Abstract
Structured functional RNAs are conserved on the level of secondary and tertiary structure, rather than at sequence level, and so traditional sequence-based searches often fail to identify them. Structure-based searches are increasingly used to discover known RNA motifs in sequence databases. We describe the application of the program RNABOB, which performs such searches by allowing the user to define a desired motif's sequence, paired and spacer elements and then scans a sequence file for regions capable of assuming the prescribed fold. Structure descriptors of stem-loops, internal loops, three-way junctions, kissing loops, and the hammerhead and hepatitis delta virus ribozymes are shown as examples of implementation of structure-based searches.
Keywords: Motif search, RNA structure, Base-pairing, Sequence covariance, Aptamer, Ribozyme, HDV, Hammerhead
1. Introduction
1.1. Non-coding RNA
Non-coding RNAs (ncRNAs) are involved in a wide array of catalytic and regulatory functions [1–3]. The structures of these folded functional RNAs are dominated by base-paired helical elements and by conserved single-stranded regions that often form active sites or binding pockets [4]. Since a helix can form between any set of complementary bases, helical requirements translate into base-pair (bp) covariation and not into specific sequence conservation; therefore molecules exhibiting the same function can be unrecognizable on a sequence level.
The population of known ncRNAs has grown dramatically in recent years and the current dataset is still thought to represent only a fraction of the total ncRNAs present in a cell [5]. Traditionally, identification of these molecules has relied on detailed analysis of specific genes or viral genomes, expression profiling, and sequencing of transcriptomes or in vitro selected pools. Because functional RNAs are typically not conserved on the sequence level, but rather on the level of secondary and tertiary structure, sequence-based bioinformatics searches are not as successful at finding functional RNAs as they are at finding functional polypeptides. In addition, alignment of sequences, particularly short ones, is greatly assisted by genome annotation, which is in turn informed by expression data and known regulatory elements. Protein-coding sequences are preceded by predictable transcription start sites, defined by translation start and stop codons, and in some cases by splicing data. On the other hand, ncRNAs do not always benefit from comparable genome annotation and are consequently harder to recognize. Therefore, to discover known ncRNAs computationally in a systematic and unbiased way, a method is required that is based on the molecular structure.
1.2. ncRNA motifs
A number of ncRNA motifs have been identified, but for our structure-based searches, we will concentrate on aptamers and ribozymes as examples. A number of aptamers, RNA motifs that bind target molecules, have been uncovered through the use of in vitro selections (or SELEX) [6,7], and genetic analysis of mRNAs of metabolic genes [8]. The small-molecule targets of the in vitro selected aptamers include adenosine [9], guanosine triphosphate [10], citrulline and arginine [11,12], a variety of organic dyes [6,13,14], theophylline, and caffeine [15]. There are several classes of metabolite binding RNAs present in genomes of bacteria, fungi, and plants [16]. These RNAs, termed riboswitches, regulate gene expression through interactions induced by ligand-binding. They have been shown to bind a multitude of metabolites, including amino acids [17,18], vitamin B12 [19,20], metabolic intermediates [21–23], and nucleobases [24,25], and are also involved in sensing Mg2+ [26] and temperature [27].
Ribozymes represent another subset of ncRNAs and have been identified in the ubiquitous RNPs, like the ribosome and RNase P, which are found in all living cells, and the eukaryotic spliceosome. Together with group I and group II introns and a plethora of small self-cleaving ribozymes, these ribozymes perform chemical transformations during protein synthesis or during processing of cellular and viral transcripts and RNA replicons [28]. The small self-cleaving ribozymes are represented by a variety of catalytic motifs, such as the hammerhead [29], HDV [30,31] and hairpin ribozymes [32], the Neurospora Varkud satellite (VS) motif [33], and the bacterial GlmS ribozyme [34].
1.3. Biochemical structure characterization
Structure-based searches rely upon detailed characterization of the RNA under scrutiny. This is most easily accomplished using comparative genomics, however, a large number of natural variants are required for this method. In a folded, functional RNA, binding- or active-site residues are largely invariant, helical regions tend to co-vary, and single-stranded spacer regions show almost no conservation [35]. By aligning the multiple sequences of a particular RNA, these regions of differing variability can be identified and a secondary structure proposed. Comparative genomics was originally used to solve the 5S ribosomal RNA secondary structure in Escherichia coli [36], and it has also been employed to define paired positions in a number of ribozymes and aptamers (e.g. Refs. [9,10,37,38]).
For RNAs lacking multiple sequence alignments, structure characterization relies on biochemical reagents that react preferentially with a particular base, specific groups in the sugar–phosphate backbone, or RNA conformation [39–42]. Reactivity for many of these molecules requires the nucleotide of interest to be exposed and not base-paired. Similarly, digestion with various RNases helps to determine whether a given position in the sequence is paired or single-stranded. Finally, hydroxyl radical footprinting, iodoethanol cleavage of phosphorothioates, and 2′ alkylation can modify exposed segments of the sugar–phosphate backbone, thus reporting the solvent-accessibility of various parts of an RNA [43,44]. By combining these various techniques, detailed secondary structure models can be inferred for a newly discovered RNA [45].
