and 2 controls; 729 kb: 19,063 – 19,792 kb; 156 kb: 17,258 kb – 17,434 kb) were atypical. For chromosome 7, although found in 2 cases and one cytogenetic study, also 2 controls are identified to have a duplication at this site. Chromosome 5q (168,385,055 – 169,072,475 bp) is a possible novel region, indicated by clustering of cytogenetic abnormalities and CNV findings. However, the only investigated candidate genes residing in this locus are SLIT3, GABRP, and FGF18 with inconsistent results (www.schizophreniaforum.org/res/sczgene).
Figure 2. Copy Number Variations in 834 Dutch schizophrenia cases compared with cytogenetic aberrations from literature.
On the left side of each chromosome, chromosomal aberrations from literature are indicated. On the right side of each chromosome, copy number variants in 834 Dutch schizophrenia cases are indicated. Blue bars represent duplications or gains, red bars represent deletions or losses, and green bars represent balanced translocations/inversions. Blue boxes indicate regions where at least one chromosomal aberration and two copy number cases overlapped. The regions 1q42.3-q43 and 22q11.2 were previously indicated by linkage (54;55) or association studies (56;57) and often supported by CNV findings from literature. The regions on chromosome 5q35 and on chromosome 7q21.12-q21.13 are potentially novel regions, not indicated in association or linkage studies (54–57). For the exact basepair boundaries of overlap regions we refer to Table S1 and S4 in the Supplement.
An overview of all CNV studies from literature can be found in Table S3 in the Supplement. Lack of information about the real number of CNVs occurring in cases and controls and unclear diagnostic criteria limited our ability to give a complete and correct overview. However, comparison with our own CNV data and schizophrenia-associated genes from CNV literature indicates some clustered regions of interest, e.g. on chromosome 1q, chromosome 15q and 22q11 (Table S3, Figure S1 in the Supplement).
Discussion
We performed a genome-wide analysis of CNVs in 834 schizophrenia cases and 672 unaffected controls from The Netherlands. The most apparent observation is that cases showed significantly more deletions for all size categories, an effect not seen for duplications. When focussing on previously reported schizophrenia-candidate CNV loci, a similar effect was observed with cases showing significantly more deletions compared to controls. These findings suggest that deletion CNVs may be more prevalent in individuals with schizophrenia and therefore confer a higher risk factor for pathogenicity. This is in line with previous CNV studies that also suggested that duplications are genetic alterations that are better tolerated in the genome and that deletions have a higher likelihood of being pathogenic (19;28).
Additionally, we reviewed all cytogenetic and chromosomal abnormalities described for schizophrenia patients (Table S4 in the Supplement). The integrated identification of CNV regions and cytogenetic and chromosomal loci highlight the following regions of interest (CROIs): 1q42, 2p25, 15q13, and 22q11 as loci previously described. Secondly, we identified 4q35 (increased number of deletions in cases compared to controls) and 17q25 (increased number of duplications in cases compared to controls) as CROIs from our CNV analysis. An interesting region from the integration of CNVs and cytogenetic abnormality reports is 5q35. Furthermore, we see regions of genomic variation both existing in cases and controls. These CNVs are very difficult to interpret in the absence of further correlative data.
It is probable that a combination of both common and rare risk variants is involved in schizophrenia etiology. Common CNVs (allele frequency >5%) are almost always inherited and comprise the majority of CNV differences between individuals (19). In two recent large GWAS studies for schizophrenia, TCF4 and ZNF804A have been found as best hits (40;41). These are transcription factors that may regulate expression of many genes. This raises the possibility that common variation may confer susceptibility to schizophrenia. However, until now, mostly rare (<0.5–1%) and large (>100 kb) CNVs have been implicated in schizophrenia (42). This study both confirms the involvement of the recurrent variants (e.g. for 1q42) and adds to the implication of rare CNVs with an elevated risk for schizophrenia (43).
Improvement of genomic array and sequencing technologies will provide higher-resolution and more accurate detection of CNVs (44). These new technologies will aid the discovery of new schizophrenia risk regions and candidate genes. In addition to technological advances there is need for development of improved algorithms and statistical methods for reliable CNV calling in existing GWAS data sets. There is still a large-scale variability between calling algorithms, so the use of multiple algorithms specific to the array platform used seems recommended (31). Moreover, it is important that studies with large sample sizes will make their CNV calls publicly available for further study and systematic meta-analyses (for example via the Database of Genomic Variants). New bioinformatic tools that combine different genomic data sources are necessary to gain better inside in this complex psychiatric disorder.
It is possible that our study is subject to various kinds of biases. First, overrepresentation of some regions in our literature study may represent availability of probes rather than significant association with schizophrenia. This is clearly the case for the 22q11 deletion, which results in reporter bias. When a specific region has been linked to schizophrenia, the locus becomes more interesting for research, resulting in more tested cases. In our effort to include previously reported cytogenetic abnormalities in schizophrenia we noticed that approximately one-third of the cases showed either mental retardation or physical anomalies. For CNV studies in literature on the other hand, this type of clinical data was not always mentioned. However, both features are associated with a high incidence of cytogenetic abnormalities (45). Therefore, the loci indicated in this study may be linked to mental retardation and dysmorphic features rather than to the schizophrenia phenotype alone.
