Abstract
Local β-actin synthesis in growth cones of developing axons plays an important role in growth cone steering; however, the mRNA binding proteins required for this process are unknown. Here we used Zbp1/Imp1−/− mice to test the hypothesis that zipcode binding protein 1 (ZBP1) is required for the regulation of β-actin mRNA transport and local translation underlying growth cone guidance. To address the biological function of ZBP1, we developed a novel in vitro turning assay with primary cortical neuron balls having axons >1 mm in length and demonstrate that growth cones of mammalian neurons exhibit protein synthesis-dependent attraction to either netrin-1 or brain-derived neurotrophic factor (BDNF). Interestingly, this attraction is lost in Zbp1-deficient neurons. Furthermore, BDNF-stimulated β-actin mRNA localization was attenuated in Zbp1-deficient neurons, which impaired enrichment of β-actin protein in the growth cone. Finally, using a photoconvertible translation reporter, we found that ZBP1 is necessary for netrin-1 stimulated local translation of β-actin mRNA in axonal growth cones. Together, these results suggest that netrin-1- and BDNF-induced growth cone attraction required ZBP1-mediated local translation of β-actin mRNA, and therefore ZBP1 regulates protein synthesis-dependent axon guidance. Thus, mRNA binding proteins regulating local translation can control spatiotemporal protein expression in response to guidance cues and directional cell motility.
Introduction
During the development of the nervous system, growth cones respond to molecular cues they encounter in their environment with a repertoire of behaviors, which facilitates pathfinding over extreme distances (Huber et al., 2003). Because axonal growth cones contain mRNAs, polyribosomes, and translational factors, cue-induced local translation is one way to directly regulate growth cone steering and axon guidance (Lin and Holt, 2007). Previous studies have demonstrated that protein synthesis is required for the well established in vitro growth cone guidance assay using Xenopus neurons (Campbell and Holt, 2001; Ming et al., 2002). In mammalian axons, the netrin-1 receptor, DCC, has been shown to colocalize with translational machinery (Tcherkezian et al., 2010). While it is becoming increasing clear that axonal mRNA translation is necessary for some forms of axon guidance (Lin and Holt, 2008), the molecular mechanisms involved remain uncharacterized. We have information on the cis-acting elements involved; however, the trans-acting factors, mRNA binding proteins, which bind to mRNAs and are required for their transport and/or local translation, have barely been examined.
Localization of β-actin mRNA is an excellent model to investigate the molecular mechanism of cue-induced growth cone guidance. For example, both antisense oligonucleotides to the 3′ untranslated (UTR) zipcode sequence and morpholinos to block new synthesis of β-actin abolished growth cone attraction in Xenopus neurons (Leung et al., 2006; Yao et al., 2006). Zipcode binding protein 1 (ZBP1) can bind this specific zipcode sequence located within the 3′UTR of β-actin mRNA (Ross et al., 1997); however, the requirement for ZBP1 in local β-actin synthesis has not been examined in primary neurons using knock-out or knockdown approaches. ZBP1 belongs to a family of highly conserved RNA binding proteins, and there are three paralogs in vertebrates: IMP1/ZBP1, IMP2, and IMP3/VgRBP (Yisraeli, 2005). Both ZBP1 and VgRBP localize to growth cones and associate with β-actin mRNA in neurons (Zhang et al., 2001; Leung et al., 2006; Yao et al., 2006). However, there are other predominantly nuclear proteins that bind β-actin mRNA and may also play a role in the cytoplasmic regulation of β-actin mRNA localization and/or translation (Gu et al., 2002; Glinka et al., 2010). These studies suggest that there may be some redundancy and/or cooperative roles played by these trans-acting factors, although the requirement of any has not been studied.
We do not yet fully understand what effect a genetic null mutation for an RNA binding protein may have on the transport or local translation of specific mRNAs necessary for protein synthesis-dependent growth cone guidance. Here we examine the genetic requirement for ZBP1 in the transport and translation of β-actin mRNA within the axon, using Zbp1/Imp1 knock-out mice, and determine the functional relevance of this mechanism for growth cone steering in response to axon guidance factors. Thus, this study provides insight into the role of RNA binding proteins, and demonstrates that they may play a role in growth cone steering via their regulation of local translation in axons.
Materials and Methods
Animals.
Timed pregnant Sprague Dawley rats were obtained from Charles River Laboratories. ZBP1 mice, with a gene trap insertion in the Zbp1 (Imp1) gene, were obtained from the Mutant Mouse Regional Resource Center. Because Zbp1−/− mice rarely survive beyond birth, Zbp1+/− mice were crossed to obtain embryos of various genotypes and of either sex. The day of the plug was counted as embryonic day 0 (E0). All experimental procedures were approved by Emory University's Institutional Animal Care and Use Committee and were in accordance with the federal Animal Welfare Act PL 89-544 (1966) and subsequent amendments and the documents entitled “Guide for the Care and Use of Laboratory Animals” and “Public Health Service Policy on Humane Care and Use of Laboratory Animals.”
Cell culture and transfection.
Cortical neuron cultures were prepared from E18 rats and E17 mice, as previously described (Kaech and Banker, 2006; Sasaki et al., 2010). Specifically, rat cortical neurons were dissected from Sprague Dawley E18 embryos of either sex. Mouse cortical neurons were obtained from E17 embryos of either sex. Cortices were trypsinized (Invitrogen), rinsed in HBSS (Cellgro), and dissociated in MEM (Cellgro) with FBS (Sigma). Cells were counted and plated on coverslips, Bioptechs Delta T open chamber dishes, or 35 mm MatTek glass bottom dishes at appropriate densities in MEM with FBS. Two to three hours following plating, the media was changed to Neurobasal (Invitrogen) with NS21 and GlutaMax (Invitrogen). NS21 was made according to the previously described protocol (Chen et al., 2008). If needed, neurons were transfected with the rat Amaxa nucleofection kit (Lonza). For Amaxa nucleofection, 5,000,000 neurons were rinsed in HBSS and then transfected using Amaxa's rat neuron nucleofection kit and associated program (O-003). Three micrograms of plasmid (mCherry-ZBP1-wt or mCherry-ZBP1-Y396F) were used for each transfection. Following nucleofection, neurons were recovered in RPMI with 10% horse serum, and then plated in RPMI (Invitrogen) with NS21 and GlutaMax. The following day, the media was changed to Neurobasal with NS21 and GlutaMax.
Neuron balls were created by diluting dissociated neurons at 15,000 neurons per 20 μl drop in Neurobasal (with GlutaMax, NS21, and 10% horse serum), as previously described (Sasaki et al., 2010). Specifically, 20 μl drops were plated on the lids of 100 mm dishes filled with 7 ml of water. Neuron balls formed within 2–3 d in the hanging drop cultures, and were then transferred to either Bioptechs open chamber Delta T or MatTek dishes containing Neurobasal (with GlutaMax and NS21) and 2% horse serum. The following day, the media was changed to Neurobasal with 3 μm cytosine β-d-arabinofuranoside hydrochloride (Sigma). Neurons extended long axons, usually ≥1 mm in length.
Axon outgrowth and filopodial dynamics.
Axon outgrowth and filopodial dynamics were imaged on a Nikon TE-2000 microscope with a Cascade 512B EMCCD camera (Photometrics). Dissociated neurons were used for these two experiments at 3 d in vitro (DIV). Thirty minutes before imaging, the media was changed to Neurobasal without phenol red (Invitrogen), and supplemented with NS21 and GlutaMax as described above. Neurons were kept at 37°C during experimentation through the use of a Bioptechs FCS2 closed chamber system. Axon outgrowth was imaged every 10 min for 60 min, using a 20× objective (Nikon Plan Fluor, NA 0.50) in combination with the 1.5 multiplier built into the microscope (total magnification of 30×). Filopodial dynamics was imaged every 5 min for 30 min, using a 60× objective (Nikon Plan Apo, NA 1.40) in combination with the 1.5 multiplier built into the microscope (total magnification of 90×). Phase images were captured using NIS-Elements software (Nikon) and analyzed using ImageJ (NIH). Axon outgrowth was analyzed by measuring the distance between the center of the growth cone at t = 0 min to t = 60 min. Only axons that grew >5 μm over the course of the experiment were included in the analysis. Total axon and dendrite length was measured using the NeuronJ plugin for ImageJ (Meijering et al., 2004).