Ultimately, the most reliable data come from atomic-resolution structures. For aptamers, which tend to be shorter than ribozymes, nuclear magnetic resonance (NMR) has been used with success. NMR structures exist for the adenosine [46,47], citrulline and arginine [48], theophylline [49], flavin mononuclotide [50], and GTP aptamers [51], among others. For larger ribozymes and riboswitches, X-ray crystallography has been employed effectively, often incorporating crystallization modules to improve crystal quality [16,52,53].
1.4. In silico structure characterization
Structural data derived from experimental methods are supplemented by models created by software programs that predict folding patterns of RNAs. Among these is the ViennaRNA package, which contains a set of structure predicting algorithms [54]. With the RNAfold algorithm, emphasis is placed on a free energy minimization routine [55]. This program seeks to maximize paired regions, which for RNAs that form only stem-loop structures has proven sufficient to reliably predict secondary structure. However, many structured functional RNAs contain bulged regions and pseudoknot structures that are difficult to encapsulate with just base-pair maximization. Incorporation of experimentally derived structural constraints improves the predictive power of the software, especially when the proposed structures are subsequently compared to each other. RNA folding programs produce a large number of potential structures and these can be examined in a manner analogous to that used in comparative genomics. In the ViennaRNA package, RNAalifold uses a tree diagram to accomplish this [56]. Internal helices in a predicted structure are defined as “internal nodes,” while bulged or unpaired regions branch from these and are “leaf nodes.” This tree method provides a rapid way to computationally relate varied structures, but it too has difficulty representing pseudoknots. The Structural RNA Motif Search program (STRMS) overcomes many of the challenges associated with pseudoknots by allowing the user to specify domains in the tree model that may form such structures [57]. The modeling capabilities of STRMS are coupled to a structure-based search algorithm that has proven robust at identifying small riboswitches whose secondary structure is accurately predicted by the software.
STRMS represents a class of search algorithms capable of “learning” a secondary structure for use in fold-based searches from a subset of structurally related RNAs. Others members of this class include the COVE suite and the ERPIN program [58,59]. Because these programs independently determine regions of structure, the user needs only to input a set of sequences that fold into the desired motif before searches can be performed. However, to efficiently train the software for a structured RNA, a number of natural variants must exist for the RNA in question. Thus, this application is limited to RNAs that have already been identified at a large number of loci.
1.5. Structure-based search programs
Less common RNAs or those with more convoluted secondary structures require a combination of modeling data and experimentally confirmed structural motifs to generate user-defined descriptors for structure-based searches. Early forays into programs capable of structure-based searches of RNAs were limited to stem-loop motifs in which the nucleotide composition of the stem needed to be specified, but the loop itself could vary [60]. Requiring defined helices severely hampered the utility of these searches, as only a small subset of the total helix-forming structures present in a genome could be obtained with just a single descriptor file. RNAMOT resolved this by permitting covariation in helical regions [61]. This “pattern searching-alignment” tool allowed users to define domains of a folded RNA and specify whether only Watson–Crick pairings were permissible or if mismatches and non-canonical pairings could occur in the structure. Furthermore, because the fold was specified by the user, pseudoknots could be incorporated into the structure descriptor, which facilitated the identification of the intricate group I intron motifs [61]. Refinements to the RNAMOT algorithm offered improved search times [62], and demonstrated that structure-based searches could identify functional RNA motifs from raw genomic sequences [63–66].
Since the development of RNAMOT, a number of other descriptor-based search algorithms has arisen. The Palingol program allows great flexibility in descriptor design by using the formal computer programming language to define motifs, while the FastR suite sought to further improve on the computational time of structure-based search tools by employing a two-step search method [67,68]. With FastR, a genome is filtered first based on the ability of a particular sequence to fit elements of the query fold, and then the remaining sequences are rated on their ability to assume the entire query fold. Other descriptor-based search tools include PatScan, which succeeded at identifying hammerhead ribozymes in the Arabidopsis thaliana genome [69,70].
The simplicity and success of RNAMOT led to additional refinements that produced RNAMotif and RNABOB [71,72]. These programs offer similar functionality while allowing a high degree of secondary structure to be defined in a user-friendly manner that does not require extensive programming experience. Furthermore, both algorithms are efficient in their search routines. The main difference between the two is the syntax used to specify a structure when creating a descriptor file. RNAMotif employs a nested descriptor language in which a given motif is sequential with the next. Each factor is thus individually declared, with the 5′ end of a helix specified as h5 and the 3′ end is termed h3. As the descriptor is built, each h3 designation is by default related to the last h5 designation that has not yet been paired to an h3 with single-stranded elements listed in between as needed. To describe pseudoknots, wherein 5′ and 3′ strands of a helix can be separated by one or several intervening paired regions, a tag must be applied to the h5 and h3 specifications that associates the non-sequentially spaced helical edges [72].
In RNABOB, each paired region is given a unique designation, i.e. h1, h2, and 5′ and 3′ strands of the helix are specified explicitly. We have chosen to focus on the RNABOB program because we found defining complex secondary structures to be easier with this tool. To demonstrate this descriptor-based secondary structure search approach, we will examine common aptamer and riboswitch motifs, the hammerhead ribozyme, and the nested double-pseudo-knot of the HDV ribozyme.