For complex traits like schizophrenia it is important to consider all classes of variation: cytogenetic abnormalities as well as SNPs and CNVs (46). This study provides a comprehensive overview of all CNVs ≥50 kb found in a homogeneous Dutch sample of schizophrenia cases and unaffected controls. Deletions are more prevalent in schizophrenia patients and may confer a higher risk factor than duplications. We combined this data with a systematic overview of cytogenetic and CNV findings from the literature. Our results confirm the already known results for e.g. the 15q13 and 22q11.2 deletions and highlight novel candidate regions. These regions indicate how classical cytogenetic findings and our current understanding of genomic variation can be combined and may contain loci with rare variants contributing to disease susceptibility. By reporting our complete CNV data set and systematically reviewing the literature, we hope this study is a step towards understanding the role of genomic chromosomal variation in schizophrenia etiology. Future studies will include genome-wide sequencing so that the whole spectrum of genomic and genetic variation can be examined for involvement in schizophrenia susceptibility.
Supplementary Material
Acknowledgments
We thank Thomas Zhangzy for helping with the CNV calling procedures and Flip Mulder for technical support. We acknowledge members of the department of Medical Genetics and the department of Psychiatry for critically reading the manuscript.
The GROUP project was supported by a grant from ZON-MW, within the Mental Health program (project number: 10.000.1001). In addition, genome-wide SNP genotyping of the Dutch schizophrenia sample was funded by the National Institute of Mental Health grant RO1 MG078075 to R.A.O.
GROUP Consortium members: René S. Kahn (Rudolf Magnus Institute of Neuroscience, Department of Psychiatry, University Medical Centre Utrecht, Utrecht, The Netherlands), Don H. Linszen (Academic Medical Centre University of Amsterdam, Department of Psychiatry, Amsterdam, The Netherlands), Jim van Os (Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands), Durk Wiersma (University Medical Centre Groningen, Department of Psychiatry, University of Groningen, The Netherlands), Richard Bruggeman (University Medical Centre Groningen, Department of Psychiatry, University of Groningen, The Netherlands), Wiepke Cahn (Rudolf Magnus Institute of Neuroscience, Department of Psychiatry, University Medical Centre Utrecht, Utrecht, The Netherlands), Lieuwe de Haan (Academic Medical Centre University of Amsterdam, Department of Psychiatry, Amsterdam, The Netherlands), Lydia Krabbendam (Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands), Inez Myin-Germeys (Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands).
Footnotes
Supplementary information is available on the Biological Psychiatry website.
Financial Disclosures
Dr. Ophoff received a grant from the National Institute of Mental Health for the genome-wide SNP genotyping of the Dutch schizophrenia sample. The GROUP project was supported by a grant from ZON-MW, within the Mental Health program. The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Web Resources
The URLs for data listed herein are as follows:
-
UCSC genome browser: http://genome.ucsc.edu/Database of Genomic Variants: http://projects.tcag.ca/variation/QuantiSNP: http://www.well.ox.ac.uk/QuantiSNP/
Reference List
- 1.Gottesman I. Schizophrenia epigenesis: past, present, and future. Acta Psychiatr Scand. 1994;90:26–33. doi: 10.1111/j.1600-0447.1994.tb05887.x. [DOI] [PubMed] [Google Scholar]
- 2.Jablensky A. Epidemiology of schizophrenia: the global burden of disease and disability. Eur Arch Psychiatr Clin Neurosci. 2000;250:274–285. doi: 10.1007/s004060070002. [DOI] [PubMed] [Google Scholar]
- 3.Andreasen N. Symptoms, signs, and diagnosis of schizophrenia. Lancet. 1995;346:477–481. doi: 10.1016/s0140-6736(95)91325-4. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister M, McInnis M, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527–540. doi: 10.1038/nrg2381. [DOI] [PubMed] [Google Scholar]
- 5.Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry. 2000;47:210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan P. The genetics of schizophrenia. PLoS Med. 2005;2:e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan P, Kendler K, Neale M. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 8.Tam G, Redon R, Carter N, Grant S. The role of DNA copy number variation in schizophrenia. Biol Psychiatry. 2009;66:1005–1012. doi: 10.1016/j.biopsych.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 9.International Schizophrenia Consortium. Purcell S, Wray N, Stone J, Visscher P, O'Donovan M, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada K, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H, Rujescu D, Cichon S, Pietiläinen O, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, et al. GROUP Consortium. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Med Genet Part B. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh T, McClellan J, McCarthy S, Addington A, Pierce S, Cooper G, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 15.Xu B, Roos J, Levy S, van Rensburg E, Gogos J, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 16.Choy K, Setlur S, Lee C, Lau T. The impact of human copy number variation on a new era of genetic testing. BJOG. 2010;117:391–398. doi: 10.1111/j.1471-0528.2009.02470.x. [DOI] [PubMed] [Google Scholar]