Filopodial length was measured by measuring the length of each filopodium on the growth cone from the central domain out to the tip, and the lengths of all the filopodia on one growth cone were averaged. Filopodial number was calculated by counting the number of filopodia on the growth cone at each time point. Filopodial data were tested for statistical significance using a repeated-measures ANOVA. However, filopodia are dynamic structures that can move in and out of the image plane, which can result in missing measurements for certain time points. If a single time point measurement is missing in a case, a repeated-measures ANOVA discards the entire case. Therefore, the full dataset could not be included in the statistical test. The graphs and the statistical tests represent only that data for which a growth cone has a measurement at every time point (0, 5, 10, 15, 20, 25, and 30 min). However, the mean is comparable between the two datasets (dataset 1: only growth cones that have a measurement at every time point; dataset 2: all growth cones). For example, in Figure 1E, the mean of ZBP1+/+ in dataset 1 is 6.51 and in dataset 2 is 6.13. The same holds true for ZBP1−/− in the same figure, as the mean of dataset 1 is 4.96 and the dataset 2 is 5.04. Therefore, although cases were removed to perform statistical analysis of the filopodial dynamics, this did not meaningfully change the data in any way.
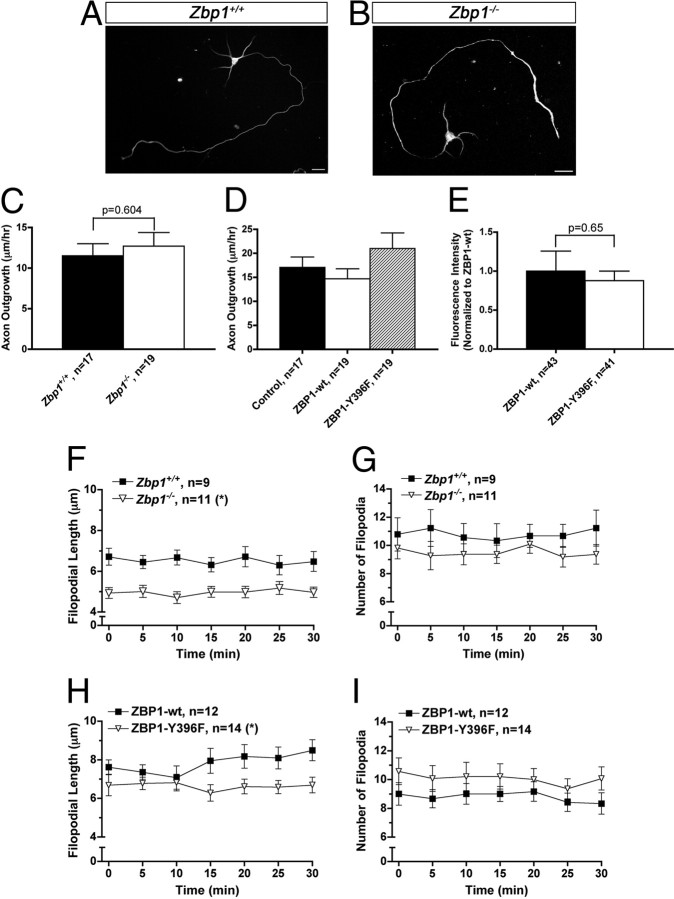
Figure 1.
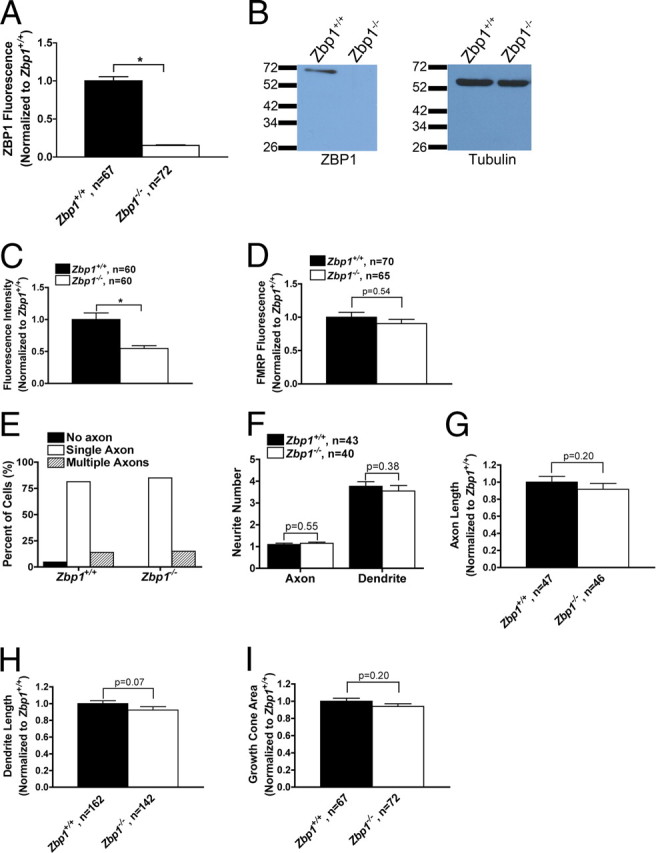
Zbp1-deficient embryos have reduced levels of ZBP1, but extend axons and dendrites normally. A, QIF with a specific ZBP1 antibody demonstrates only background expression of ZBP1 in the soma of neurons cultured from Zbp1-deficient embryos. *p ≤ 0.05, Mann–Whitney. B, No visible protein levels of ZBP1 are seen in the cortical lysate from Zbp1−/− embryos (ZBP1, 70 kDa). β-Tubulin demonstrates that equal amounts of lysate were loaded in each lane (β-tubulin, 55 kDa). C, QIF with a specific ZBP1 antibody demonstrates significantly reduced levels of ZBP1 in the growth cones of Zbp1−/− embryos. *p ≤ 0.05, Mann–Whitney. D, QIF with an anti-FMRP antibody demonstrates that it is not changed in growth cones of Zbp1−/− embryos. Mann–Whitney. E–H, Neurons cultured from Zbp1-deficient mice have numbers of axons (E), proportions of axons and dendrites (F), and total axon (G) and dendrite (H) length similar to those of their wild-type counterparts from the same pregnant mother. Mann–Whitney. I, Growth cone area is not significantly different between Zbp1−/− mice and their wild-type counterparts. Student's t test.
In vitro turning assays.
Axons extending out from neuron balls were exposed to either netrin-1 (5 μg/ml; R&D Systems) or BDNF (10 μg/ml; PeproTech) applied in a pulsatile fashion from a micropipette (1 μm opening). Micropipettes were pulled on a Narishige PC-10 pipette puller. Micropipettes were placed 80–100 μm away and at a 45° angle to the direction of the migrating growth cone and the appropriate solution was applied using a Picospritzer II (General Valve Corporation) in combination with a square pulse stimulator (SD9; Grass Technologies), using the following parameters: 3 psi, 2 Hz, 20 ms pulse duration. Experiments were imaged using the same microscope and parameters as described above, except that neuron balls were kept at 37°C through the use of a Bioptechs Delta T open chamber system, and images were acquired every 5 min for 30 min using a 20× objective (Nikon Plan Fluor, NA 0.50) in combination with the 1.5 multiplier built into the microscope (total magnification of 30×). Only growth cones that grew >10 μm during the 30 min experiment were included in the analysis. ImageJ was used to measure the turning angle. The turning angle was calculated by measuring the angle between the growth cone's initial direction at 0 min and its final position at 30 min. When needed, anisomycin (40 μm; Sigma) or vehicle control (DMSO; Sigma) was added to the media 20 min before the start of the experiment.
Quantitative fluorescent in situ hybridization and immunofluorescence.