2. Methods
2.1. RNABOB syntax
In designing a descriptor, both sensitivity, the fraction of active sequences out of those identified, and specificity, the number of found active sequences relative to the number of missed sequences, should be considered [73]. Specificity is difficult to estimate when attempting to uncover motifs at new genomic loci, as it is often not known how much sequence drift is permissive for a given motif. An overly permissive descriptor yields many sequences, potentially discovering all active ones, but it leads to low sensitivity as too many results are returned and active RNAs are obscured by false positives.
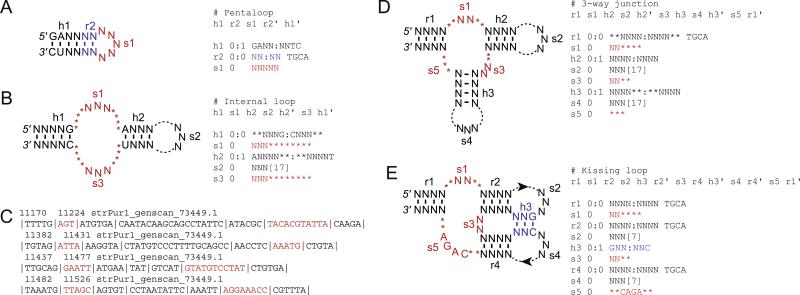
To design a descriptor for RNABOB, it is necessary to first specify each aspect of secondary structure. To do this, RNABOB uses three letters, “s”, “h”, and “r”, representing a “single-stranded”, “helix”, or “relational” element, respectively. Using this syntax, each line in a descriptor file contains all the information needed to define one element of secondary structure. By default, RNABOB will include G–U wobble-pairs in helices defined with the “h” designation. To allow for strict Watson–Crick or non-canonical base-pairing, the “r” label is used. The relational “r” designation allows the user to specify what residue each nucleotide, in ACGT order, is capable of pairing with. Thus, in a strict Watson–Crick helix, the pairing would be defined as “TGCA”. However, if AGT were required to form Watson–Crick pairs, but C was allowed to appear opposite of any nucleotide, pairing in the relational element would be defined as “TNCA”. Mismatches in helices are denoted by stating their maximum number to the right of a colon that follows the “h” or “r” term. A number placed on the left of the colon indicates mismatches permitted to a defined sequence, like in conserved, single-stranded regions. To define single-stranded regions of variable length, an “*” or a bracketed number can be used. Paired regions of variable length can also be designated by asterisks (Fig. 1), however, the program does not allow for a single-nucleotide insertion within a paired region. To do that, a different descriptor must be written for each position in a paired region where an insertion is allowed. This makes the searches somewhat cumbersome, because single-nucleotide insertions, and to some extent deletions (which are formally insertions in the opposite strand of a shorter paired region), are commonly found in secondary structures of functional RNAs.
Fig. 1.
(A) Descriptor for a hairpin motif specifying the first two base-pairs and allowing one mismatch in the first four base-pairs of the helix but requiring strict Watson–Crick pairing for the last two. (B) Descriptor for an internal loop allowing for variable s1, s2, and s3 regions, and one mismatch in h2. Note that because any nucleotide was allowed in these regions, a minimum of three nucleotides can appear at any position in the loops. The maximum is set by the sum of Ns and asterisks or the sum of Ns and the bracketed number. Here, the s1 element allows 3–11 nt of any sequence and s2 allows 3–20 nt. (C) Example of an RNABOB output for a search of the S. purpuratus genome using the internal loop descriptor with the s1 and s3 sequences shown in red. (D) Descriptor for a three-way junction with variable single-stranded regions. (E) Descriptor for a kissing loop allowing for one mismatch in the pseudoknot-forming h3 helix.
After each element has been specified, their relation to one another must be defined. This is done at the top of the descriptor file by listing each motif in the order in which they will be searched in the sequence file. Although each “s” term will appear only once in this line, the “h” and “r” labeled motifs are required to appear twice. Their first appearance specifies the upstream strand of the helix, while the second appearance signifies the downstream portion and is denoted by an apostrophe following the tag.
2.2. Database preparation
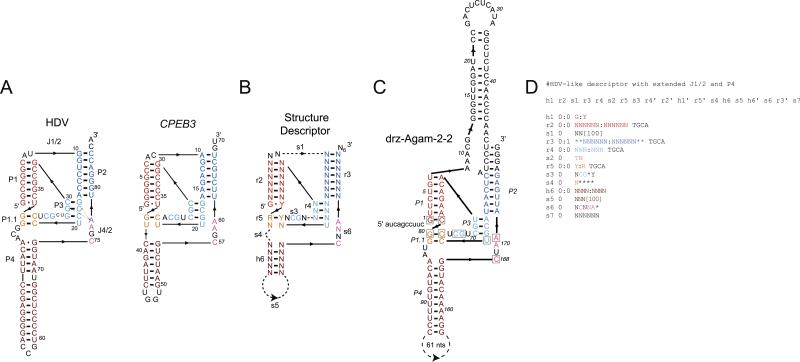
RNABOB is capable of reading the variety of standard file formats (e.g. FASTA, GenBank) available for downloadable genomic databases. These files should only be opened using a simple text editor, as more advanced word processing programs (e.g. MS Word) will insert hidden characters at the ends of lines that are recognized by RNABOB and produce a fatal error in the search routine. Search times are largely independent of database format and linearly dependent on the length of the sequence file. A search for the HDV ribozyme fold shown in Fig. 3 on a MacBook (2.26 GHz, 2 GB 1067 MHz DDR3) through a single random sequence of 107 nt takes ~7 min. The same descriptor tested with the human PhastCons elements in both sense and antisense directions (~26 × 107 nt) on a single node of a Dual Opteron Powerwulf cluster running Fedora Core 4 Linux takes ~7 min to run.