- 17.Mefford H, Sharp A, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Need A, Ge D, Weale M, Maia J, Feng S, Heinzen E, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassett A, Scherer SW, Brzustowicz L. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iafrate A, Feuk L, Rivera M, Listewnik M, Donahoe P, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 21.Vorstman JAS, Staal WG, van Daalen E, van Engeland H, Hochstenbach PFR, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 22.Bray N. Gene expression in the etiology of schizophrenia. Schizophr Bull. 2008;34:412–418. doi: 10.1093/schbul/sbn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore J, Williams S. Epistasis and its implications for personal genetics. Am J Med Genet Part A. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colella S, Yau C, Taylor J, Mirza G, Butler H, Clouston P, et al. QuantiSNP: an objective bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant S, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dellinger A, Saw S, Goh L, Seielstad M, Young T, Li Y. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nuc Acids Res. 2010;38:e105. doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct Genomic Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 28.Lee C, Iafrate A, Brothman A. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet Suppl. 2007;39:S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 29.Muir W, Pickard B, Blackwood D. Chromosomal abnormalities and psychosis. Br J Psychiatry. 2006;188:501–503. doi: 10.1192/bjp.bp.106.023895. [DOI] [PubMed] [Google Scholar]
- 30.Marenne G, Chanoch S, Rothman N, Rodriguez-Santiago B, Rico D, Pita G, et al. CNV assessment using Illumina Infinium 1M platform: Agreement according to algorithm and source of DNA. Ann Hum Genet. 2009;73:658–669. [Google Scholar]
- 31.Tsuang D, Millard S, Ely B, Chi P, Wang K, Raskind W, et al. The effect of algorithms on copy number variant detection. PLoS ONE. 2011;5:e14456. doi: 10.1371/journal.pone.0014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey J, Kidd J, Eichler E. Human copy number polymorphic genes. Cytogenet Genome Res. 2008;123:234–243. doi: 10.1159/000184713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freeman J, Perry G, Feuk L, Redon R, McCarroll S, Altshuler D, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 34.Sharp A, Locke D, McGrath S, Cheng Z, Bailey J, Vallente R, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackwood D, Fordyce A, Walker M, St Clair D, Porteous D, Muir W. Schizophrenia and affective disorders - cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoogendoorn M, Vorstman J, Jalali G, Selten J, Sinke R, Emanuel B, et al. Prevalence of 22q11.2 deletions in 311 Dutch patients with schizophrenia. Schizophr Res. 2008;98:84–88. doi: 10.1016/j.schres.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karayiorgou M, Gogos J. The molecular genetics of the 22q11-associated schizophrenia. Mol Brain Res. 2004;132:95–104. doi: 10.1016/j.molbrainres.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 38.Weksberg R, Stachon A, Squire J, Moldovan L, Bayani J, Meyn S, et al. Molecular characterization of deletion breakpoints in adults with 22q11 deletion syndrome. Hum Genet. 2007;120:837–845. doi: 10.1007/s00439-006-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaikh T, Kurahashi H, Saitta S, Mizraha O'Hare A, Hu P, Roe B, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 40.O'Donovan M, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 41.Stefansson H, Ophoff R, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manolio T, Collins F, Cox N, Goldstein D, Hindorff L, Hunter D, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan J, Sanders A, Gejman P. Genome-wide approaches to schizophrenia. Brain Res Bull. 2010;83:93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alkan C, Kidd J, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riegel M, Baumer A, Jamar M, Delbecque K, Herens C, Verloes A, et al. Submicroscopic terminal deletions and duplications in retarded patients with unclassified malformation syndromes. Hum Genet. 2001;109:286–294. doi: 10.1007/s004390100585. [DOI] [PubMed] [Google Scholar]
- 46.Conrad D, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Santiago B, Brunet A, Sobrino B, Serra-Juhé C, Flores R, Armengol L, et al. Association of common copy number variants at the glutathione S-transferase genes and rare novel genomic changes with schizophrenia. Mol Psychiatry. 2009;15:1023–1033. doi: 10.1038/mp.2009.53. [DOI] [PubMed] [Google Scholar]
- 48.Guilmatre A, Dubourg D, Mosca A, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 50.Friedman J, Vrijenhoek T, Markx S, Janssen I, van der Vliet W, Faas B, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 51.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 52.Ingason A, Rujescu D, Cichon S, Sigurdsoon E, Sigmundsson T, Pietiläinen O, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Badner J, Gershon E. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 55.Lewis C, Levinson D, Wise L, DeLisi L, Straub R, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen N, Bagade S, McQueen M, Ioannidis J, Kavvoura F, Khoury M, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 57.Wall J, Pritchard J. Assessing the performance of the haplotype block model of linkage disequilibrium. Am J Hum Genet. 2003;73:502–515. doi: 10.1086/378099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.