For the quantitative fluorescent in situ hybridization (Q-FISH) experiments, neurons were starved in Neurobasal without NS21 for 3 h before the start of the experiment. Following starvation, neurons were stimulated with BDNF or vehicle control for 15 min and then fixed. One digoxigenin-labeled oligonucleotide probe was used to detect mouse β-actin mRNA. The sequence of this probe was 5′-TGTTCAATGGGGTACTTCAGGGTCAGGATACCTCTCTTGCTCTGGGCCTCGTC-3′. Q-FISH was performed as previously described (Sasaki et al., 2010). Specifically, cells were fixed in 4% paraformaldehyde, rinsed three times in PBS/MgCl2, and equilibrated in 1× SSC. They were then briefly incubated in 40% formamide/1× SSC and prehybridized with hybridization buffer alone (10% dextran sulfate, 4 mg/ml bovine serum albumin, 2× SSC, 20 mm ribonucleoside vanadyl complex, 1× PBS) for 1.5 h at 37°C. The coverslips were then incubated overnight with DIG-labeled oligonucleotide probe in hybridization buffer overnight at 37°C. Cells were rinsed with 40% formamide/1× SSC 2 times (20 min each) at 37°C, and then rinsed with 1× SSC alone 5 times (2 rinses for 15 min each and 3 rinses for 5 min each) at room temperature. β-Actin mRNA was then visualized using a Cy3-labeled mouse anti-DIG primary antibody (Jackson ImmunoResearch; 1:1000). As controls, DIG-labeled scrambled oligonucleotide probe and DIG-labeled GFP oligonucleotide probe were used. These controls did not show specific staining. Q-FISH images were deconvolved using AutoQuant (Media Cybernetics).
For the quantitative immunofluorescence (QIF) experiments, neurons were fixed in 4% paraformaldehyde, rinsed 3 times in PBS with MgCl2, permeabilized in 0.3% Triton X-100, and then incubated in blocking buffer for 1 h. Cells were exposed to primary antibody for 1 h at room temperature, which was followed by multiple washes. Cells were then exposed to the secondary antibody for 30 min at room temperature, followed by multiple washes, and then mounted. The following primary antibodies were used: guinea pig anti-ZBP1 (1:300; Sasaki et al., 2010), mouse anti-FMRP (1C3, Millipore; 1:200), mouse anti-tau (Millipore; 1:1000), rabbit anti-MAP2 (Millipore; 1:500), mouse anti-tubulin (E7, Developmental Studies Hybridoma Bank; 1:1000), rabbit anti-tubulin (Sigma; 1:1000), and mouse anti-β actin (AC15, Abcam; 1:1500).
Q-FISH and QIF experiments were imaged on a Nikon Eclipse Ti microscope, in combination with a Nikon Intensilight excitation source and Photometrics Coolsnap HQ2 camera. Images were acquired with NIS-Elements software (Nikon) using a 60× objective (Nikon Plan Apo, NA 1.40) with the 1.5 multiplier built into the microscope (total magnification of 90×). To quantify fluorescent intensity, all experimental and image acquisition parameters were kept constant throughout each experiment. Using ImageJ, the fluorescent intensity was measured as the total fluorescent intensity in the growth cone and divided by the area to normalize for differences in growth cone size. This involved selecting a region of interest that encompassed the entire perimeter of the growth cone, including the central domain, lamellipodia, and filopodia.
For the β-actin QIF experiments, neurons were starved in Neurobasal without NS21 for 3 h before the start of the experiment. Following starvation, neurons were stimulated with BDNF (100 ng/ml) or vehicle control for 30 min and then fixed and processed for IF as described above. In this experiment, the fluorescent intensity in the growth cone was measured in two separate regions of interest. Using DIC images, the peripheral region was defined as the region encompassing the lamellipodia and filopodia and the central region was defined as the thickened center region of the growth cone. Hot spots or enriched regions were defined as a region of increased fluorescence intensity that was 1.5 times the fluorescence of an adjacent, similarly sized region. In all experiments, the mean fluorescent intensity was background subtracted by measuring the mean fluorescent intensity in a region near, but not encompassing, the growth cone.
Granule tracking imaging and analysis.
Cells were transfected using Amaxa Nucleofector technology (see above) and allowed to express for 2–3 d before imaging. Images were acquired using NIS-Elements at a rate of either 2 or 4 fps on a Nikon A1R confocal microscope using the 561 nm laser and a 60× objective (Nikon Apo TIRF, NA 1.49). Neurons were kept at 37°C throughout the experiment via the use of an environmental chamber (In Vivo Scientific). Thirty minutes before imaging, the media was changed to Neurobasal without phenol red (Invitrogen), and supplemented with NS21 and GlutaMax as described above. Only actively moving granules in the distal axon and growth cone were included in the analysis. Analysis was performed using the spot tracking feature in the software package Imaris (Bitplane).
Lentivirus production and Dendra2 translation reporter.
Lentivirus constructs used were based on pFUGW (Lois et al., 2002) and lentivirus was produced by the Viral Vector Core at Emory University. The sequence encoding eGFP was removed, and two repeats of the GAP-43 palmitoylation sequence, dendra2, and the mouse β-actin 3′UTR were inserted into the pFUGW vector to create pFUGW-Palx2-Dendra2 and pFUGW-Palx2-Dendra2-β-actin 3′UTR. Viral stocks of pFUGW-PalX2-Dendra2 and pFUGW-PalX2-Dendra2-β-actin 3′UTR were added to dissociated neurons immediately before neuron balls were created. After neuron balls formed, they were plated onto 35 mm MatTek glass bottom dishes. Neuron balls were used for experiments 2–3 d following plating, when neurons had extended long axons. Neurons to be used for experimentation were starved for 3 h in Neurobasal without NS21 and phenol red. Neurons were then exposed either to netrin-1 (500 ng/ml; R&D Systems) or vehicle control. For some experiments, neurons were pretreated with anisomycin (40 μm) or vehicle control (DMSO) for 20 min before the start of the experiment. Growth cones and a portion of the distal axon were photoconverted using the 405 laser of the Nikon A1R confocal microscope, treated with netrin-1 (or vehicle control), and then imaged using the 488 and 561 nm laser every 5 min for 60 min. Growth cones were imaged using NIS-Elements software, a 60× objective (Nikon Apo TIRF, NA 1.49) and kept at 37°C throughout the experiment via the use of an environmental chamber (In Vivo Scientific). In order for a growth cone to be included in the analysis, the growth cone could not change in area over the course of the experiment by >50%. Total fluorescent intensity was measured in the entire growth cone at each time point, divided by area, and background subtracted. These intensity measurements were then normalized to the first image acquired following addition of netrin-1 and graphed as ΔF/F0 × 100.
Statistical analysis.
Each experiment was repeated a minimum of three independent times. A variety of statistical tests were performed using SPSS (IBM), depending on whether the data were parametric or nonparametric, and included Student's t tests, one-way ANOVA, repeated-measures ANOVA, Kruskal–Wallis, Mann–Whitney, and Bonferroni tests. The specific statistical test used in each experiment is given in its figure legend. Significance was set as p ≤ 0.05. When significance was adjusted, it is referred to as α. All data are graphed as mean ± SEM.
Results
ZBP1 regulates filopodial dynamics in axonal growth cones, but not axon outgrowth
To examine novel functions of ZBP1, we investigated the requirement for ZBP1 in developing neurons. ZBP1 is expressed in growth cones of developing chick forebrain neurons (Zhang et al., 2001) and rat cortical neurons (Sasaki et al., 2010); however, no study has yet examined the requirement of ZBP1 in developing axons. Previous studies have used acute antisense application to the zipcode sequence of β-actin mRNA, which does not determine the mRNA binding proteins involved because it can reduce the binding of multiple RNA binding proteins and may also perturb other mRNAs that contain a similar zipcode. The requirement of a single RNA binding protein that is known to bind β-actin mRNA has not yet been studied. Thus, in the current experiments we used a genetic model organism, Zbp1/Imp1 knock-out mice (Hansen et al., 2004). Although these mice have a low survival rate and gross abnormalities in tissue development, potential defects in neuronal development have not been examined. We first confirmed that ZBP1 is undetectable in the cortex of Zbp1−/− embryos by quantitative immunofluorescence (QIF) and Western blot (Fig. 1A,B). Cultured cortical neurons from Zbp1−/− embryos also show significantly reduced levels of ZBP1 in the growth cone (Fig. 1C). QIF demonstrated that levels of ZBP1 in knock-out neurons were reduced by 85% in the soma (Fig. 1A), but only 48% in the growth cone (Fig. 1C), which was due to more prominent background staining of the antibody in growth cones. It is important to note that the reduction was significant in both cases, and Western blot (Fig. 1B) unequivocally demonstrates the loss of ZBP1 protein in Zbp1−/− embryos. Importantly, another unrelated RNA binding protein, Fragile X mental retardation protein (FMRP), did not show reduced growth cone localization in Zbp1−/− mice (Fig. 1D).