Fig. 3.
(A) Secondary structures of the genomic HDV and the concensus mammalian CPEB3 ribozymes. (B) Structure descriptor for a minimal HDV-like self-cleaving sequence. (C) The drz-Agam-2-2 ribozyme, containing an extended J1/2 region, and (D) the RNABOB descriptor used to identify this ribozyme. Boxed nucleotides in the drz-Agam-2-2 secondary structure correspond to the explicitly defined nucleotides seen in (B).
3. Aptamers
3.1. Aptamer folds
One application of structure-based searches is in analysis of in vitro selected nucleic acids. In vitro selection experiments start with diverse DNAs in the form of random pools, mutagenized clones, or genome-derived sequences. Typically, selection experiments are designed to yield aptamers or ribozymes which bind target molecules or catalyze a chemical transformation as part of the selection step, respectively [74]. The experiments yield pools with much reduced sequence diversity that are enriched for molecules that fulfill the selection criteria. When an appreciable fraction of the pool exhibits the desired activity, it is sequenced to establish whether a particular motif dominates the population. If the sequenced population is not dominated by one conserved sequence element, either the selection is continued under more stringent conditions to enrich the pool for a dominant sequence, or a more careful analysis of the sequence information is required. A structure-based search may uncover a family of conserved functional molecules that sequence alignment would not.
Aptamers, particularly small-molecule binding RNAs, which include many riboswitches, exhibit conserved secondary structures that allow the formation of specific tertiary folds to facilitate ligand-binding. In motifs with no obvious sequence conservation, secondary structure analysis helps to narrow the search for conserved elements to the single-stranded regions of loops and junctions. Therefore, it may be beneficial to sort the in vitro selected sequences according to their ability to form basic aptamer secondary structures. Here we will describe four of the most common RNA folds that are found in small-molecule aptamers and riboswitches.
The simplest RNA structures that have been found in aptamers are: (1) stem-loops, (2) internal loops and bulges, (3) three-way junctions, and (4) pseudoknots and kissing loops. As stated before, the strategy is to identify short conserved elements interspersed with paired regions of low conservation. The exercise is based on the assumption that presorting a selected pool of RNAs or DNAs according to the secondary structure they may form can lead to subsequent identification of conserved short single-stranded regions.
3.2. Stem-loops and internal loops
Stem-loop structures are the easiest to predict and the most likely structures identified by sequence alignment. They are also the easiest to define (Fig. 1A), requiring just one paired element and one single-stranded element. We find that a six base-pair helix with a single mismatch is often stabilizing enough to support the loop that presumably forms the ligand-binding pocket. Here a stem starting with a GA sequence on the 5′ strand is specified. To prevent the mismatch from occurring at the loop side of the helix, the first one or two base-pairs next to the loop are specified separately, e.g. as a 2 bp relational element with no mismatches and no G–U pairs, and the rest of the paired region is specified by a separate element of 4 bp with a single allowed mismatch. The loop length in the stem-loop structure must also be defined in the descriptor. Because tetraloops are sufficient to cap a helix and to recognize an RNA or protein ligand specifically, the minimum length of the loop is usually set to 4. Any loop length can be allowed (by defining the loop as “s1 0 NNNN [16]” RNABOB will look for single-stranded regions of four to 20 nt long), however, creating separate descriptors for different loop lengths results in a more tractable output file. Moreover, long, conserved loops are more likely to be recognized by alignment-based searches, therefore the highest utility of structure-based searches may be in identifying short loops and then aligning the loop sequences separately.
Fig. 1A and B show examples of secondary structures and RNABOB descriptors for a simple pentaloop and an internal loop, respectively. Stem-loops and internal loops are some the most common structures among small-molecule binding aptamers. For example, of the eleven known classes of aptamers that bind GTP, four form stem-loops and five form internal loops [10]. The internal loop descriptor defines two helical elements, h1 and h2, which flank an internal loop wherein each strand, s1 and s3, can vary between 3 to 11 nt in length. In this example, s1 and s3 contain no defined bases, therefore the arrangement of asterisks and bases is inconsequential. However, if the RNABOB algorithm is searching for a specific single-stranded 3 nt sequence, then for a helical element appearing either 0 or 8 nt downstream of it, the order spec-ified in s1 does matter (i.e. “s1 0 ACA********” will give different results from “s1 0 ****ACA****”). Consequently, care must be taken to position regions of variability when searching for variable loops containing an explicit sequence. The s2 element, here 3–20 nt long, serves to connect the two strands of the structure and may or may not show conservation. The outer helix, h1, can be 5 or 6 bp long and ends with a G–C base-pair, while the inner helix, h2, is 4–6 bp long, starts with an A–U base-pair and allows for a single mismatch. Because no sequence mismatches are allowed in h2, but a single pairing mismatch can occur (“h2 0:1”), the element always starts with an adenosine, but its pairing partner need not be a uridine.