We first examined whether total neurite outgrowth or the establishment of neuronal polarity and differentiation would be affected by the loss of Zbp1. We analyzed the number of axons and dendrites per neuron, the total axon and dendrite length, and the growth cone area at 4 DIV and found no significant difference between dissociated cortical neurons cultured from Zbp1−/− mice and wild-type embryos from the same pregnant mouse (Fig. 1E–I). Furthermore, cortical neurons cultured from Zbp1−/− mice for 3 DIV were morphologically indistinguishable from their wild-type counterparts at a gross level (Fig. 2A,B). Thus, we next used live cell imaging to determine whether the basal axon outgrowth rate of dissociated cortical neurons would be affected by the loss of Zbp1. The basal axon outgrowth rate over 1 h was not significantly different between these two groups (Fig. 2C). We also examined whether stimulation with BDNF would affect the axon outgrowth rate of these neurons. BDNF stimulation (100 ng/ml) did increase the axon outgrowth rate over basal rates; however, it increased the rate in both wild-type and Zbp1−/− neurons. Interestingly, the axon outgrowth rate over 1 h after BDNF stimulation (100 ng/ml) was not significantly different between wild-type and Zbp1−/− neurons (15.8 ± 1.7 for Zbp1+/+ and 15.1 ± 1.7 for Zbp1−/−; p = 0.65, Mann–Whitney; data not shown). In addition to using Zbp1 knock-out mice, in a separate experiment, we overexpressed a nonphosphorylatable mutant form of ZBP1, termed ZBP1-Y396F, in wild-type rat cortical neurons. This mutant form of ZBP1 cannot be phosphorylated by Src kinase at the tyrosine 396 residue and therefore has a strong repressive effect on β-actin mRNA translation (Hüttelmaier et al., 2005). Under physiological conditions, this mechanism allows ZBP1 to repress β-actin mRNA translation during its transport to the growth cone, where phosphorylation of ZBP1 by Src kinase in the growth cone stimulates local translation (Sasaki et al., 2010). The axon outgrowth rate over 1 h was not significantly different between neurons overexpressing ZBP1-Y396F, ZBP1-wt, and untransfected neurons (Fig. 2D). Together, these results with ZBP1 knock-out or functional perturbation demonstrate that ZBP1 does not regulate axon outgrowth or the polarity and differentiation of developing neurons.
Figure 2.
Perturbation of ZBP1 affects filopodial length, but does not affect axon outgrowth. A, B, Cortical neurons cultured for 3 DIV from E17 Zbp1+/+ mice (A) and stained with β-tubulin are morphologically indistinguishable from Zbp1−/− neurons (B). Scale bar, 20 μm. C, D, Axon outgrowth measured over 1 h is not significantly different between Zbp1−/− neurons and their wild-type counterparts (C) or between untransfected rat cortical neurons and those overexpressing ZBP1-wt or ZBP1–Y396F, a nonphosphorylatable mutant form of ZBP1 (D). Student's t test (C) and one-way ANOVA (D) demonstrate no significant difference between groups. E, Overexpression of either ZBP1–Y396F or ZBP1-wt in rat cortical neurons results in similar expression levels in the growth cone. Mann–Whitney. F–I, Growth cones from Zbp1−/− mice or those overexpressing ZBP1–Y396F have significantly shorter filopodia than their wild-type counterparts (F, H), but filopodial number is not different between the two groups (G, I). *p ≤ 0.05, repeated-measures ANOVA.
Because ZBP1 binds the β-actin mRNA zipcode, and ZBP1 and β-actin mRNA are localized to growth cones and filopodia (Zheng et al., 2001; Leung et al., 2006; Yao et al., 2006), we examined a possible function of ZBP1 in regulating filopodial dynamics. Filopodia, composed of bundled actin, are dynamic structures that continuously extend and retract and act as the sensing unit of the growth cone (Kater and Rehder, 1995). Using live cell imaging, the length of filopodia and the number of filopodia per growth cone were examined over 30 min. Here, we find that cortical neurons cultured from Zbp1−/− mice or those overexpressing ZBP1-Y396F had significantly shorter filopodia than those from their wild-type counterparts (Fig. 2F,H). Overexpression of ZBP1-Y396F did not appear to have as dramatic of an effect on filopodial length as complete loss of Zbp1, especially for the first 10 min of the time course. However, it is important to note that filopodia are dynamic structures that rapidly change over time and the data were nonetheless statistically significant (Fig. 2H). The number of filopodia per growth cone was not affected by these perturbations (Fig. 2G,I). Importantly, we ensured that any differences between neurons overexpressing ZBP1-Y396F and ZBP1-wt were not due to variable expression levels between these two constructs (Fig. 2E). Together, these data suggest that ZBP1 regulates filopodial dynamics, perhaps through its local regulation of β-actin mRNA. Because filopodia are essential to growth cone pathfinding (Bentley and Toroian-Raymond, 1986; O'Connor et al., 1990; Chien et al., 1993; Zheng et al., 1996), the current data suggest that ZBP1 may regulate axon guidance.
Cortical mammalian neurons demonstrate protein synthesis-dependent growth cone turning, which is lost in Zbp1−/− embryos
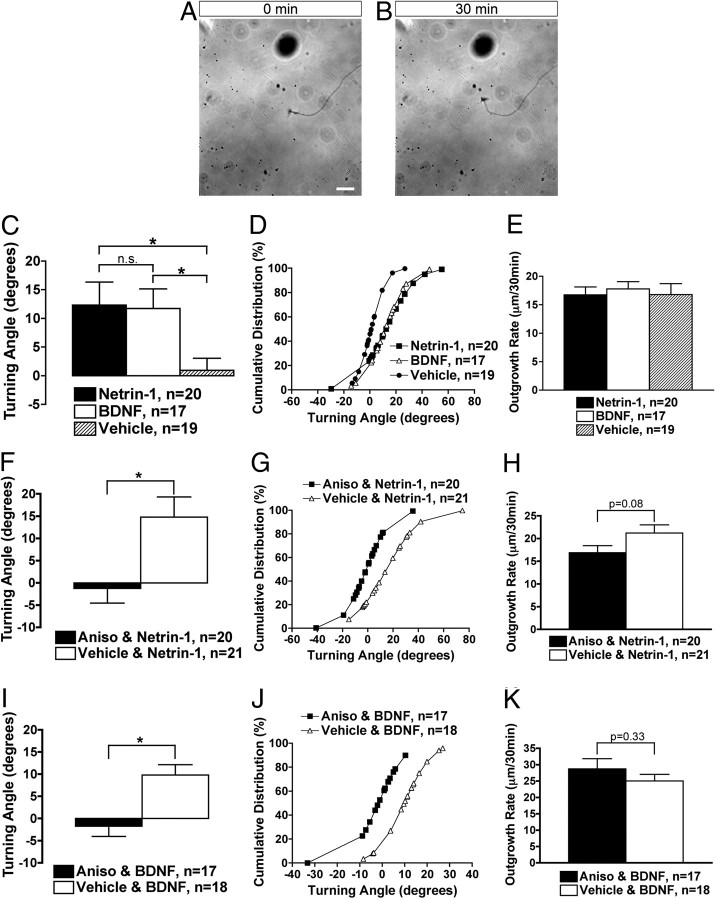
Although in vitro turning assays, which are commonly used as a model for growth cone guidance, have been well established using Xenopus spinal and retinal neurons (Lohof et al., 1992; de la Torre et al., 1997), mammalian neurons have infrequently been used in this type of turning assay [rat cerebellar and dorsal root ganglion (DRG) neurons (Xiang et al., 2002; Li et al., 2005; Gasperini et al., 2009; Murray et al., 2009)]. Furthermore, it has yet to be examined whether growth cone turning of mammalian neurons is protein synthesis dependent, as has been demonstrated in Xenopus (Campbell and Holt, 2001). As such, we applied the well established Xenopus turning assay to rat cortical neurons, cultured as aggregated neuron balls and having easily identifiable axons >1 mm in length (see Materials and Methods) (Sasaki et al., 2010). We used two factors, netrin-1 and BDNF, which are commonly used in turning assays and are known to signal synthesis of β-actin protein (Leung et al., 2006; Yao et al., 2006; Sasaki et al., 2010). Netrin-1 is well established as a guidance cue for a variety of neuronal types both in vitro and in vivo, and although BDNF acts as a guidance cue in vitro, its role in vivo has been described primarily as a survival and growth factor (Huber et al., 2003). Importantly, the receptor for each of these factors (DCC and TrkB, respectively) is expressed in embryonic cortical neurons (Cheng and Mattson, 1994; Ren et al., 2004). Following 30 min of repetitive application via a micropipette, rat cortical axonal growth cones showed a significant attractive turning response to either netrin-1 or BDNF (Fig. 3A–D, Movie 1). Growth cones exposed to netrin-1 or BDNF demonstrated mean attractive turning angles of 12.3 ± 4.0 and 11.7 ± 3.4, respectively, as compared to 0.94 ± 2.1 for vehicle control (Fig. 3C). Yet, the axon outgrowth rate over 30 min was not significantly different between these groups (Fig. 3E). We next examined whether these attractive turning responses were protein synthesis dependent, via preincubation with a protein synthesis inhibitor, anisomycin, for 20 min before application of the guidance cue. We found that anisomycin abolished the attractive turning responses to both netrin-1 and BDNF (Fig. 3F,G,I,J, Movie 1), but did not affect the axon outgrowth rate over 30 min (Fig. 3H,K). Previous studies have demonstrated the necessity of protein synthesis for growth cone turning in Xenopus (Campbell and Holt, 2001). Here we show that mammalian neurons also demonstrate protein synthesis-dependent turning in response to an attractive guidance cue.
Figure 3.
Rat cortical neurons demonstrate a protein synthesis-dependent attractive turning response to either netrin-1 or BDNF. A, B, Images of axonal growth cones, cultured from rat cortical neuron balls, before and after 30 min of exposure to a netrin-1 gradient (A, 0 min; B, 30 min). The tip of the micropipette can be seen in the upper left hand corner of the image. Axons subjected to turning assays were >1 mm in length (see Materials and Methods). Scale bar, 20 μm. C, Growth cones exposed to netrin-1 or BDNF exhibit positive turning angles, as compared to vehicle control. *p ≤ 0.017, Kruskal–Wallis and Mann–Whitney post hoc with Bonferroni correction (α = 0.017). D, Cumulative distribution plot of turning angles representing the individual growth cones in C. E, The outgrowth rate over the time course of the experiment was not significantly different between the groups. One-way ANOVA. F, I, Preincubation with a protein synthesis inhibitor, anisomycin (Aniso, 40 μm), abolishes the attractive turning response of growth cones to netrin-1 (F) and BDNF (I). *p ≤ 0.05, Mann–Whitney. G, J, Cumulative distribution plot of turning angles representing the individual growth cones in F and I. H, K, The outgrowth rate over the time course of the experiment was not significantly different between the groups. Student's t test.
Movie 1.
Examples of turning assays. A, Rat cortical neurons show an attractive turning response to netrin-1 (5 μg/ml). Images were acquired by time-lapse phase microscopy. Frames were taken every 50 s for 20 min. B, Rat cortical neurons exhibit protein synthesis-dependent attractive turning to netrin-1. Preincubation of rat cortical neurons with anisomycin (40 μm) prior to application of netrin-1 (5 μg/ml) results in inhibition of the attractive turning response. Images were acquired by time-lapse phase microscopy. Frames were taken every 5 min for 30 min. C, Mouse cortical growth cones from Zbp1+/+ embryos exhibit attractive turning in response to netrin-1 (5 μg/ml). Images were acquired by time-lapse phase microscopy. Frames were taken every 5 min for 30 min. D, Mouse cortical growth cones from Zbp1-deficient embryos lose their attractive turning response to netrin-1 (5 μg/ml). Images were acquired by time-lapse phase microscopy. Frames were taken every 5 min for 30 min.
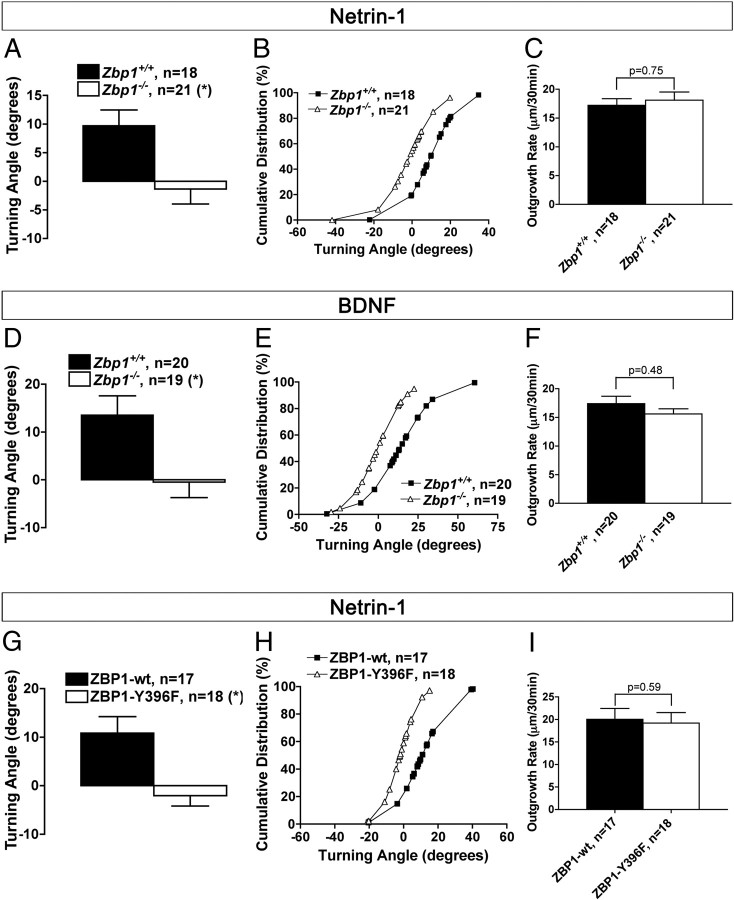
Our next step was to investigate the role of the RNA binding protein, ZBP1, in the process of axon guidance in culture. Interestingly, we found that neurons cultured from Zbp1−/− embryos do not show the attractive turning response to both netrin-1 (Fig. 4A,B, Movie 1) and BDNF (Fig. 4D,E). Outgrowth rate of axons, however, was not affected (Fig. 4C,F). We found a similar result when we overexpressed the nonphosphorylatable mutant form of ZBP1, ZBP1-Y396F, in rat cortical neurons. Growth cones overexpressing ZBP1-Y396F do not respond to netrin-1, as compared to those overexpressing ZBP1-wt, but outgrowth rate was not affected under these conditions (Fig. 4G–I). These results suggest that ZBP1 and the phosphorylation of ZBP1 are essential for guidance cue-induced growth cone steering of mammalian cortical neurons.
Figure 4.