The RNABOB output for the internal loop descriptor shows several examples of potential internal loop structures from the genome of the purple sea urchin ( Strongylocentrotus purpuratus) (Fig. 1C). The position of each sequence within the searched file is indicated explicitly, followed by the name of the file. Individual elements that were defined in the descriptor are indicated by the vertical lines in the output. This leads to easy identification of core elements in the structure and allows for straightforward parsing of the sequences into tables (e.g. using the text-to-column function in Excel) where each column of the parsed output file contains sequences from the same element of the RNA structure. Thus, one column contains all s1 sequences and another all s3 sequences, together comprising the internal loops. Subsequently, the parsed sequences can be analyzed in part, for example by aligning only the loop sequences, to gauge the variability within a group or to measure sequence drift in the paired regions for evolutionary analysis of the molecules, or in full. For complex functional RNAs, alignment of individual regions tends to be more informative than alignment of entire sequences. Moreover, extensive peripheral regions can be directly submitted into secondary structure prediction programs to estimate whether they form motifs compatible with the rest of the molecule. For example, if a stem-loop structure is predicted for the peripheral regions, the described core may be stabilized more so than if the peripheral region is predicted to be unstructured domain or form an alternative secondary structure.
3.3. Three-way junctions and kissing loops
Three-way junctions consist of three helical elements that are connected by three single-stranded regions (Fig. 1D). For the descriptor shown in Fig. 1D, the helices are capped by single-stranded segments of 3–20 nt, defined using the bracket notation (s2 and s4 in Fig. 1D). The interhelical loops are shown with unequal lengths of 2–6, 2–4, and 0–3 nt for s1, s3, and s5, respectively.
A kissing loop structure that stabilizes a three-way junction is shown in Fig. 1E. The two loops that pair to form the kissing structure are defined by s2, s4, and h3, which is allowed to have a single mismatch. This is also the first example of a pseudoknot, where a loop of one stem-loop (h2–s2) pairs with another single-stranded region, forming the h3 region. The s5 region is shown with a specific sequence (CAGA) flanked on both sides by potential inserts of up to 2 nt. Because a specific sequence is required, the positions of the allowed inserts have to be defined, unlike in previous examples where no sequence was specified in the loops and the descriptors only needed to account for the minimum and maximum length of the loop.
4. Hammerhead ribozyme
4.1. Hammerhead ribozyme secondary structure
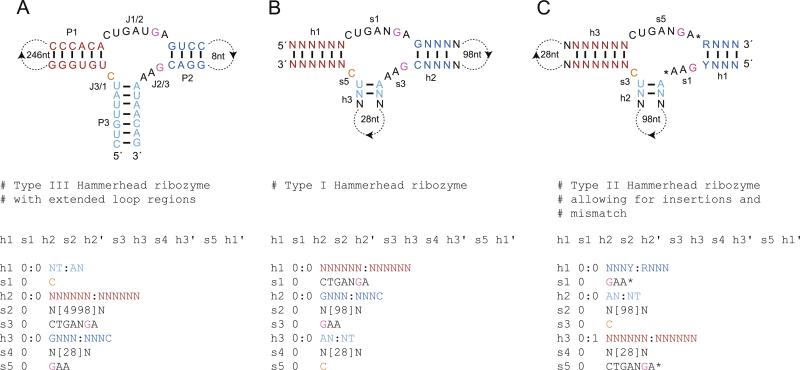
The minimal hammerhead motif is characterized by three helical structures, P1, P2, and P3, of variable length that are joined sequentially by single-stranded regions J1/2, J2/3, and J3/1 of seven, three, and 1 nt in length, respectively (Fig. 2A) [38,53,75–78]. The helices are arranged in a plane with the cleavage site located between the P1 helix and J3/1 region [79]. Of the 11 nt that form the active-site of the molecule, nine are conserved [80]. The residues that form the closing pairs of P2 and P3 helices also show conservation, with a purine–pyrimidine base-pair at the terminus of P2 and an A–U pair at the P3 terminus. Minimal hammerhead motifs have been obtained in vitro, where they eventually dominate the sequence space during selections for moderately-fast self-cleaving RNAs [81]. A hammerhead ribozyme can be embedded in surrounding sequences through any of the three helices, with type I, II, and III referring to those embedded through P1, P2, and P3, respectively. In selection experiments, type III hammerheads appear most frequently and genomic hammerheads are usually types I and III, but to fully explore all potential hammerhead motifs bioinformatically, a secondary structure descriptor is required for each type of topology.
Fig. 2.
(A) The discontinuous CLEC2 hammerhead ribozyme and the descriptor used to isolate it by Martick et al. [82]. The cleavage site is 3′ to the orange nucleotide and the catalytic residues are colored pink. (B) Descriptor for a type I hammerhead ribozyme subject to the same constraints but with different-length capping loops. (C) Descriptor for a type II hammerhead ribozyme that allows for elongated J1/2 and J2/3 regions and one mismatch in the P1 helix.