ZBP1 is required for netrin-1 and BDNF-induced attractive turning. A, D, Axonal growth cones from Zbp1−/− neuron balls do not show the attractive turning response to netrin-1 (A) and BDNF (D) exhibited by wild-type neurons. *p ≤ 0.05, Mann–Whitney. B, E, Cumulative distribution plot of turning angles representing the individual growth cones in A and D. C, F, The outgrowth rate over the time course of the experiments was not significantly different between the groups. *p ≤ 0.05, Mann–Whitney. G–I, Overexpression of ZBP1–Y396F in rat cortical neurons results in the loss of netrin-1-induced attractive turning demonstrated by those neurons overexpressing ZBP1-wt (G, H); however, the outgrowth rate is not significantly different between the two groups (I). *p ≤ 0.05, Mann–Whitney.
Anterograde and retrograde traffic of ZBP1 throughout the axon and growth cone
In neurons, mRNA binding proteins transport mRNA in both anterograde and retrograde fashions (Kiebler and Bassell, 2006). Complexes of RNA binding proteins, mRNAs, and associated proteins are termed mRNP granules and are actively transported along microtubules in neuronal processes at the rate of fast axonal transport. We investigated whether ZBP1-containing granules exhibit fast axonal transport specifically within the growth cones of cortical neurons. Although we have previously demonstrated fluorescently labeled ZBP1 granules moving in the neurites of chick neurons (Zhang et al., 2001), movements of ZBP1 in a dynamic fashion into and out of growth cones would suggest a direct role for ZBP1 in the regulation of growth cone motility.
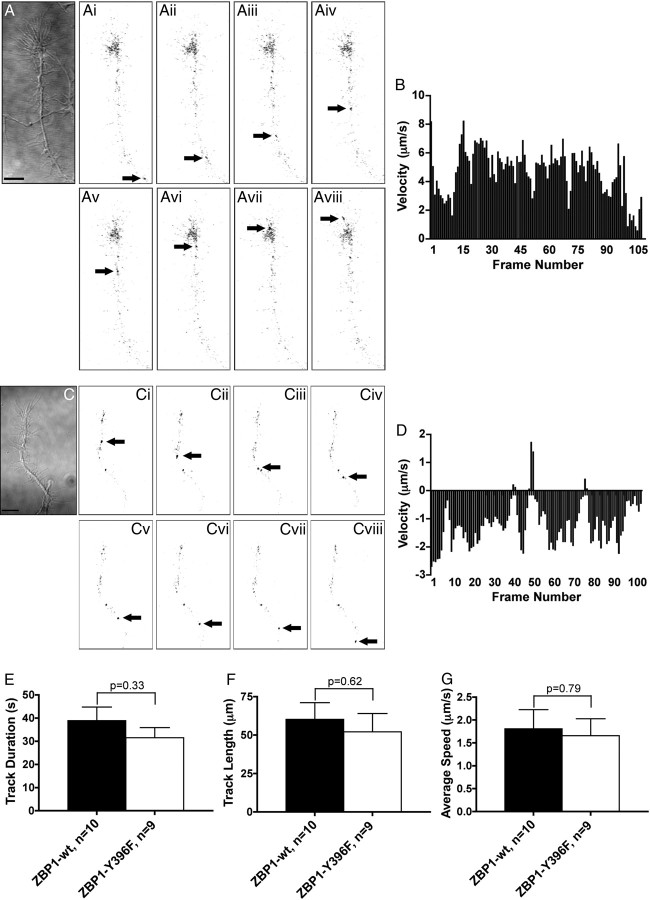
Using live cell imaging, monomeric red fluorescent protein mCherry-tagged ZBP1 exhibited bidirectional movements throughout the axon, as well as within the axonal growth cone. Although many granules were stationary throughout the acquisition period, trafficking of ZBP1-wt granules in growth cones ranged from oscillatory to long trajectories. ZBP1-wt granules exhibited both anterograde (Fig. 5A,B) and retrograde movements (Fig. 5C,D) throughout the distal axon and growth cone (Movie 2). We found that 55% of all ZBP1-wt granules analyzed were moving in an anterograde fashion, whereas 45% were exhibiting retrograde movements. mCherry-ZBP1-Y396F, the nonphosphorylatable mutant form of ZBP1, also exhibited anterograde and retrograde trafficking throughout the distal axon and growth cone (Movie 2). Both ZBP1-wt and ZBP1-Y396F exhibited long track durations and length, and the average speed (1.8 ± 0.4 μm/s for ZBP1-wt and 1.7 ± 0.4 μm/s for ZBP1-Y396F) was slightly faster than the rate of ZBP1 transport previously reported in chick forebrain neurites (Fig. 5E–G) (Zhang et al., 2001). Here we demonstrate that ZBP1-wt traffics dynamically in and out of the distal axon and growth cone, thus suggesting a role for ZBP1 in β-actin mRNA regulation underlying growth cone steering.
Figure 5.
mCherry-ZBP1-wt exhibits dynamic anterograde and retrograde movements within the axonal growth cone and distal axon. A–Aviii, mCherry-ZBP1-wt moves in an anterograde fashion into an axonal growth cone. DIC image shown in A and inverted fluorescent images shown in Ai–Aviii. The arrow marks the granule in each image. The images in Ai–Aviii are 2.5 s apart. Scale bar, 10 μm. B, Instantaneous velocity histogram of granule depicted in A (positive values are anterograde and negative values are retrograde). Although only part of the granule track is shown in A, the entire track is represented in the histogram in B. C–Cviii, mCherry-ZBP1-wt moves in a retrograde fashion out of a growth cone and into the distal axon. DIC image shown in C and inverted fluorescent images shown in Ci–Cviii. The arrow marks the granule in each image. The images in Ci–Cviii are 5 s apart. Scale bar, 10 μm. D, Instantaneous velocity histogram of the granule depicted in C (positive values are anterograde and negative values are retrograde). Although only part of the granule track is shown in C, the entire track is represented in the histogram in D. E–G, The track duration, track length, and average speed of ZBP1-wt and ZBP1-Y396F granules are not significantly different from one another. Student's t test.
Movie 2.
Anterograde and retrograde traffic of ZBP1 throughout the axon and growth cone. A, Expression of mCherry-ZBP1-wt (green) in rat cortical neurons demonstrates fast anterograde and retrograde movements of ZBP1-wt into and out of the growth cone. Images were acquired by time-lapse confocal microscopy. Frames were taken every 1 s for 100 s. B, Expression of mCherry-ZBP1-Y396F (green) in rat cortical neurons demonstrates fast anterograde and retrograde movements of ZBP1-Y396F into and out of the growth cone. Images were acquired by time-lapse confocal microscopy. Frames were taken every 0.5 s for 90 s.
Stimulation-induced increases of β-actin mRNA and protein levels in the growth cone are dependent on ZBP1
We have previously demonstrated that neurotrophin stimulation results in an increase in β-actin mRNA and protein localization within the growth cone (Zhang et al., 1999). Although our previous study demonstrated that acute antisense to the 3′UTR of β-actin resulted in the loss of this neurotrophin-induced enrichment of endogenous β-actin mRNA and protein in growth cones (Zhang et al., 2001), this antisense paradigm can affect the binding of multiple mRNA binding proteins. As such, we used the Zbp1 knock-out mouse to determine whether the loss of the mRNA binding protein, ZBP1, results in altered BDNF-induced localization of β-actin mRNA and protein.
In the current experiments, neurons were cultured from Zbp1−/− mice or wild-type embryos from the same pregnant mouse under normal conditions for 3 d. Then, these neurons were starved for 3 h by depriving them of their normal growth factors and supplements. Following starvation, neurons were stimulated with BDNF for 15 min, due to its known role to stimulate mRNA localization (Zhang et al., 1999; Leung et al., 2006; Yao et al., 2006). Following stimulation, neurons were fixed and quantitative fluorescent in situ hybridization (Q-FISH) for β-actin mRNA was performed. Starvation of neurons from wild-type or knock-out mice, followed by vehicle treatment, resulted in very little localization of β-actin mRNA in the growth cone (Fig. 6A,C). Furthermore, vehicle stimulation did not result in a difference in β-actin mRNA fluorescence in the growth cone between Zbp1+/+ and Zbp1−/− neurons (Fig. 6E). However, Zbp1+/+ neurons that were starved and then stimulated with BDNF showed a 66% increase in fluorescent signal for β-actin mRNA over control (Fig. 6B,F). In comparison, growth cones from Zbp1−/− embryos that were starved and stimulated with BDNF demonstrated only a 15% increase in β-actin mRNA fluorescent signal over control (Fig. 6D,F). Importantly, we confirmed that growth cone area was not significantly different between the groups (data not shown). Thus, we found that loss of Zbp1 resulted in a significant reduction of BDNF-stimulated localization of β-actin mRNA to the growth cone (Fig. 6F).