4.2. Hammerhead ribozyme descriptor design
Hammerhead ribozymes have been detected via structure-based bioinformatics on several occasions. RNAMOTIF uncovered hammerheads in the schistosome satellite DNA and was used to estimate their appearance in the GenBank database [65,66], Patscan found hammerhead ribozymes in A. thaliana [70], and most recently an RNABOB search using a minimal hammerhead ribozyme core with permissive peripheral regions revealed the presence of a type III hammerhead ribozyme containing a long intervening sequence in the P1 loop in the rodent C-type lectin type II (CLEC2) genes (Fig. 2A) [82]. The descriptors employed by Martick et al. contained only those constraints crucial to hammerhead self-cleavage and thus remained specific while minimizing false positives. The CLEC2 ribozymes represent a family of type III hammerheads that can be readily identified with a strict secondary structure descriptor (Fig. 2A). This descriptor can easily be modified into a strict type I motif simply by reordering the relational line and the positions of the extended loops (Fig. 2B). For type II hammer-heads, which appear to be much less common than types I and III, a strict descriptor runs the risk of being too specific, therefore it is often beneficial to loosen some of the requirements. Several changes can be made to the original descriptors that will retain a hammerhead-like self-cleavage functionality. Mutagenesis has indicated that the G–C pair at the close of the P2 helix can be substituted with an A–U base-pair [80]. In addition, the lengths of J1/2 and J2/3 segments can each be extended by a single nucleotide without abolishing activity [29,83]. Mismatches in the longer helical regions are tolerated as well, however, care must be taken when allowing these as the number of putative results can grow rapidly without a corresponding increase in active sequences (Fig. 2C) [80].
5. HDV ribozyme
5.1. HDV ribozyme secondary structure
The HDV ribozyme contains a significantly more complex secondary structure than the hammerhead motif. This ribozyme is formed around five helices (P1, P2, P3, P1.1, and P4), that form a nested double-pseudoknot structure (Fig. 3). The P1 and P1.1 helices stack upon the P4 helix, P2 and P3 are also coaxial, and the J1/2 and J4/2 regions serve as joining strands [84–86]. The catalytic core of the ribozyme is formed from the L3, P1.1, and J4/2 sections of the molecule, with the J4/2 region providing the active-site cytosine that most likely acts as a general acid during the cleavage reaction and an adenosine residue that forms an A-minor tertiary interaction with the P3 helix [84,87]. Cleavage occurs at the base of the P1 helix, which requires a guanosine–pyrimidine nucleotide pair for activity. Mutation and selection studies have demonstrated that of the approximately 60 nt required to form the minimal structure, only six are invariant on a sequence level [88,89].
5.2. HDV ribozyme descriptors design
Unlike hammerhead ribozymes, which have been identified on several occasions through bioinformatic searches, HDV-like ribozymes have only recently been found using this methodology [90]. The creation of robust secondary structure descriptors for the HDV motif was greatly facilitated by the identification of the mammalian CPEB3 ribozyme, which shares the same secondary structure (Fig. 3A). In our experience, the six invariant positions, with several more partially-defined positions (purines or pyrimi-dines), and a rigorously-defined double-pseudoknot are sufficient to identify active HDV-like ribozymes in many genomes [90]. More loosely described structures result in poor sensitivity and yield many sequences of low complexity. These sequences are generally found in genomic repeats and contain long runs of AUs or GCs (or GUs) that have the ability to fold into any secondary structure, but do not form specific folds and thus represent inactive sequences. Using a descriptor for the HDV-like ribozymes shown in Fig. 3 yields many eukaryotic ribozymes, in some cases belonging to multiple sequence families in a single organism.
As in the case of the hammerhead ribozyme, allowing for variable length peripheral domains identifies sequences with extended structures in these regions. The HDV-like ribozymes have such variable regions in the P4 helix and the L4 loop [91–93]. The fact that the loop is not essential for the ribozyme activity led to its replacement with the U1A binding loop and successful co-crystallization of the genomic HDV ribozyme with the U1A protein [84]. Permitting long inserts in this region yields active ribozymes with extended helices and the predicted stability of this peripheral domain correlates with the in vitro cleavage rate constant of closely-related sequences from Pristionchus pacificus, suggesting that the ribozyme kinetics are dominated by folding (in an experiment initiated by addition of Mg2+) [90].
The J1/2 region of the HDV-like ribozymes is not directly involved in formation of the ribozyme core and appears to play a strictly structural and topological role, connecting P1 with P2 [84]. The descriptor for HDV-like ribozymes allows for variable sequence to be inserted in J1/2, potentially creating another peripheral domain of the ribozyme (Fig. 3C). While the HDV and CPEB3 ribozymes do not contain any additional domain in J1/2, new ribozymes have been identified in Anopheles gambiae with long inserts in this region [90]. The inserts potentially form extended secondary structure elements and likely stabilize the overall structure because the large ribozymes are among the fastest in the HDV family (drz-Agam-2-1 has a kobs = 7 min-1 at 37 °C and 1 mM Mg2+). These examples show that when the functional core of an RNA is well defined, a structure-based search can uncover new stabilizing motifs in the peripheral domains of the molecule, in addition to new candidate ribozymes.