Figure 6.
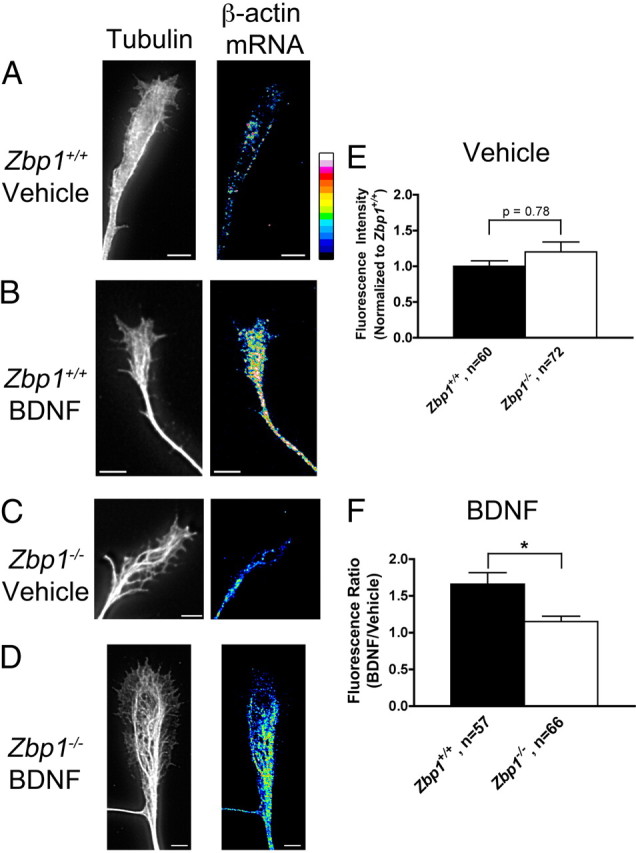
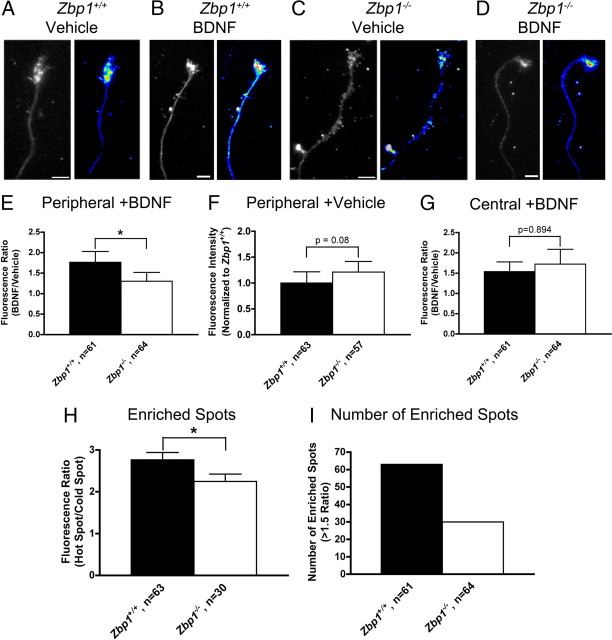
BDNF-induced localization of β-actin mRNA is significantly reduced in Zbp1-deficient mice. A, C, Neurons from Zbp1+/+ (A) and Zbp1−/− (C) embryos were starved for 3 h and then treated with a vehicle control for 15 min. Q-FISH for β-actin mRNA demonstrates only a small amount of staining in the growth cone (tubulin, left panel; β-actin mRNA, right panel). Images of β-actin mRNA are shown in false color for ease of visualization. A false-color intensity map is shown on the far right, with lowest intensities at the bottom of the bar and highest intensities on the top (low intensity pixels are black and blue, and intensities increase from there to yellow, red, and finally white). Scale bar, 5 μm. B, Neurons cultured from Zbp1+/+ embryos were starved for 3 h and then stimulated with BDNF for 15 min, which resulted in a visible increase in β-actin mRNA fluorescent signal in the growth cone (tubulin, left panel; β-actin mRNA, right panel). Scale bar, 5 μm. D, Neurons cultured from Zbp1−/− embryos were starved for 3 h and then stimulated with BDNF for 15 min, which resulted in only a modest increase in β-actin mRNA fluorescent signal in the growth cone (tubulin, left panel; β-actin mRNA, right panel). Scale bar, 5 μm. E, Using Q-FISH, there is no significant difference between Zbp1+/+ and Zbp1−/− neurons that are starved and vehicle treated in terms of the β-actin mRNA fluorescent signal in the growth cone. F, Quantification of β-actin mRNA FISH signal in neurons from Zbp1+/+ or Zbp1−/− embryos subjected to the above treatments. The fluorescent signal of growth cones treated with BDNF was normalized to vehicle-treated growth cones. Treatment of neurons from Zbp1+/+ embryos with BDNF results in significantly increased localization of β-actin mRNA fluorescent signal in the growth cone, as compared to those from Zbp1−/− embryos. *p ≤ 0.05, Mann–Whitney test.
To determine whether the loss of Zbp1 affects the BDNF-induced localization of β-actin protein, neurons were cultured and starved as described above, but then stimulated with BDNF for 30 min and fixed, and QIF for β-actin protein was performed. β-Actin protein staining appeared to be concentrated within the peripheral region of the growth cone and localized in hot spots within that region, in accordance with previous reports (Fig. 7A–D) (Spooner and Holladay, 1981; Forscher and Smith, 1988). As such, fluorescence measurements were not taken over the entire growth cone, but rather divided up into two regions. Fluorescent intensity was measured separately in the peripheral region, which encompassed the lamellipodia and filopodia, and the central region of the growth cone. Furthermore, enriched regions or hot spots of fluorescence were defined as a region of increased fluorescence intensity that was 1.5 times the fluorescence of an adjacent, similarly sized region. Using these criteria, we found that BDNF stimulation resulted in a 77% increase in β-actin protein localization in the peripheral domain of Zbp1+/+ growth cones, as compared to only a 30% increase in Zbp1−/− growth cones (Fig. 7E). Thus, BDNF stimulated β-actin protein localization was significantly increased in Zbp1+/+ growth cones over those deficient in Zbp1. Importantly, we have previously demonstrated that BDNF stimulated increases in β-actin protein in the growth cone are protein synthesis dependent (Sasaki et al., 2010).
Figure 7.
BDNF-induced localization of β-actin protein is significantly reduced in Zbp1-deficient mice. A–D, Neurons cultured from Zbp1+/+ (A, B) and Zbp1−/− (C, D) embryos were starved for 3 h and then stimulated with BDNF (B, D) or vehicle control (A, C) for 30 min. There is a larger increase in β-actin protein fluorescent signal in Zbp1+/+ growth cones (B) than in Zbp1−/− growth cones (D) following BDNF stimulation. β-Actin protein is shown in black and white (left panel) and false color (right panel) for ease of visualization. See Figure 6A legend for explanation of false color scale. Scale bar, 5 μm. E, QIF demonstrates that BDNF-induced localization of β-actin protein is significantly reduced in the peripheral region of growth cones from Zbp1−/− embryos. *p ≤ 0.05, Mann–Whitney test. F, QIF shows that there is no significant difference between Zbp1+/+ and Zbp1−/− neurons that are starved and vehicle treated in terms of the β-actin protein fluorescent signal in the peripheral region of the growth cone. G, QIF demonstrates that BDNF-induced localization of β-actin protein is not affected in the central region of growth cones from Zbp1−/− embryos. *p ≤ 0.05, Mann–Whitney test. H, I, Quantitative image analysis shows that BDNF-induced localization of β-actin protein in enriched or hot spots is significantly reduced in growth cones from Zbp1-deficient mice (H). Additionally, there is a reduction in the number of these spots in Zbp1-deficient mice (I). *p ≤ 0.05, Mann–Whitney test.