6. Discussion
The rapid growth of genomic data over the last decade has altered the landscape of molecular biology and biochemistry. The importance of ncRNAs has come to light and the identification of these molecules from the surrounding sequence space has become a task of paramount importance. In this endeavor, structure-based searches for functional RNAs have emerged as an effective tactic. These searches take advantage of the conserved secondary structure of aptamers and ribozymes and have identified such RNAs in new organisms and at additional genomic loci.
The RNABOB algorithm is one of several programs capable of structure-based searches. RNABOB allows for the rapid generation of structure descriptors for a seemingly endless variety of structural motifs. Although search times can be quite long for complex RNAs in large genomes, they still remain much shorter than the time scale required to test the candidate sequences. Alternatively, searches for simple RNA structures can be quite rapid but often lack sensitivity and return a large number of putative sequences, however, the output of the searches can be used to identify sequence conservation in individual structural elements that would not be apparent from alignments of entire sequences.
The current generation of the RNABOB algorithm possesses some limitations in its search capabilities. RNABOB is useful for identification of sequences capable of forming a defined motif, but it has no metric to determine if such a sequence would be likely to form from the surrounding sequence. This can be partially overcome by inputting RNABOB outputs and their flanking genomic regions into an RNA folding program, but the accuracy of such programs for determining structures from large tracts of genomic sequence is low. In addition, single-nucleotide insertions and deletions in specific sequences or in helices have to be specified at explicit locations and cannot be defined more broadly (e.g. one adenosine insertion per segment). Separate descriptors have to be defined for every point permissible to insertion, but such a brute force approach is only practical for short RNAs or those with few points of insertion or deletion. As single-nucleotide insertions or deletions provide some of the most common variation to an RNA molecule, there is a significant drive to overcome this limitation.
We expect that structure-based searches will grow in importance as more structured RNAs, both coding and non-coding, are described and as the amount of sequence data grows with precipitating cost of sequencing nucleic acids.
Abbreviations
- RNA
ribonucleic acid
- ncRNA
Non-coding RNA
- bp
base-pair
- nt
nucleotide
- SELEX
systematic evolution of ligands by exponential enrichment
- RNP
ribonucleoprotein
- HDV
hepatitis delta virus
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- 1.Sharp PA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedor MJ, Williamson JR. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 4.Doherty EA, Doudna JA. Annu. Rev. Biochem. 2000;69:597–615. doi: 10.1146/annurev.biochem.69.1.597. [DOI] [PubMed] [Google Scholar]
- 5.Eddy SR. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 6.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Henkin TM. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sassanfar M, Szostak JW. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 10.Carothers JM, Oestreich SC, Davis JH, Szostak JW. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famulok M. J. Am. Chem. Soc. 1994;116:1698–1706. [Google Scholar]
- 12.Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holeman LA, Robinson SL, Szostak JW, Wilson C. Fold. Des. 1998;3:423–431. doi: 10.1016/S1359-0278(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Wilson C, Szostak JW. Chem. Biol. 1998;5:609–617. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 15.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 16.Montange RK, Batey RT. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 17.Grundy FJ, Lehman SC, Henkin TM. Proc. Natl. Acad. Sci. USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 19.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Chem. Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 20.Borovok I, Gorovitz B, Schreiber R, Aharonowitz Y, Cohen G. J. Bacteriol. 2006;188:2512–2520. doi: 10.1128/JB.188.7.2512-2520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 23.Roth A, Winkler WC, Regulski EE, Lee BWK, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, Iwata-Reuyl D, Breaker RR. Nat. Struct. Mol. Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 24.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 25.Mandal M, Breaker RR. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 26.Cromie MJ, Shi YX, Latifi T, Groisman EA. Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 27.Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. Biol. Chem. 2005;386:1279–1286. doi: 10.1515/BC.2005.145. [DOI] [PubMed] [Google Scholar]
- 28.Joyce GF, Orgel LE. In: The RNA World. Gestland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Press; Cold Spring Harbor: 1999. pp. 49–77. [Google Scholar]
- 29.Forster AC, Symons RH. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 30.Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J. J. Virol. 1988;62:4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MM. Proc. Natl. Acad. Sci. USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 33.Saville BJ, Collins RA. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 34.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 35.Woese CR, Gutell R, Gupta R, Noller HF. Microbiol. Rev. 1983;47:621. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox GW, Woese CR. Nature. 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- 37.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 38.Forster AC, Symons RH. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 39.Peattie DA, Gilbert W. Proc. Natl. Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayford M, Weisblum B. EMBO J. 1989;8:4307–4314. doi: 10.1002/j.1460-2075.1989.tb08617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzer M, Wagner R. J. Mol. Biol. 1998;276:547–557. doi: 10.1006/jmbi.1997.1556. [DOI] [PubMed] [Google Scholar]
- 42.Lindell M, Romby P, Wagner EGH. RNA. 2002;8:534–541. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krol A, Carbon P. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 44.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 45.Brunel C, Romby P. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- 46.Jiang F, Kumar RA, Jones RA, Patel DJ. Nature. 1996;382:183–186. doi: 10.1038/382183a0. [DOI] [PubMed] [Google Scholar]
- 47.Dieckmann T, Suzuki E, Nakamura GK, Feigon J. RNA. 1996;2:628–640. [PMC free article] [PubMed] [Google Scholar]
- 48.Yang YS, Kochoyan M, Burgstaller P, Westhof E, Famulok M. Science. 1996;272:1343–1347. doi: 10.1126/science.272.5266.1343. [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Nat. Struct. Mol. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 50.Fan P, Suri AK, Fiala R, Live D, Patel DJ. J. Mol. Biol. 1996;258:480–500. doi: 10.1006/jmbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 51.Carothers JM, Davis JH, Chou JJ, Szostak JW. RNA. 2006;12:567–579. doi: 10.1261/rna.2251306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferre-D'Amare AR, Zhou K, Doudna JA. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 53.Fedor MJ. Annu. Rev. Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 54.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 55.Zuker M, Stiegler P. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shapiro BA, Zhang KZ. Comput. Appl. Biosci. 1990;6:309–318. doi: 10.1093/bioinformatics/6.4.309. [DOI] [PubMed] [Google Scholar]
- 57.Veksler-Lublinsky I, Ziv-Ukelson M, Barash D, Kedem K. J. Comput. Biol. 2007;14:908–926. doi: 10.1089/cmb.2007.0061. [DOI] [PubMed] [Google Scholar]
- 58.Eddy SR, Durbin R. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gautheret D, Lambert A. J. Mol. Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 60.Saurin W, Marliere P. Comput. Appl. Biosci. 1987;3:115–120. doi: 10.1093/bioinformatics/3.2.115. [DOI] [PubMed] [Google Scholar]
- 61.Gautheret D, Major F, Cedergren R. Comput. Appl. Biosci. 1990;6:325–331. doi: 10.1093/bioinformatics/6.4.325. [DOI] [PubMed] [Google Scholar]
- 62.Laferriere A, Gautheret D, Cedergren R. Comput. Appl. Biosci. 1994;10:211–212. doi: 10.1093/bioinformatics/10.2.211. [DOI] [PubMed] [Google Scholar]
- 63.Steinberg S, Gautheret D, Cedergren R. J. Mol. Biol. 1994;236:982–989. doi: 10.1016/0022-2836(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 64.Ferbeyre G, Smith JM, Cedergren R. Mol. Cell. Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferbeyre G, Bourdeau V, Pageau M, Miramontes P, Cedergren R. Genome Res. 2000;10:1011–1019. doi: 10.1101/gr.10.7.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Billoud B, Kontic M, Viari A. Nucleic Acids Res. 1996;24:1395–1403. doi: 10.1093/nar/24.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang SJ, Haas B, Eskin E, Bafna V. IEEE–ACM Trans. Comput. Biol. Bioinf. 2005;2:366–379. doi: 10.1109/TCBB.2005.57. [DOI] [PubMed] [Google Scholar]
- 69.Dsouza M, Larsen N, Overbeek R. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 70.Przybilski R, Graf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eddy SR. 2001 Available from: < ftp://selab.janelia.org/pub/software/rnabob/>.
- 72.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gautheret D, Legendre M. In: Handbook of RNA Biochemistry. Hartmann RK, Bindereif A, Schön A, Westhof E, editors. Wiley-VCH; Weinheim: 2009. pp. 577–594. [Google Scholar]
- 74.Wilson DS, Szostak JW. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 75.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 76.Epstein LM, Gall JG. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 77.Fedor MJ. Annu. Rev. Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 78.Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, Symons RH. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amiri KMA, Hagerman PJ. Biochemistry. 1994;33:13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- 80.Ruffner DE, Stormo GD, Uhlenbeck OC. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 81.Salehi-Ashtiani K, Szostak JW. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- 82.Martick M, Horan LH, Noller HF, Scott WG. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheldon CC, Symons RH. Nucleic Acids Res. 1989;17:5679–5685. doi: 10.1093/nar/17.14.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferre-D'Amare AR, Zhou K, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 85.Wadkins TS, Perrotta AT, Ferre-D'Amare AR, Doudna JA, Been MD. RNA. 1999;5:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 87.Das SR, Piccirilli JA. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 88.Legiewicz M, Wichlacz A, Brzezicha B, Ciesiolka J. Nucleic Acids Res. 2006;34:1270–1280. doi: 10.1093/nar/gkl018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nehdi A, Perreault JP. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Webb C-HT, Riccitelli NJ, Ruminski DJ, Luptak A. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Been MD, Perrotta AT, Rosenstein SP. Biochemistry. 1992;31:11843–11852. doi: 10.1021/bi00162a024. [DOI] [PubMed] [Google Scholar]
- 92.Been MD, Perrotta AT. RNA. 1995;1:1061–1070. [PMC free article] [PubMed] [Google Scholar]
- 93.Thill G, Vasseur M, Tanner NK. Biochemistry. 1993;32:4254–4262. doi: 10.1021/bi00067a013. [DOI] [PubMed] [Google Scholar]