Unexpectedly, there was no difference in basal β-actin protein fluorescence in the peripheral region of the growth cone between Zbp1+/+ and Zbp1−/− neurons as analyzed after vehicle treatment (Fig. 7F). Furthermore, BDNF stimulation resulted in a similar increase in β-actin protein localization in the central growth cone region of both Zbp1+/+ and Zbp1−/− mice (Fig. 7G), suggesting that ZBP1 may be necessary to enrich β-actin protein in peripheral microdomains within the growth cone in a stimulus-dependent manner. When we examined enriched or hot spots of β-actin protein after BDNF treatment, the fluorescence intensity was significantly decreased in Zbp1−/− mice as compared to wild-type embryos (Fig. 7H). Finally, there were many more enriched or hot spots of β-actin protein in Zbp1+/+ growth cones, as compared to those deficient for Zbp1 (Fig. 7I). Again in this experiment, we confirmed that growth cone area was not significantly different between the groups (data not shown). Thus, these data suggest that local translation of β-actin in the growth cone is sensitive to stimulation by growth and guidance factors, and regulated by ZBP1 to influence spatiotemporal levels of β-actin within the growth cone. Together, we find that although ZBP1 does not regulate β-actin mRNA and protein localization under control basal conditions, it does play an integral role in the BDNF-stimulated localization of β-actin mRNA and protein into and within the growth cone. Thus, the loss of Zbp1 can have a drastic effect on a growth cone's ability to rapidly respond to growth and/or guidance cues to enrich β-actin protein in a compartmentalized fashion.
ZBP1 is required for netrin-1-stimulated β-actin mRNA translation in the growth cone
The data shown in Figures 6 and 7 suggest that ZBP1 plays an essential role in the stimulation-induced localization and enrichment of β-actin mRNA and protein to the growth cone. However, we wanted to take these experiments a step further to determine whether ZBP1 regulates the local translation of β-actin mRNA within the growth cone, which is implied by our previous experiments (Figs. 6, 7) but not tested directly. No study has yet examined a role for ZBP1 in local β-actin synthesis in primary neurons using knock-out or knockdown approaches. In fact, our knowledge about the requirement of any RNA binding protein in the regulation of local protein synthesis within the developing axon or growth cone is limited. However, we do know that mRNA translation is regulated by the 5′ and 3′UTR of the mRNAs (Wilkie et al., 2003), and for β-actin mRNA, translation can be driven by its 3′UTR (Leung et al., 2006; Sasaki et al., 2010). We created a photoconvertible fluorescent protein translation reporter that is linked to the β-actin 3′UTR (PalX2-Dendra2-β-actin 3′UTR), in a manner similar to a previous report (Leung et al., 2006). Our translation reporter also contained a dual palmitoylation sequence from GAP-43 (PalX2), which can limit diffusion by anchoring to local membranes. We have previously used this palmitoylation sequence in translation reporters using dEGFP, and used FRAP to demonstrate negligible levels of fluorescence recovery in axons or growth cones when the β-actin 3′UTR was lacking, suggesting limited diffusion or transport of somatically synthesized reporter protein (Sasaki et al., 2010). In contrast, recovery of dEGFP bearing the membrane anchor and the β-actin 3′UTR showed robust recovery in growth cones, even after subtraction of the low levels of recovery observed in proximal axons. This suggests that over the time course of our experiment, the palmitoylation sequence will ensure that only locally translated protein will be visualized in growth cones. The photoconvertible protein used, dendra2, which has been used recently as a translation reporter (Wang et al., 2009), shows green fluorescence originally, but can be converted to its red fluorescent form by illumination with ultraviolet light. Following conversion, any new green fluorescence that is visualized is newly synthesized protein.
Neurons from either Zbp1+/+ or Zbp1−/− mouse embryos were transduced with PalX2-Dendra2-β-actin 3′UTR and then cultured as neuron balls. Neuron ball culture (see Materials and Methods) is unique in that the axons can extend >1 mm in length (Sasaki et al., 2010). Because membrane-tagged proteins have been shown to have a diffusion rate of ∼50 μm/h (Fivaz and Meyer, 2003), it is highly unlikely that proteins could be synthesized in the soma and transported down that length of axon over the short time course of the experiment. This stipulation ensures that any new green fluorescence that we visualize within the growth cone is due to local translation at that location. Before experimentation, neurons were starved for 3 h, and then dendra2 was photoconverted. Netrin-1 was applied, due it its known role as a guidance cue and in stimulating β-actin translation (Huber et al., 2003; Leung et al., 2006), and growth cones were monitored for the recovery of green fluorescence for 60 min. Green fluorescence significantly increased in Zbp1+/+ growth cones transduced with the PalX2-Dendra2-β-actin 3′UTR construct following photoconversion (Fig. 8A,C,D). However, green fluorescence did not increase over basal translation levels in Zbp1−/− growth cones transduced with the same construct (Fig. 8B–D). These data suggest that ZBP1 is necessary for netrin-1-induced local translation of β-actin mRNA within the growth cone. Importantly, green fluorescence did not increase over basal levels in either Zbp1+/+ or Zbp1−/− growth cones transduced with the PalX2-Dendra2-β-actin 3′UTR construct and given control vehicle instead of netrin-1. Also, translation of the reporter construct was not significantly different between Zbp1+/+ and Zbp1−/− neurons under these control basal conditions (Fig. 8C,D). These data suggest that ZBP1 is not required for the basal translation of β-actin mRNA within the growth cone, but that stimulation-induced translation is regulated by ZBP1. Green fluorescence also did not increase over basal levels in growth cones transduced with a control construct that did not contain the β-actin 3′UTR (Fig. 8C,D). In fact, the green fluorescence decreased due to photobleaching under these control conditions, which suggests that the netrin-1-induced increase in fluorescence may be slightly underestimated.
Figure 8.
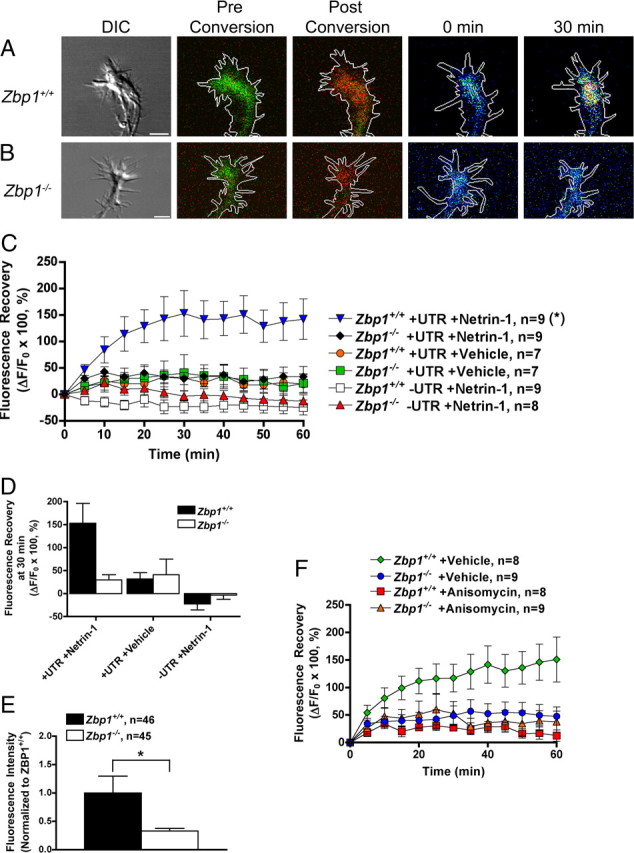
ZBP1 is required for netrin-1 stimulated local translation of a β-actin reporter in growth cones. A, B, Neuron balls from either Zbp1+/+ (A) or Zbp1−/− (B) embryos were transduced with PalX2-Dendra2-β-actin 3′UTR. Growth cones can be seen before conversion in “DIC” (leftmost panel), followed by “Pre Conversion” (overlay of green and red channels, but green fluorescence predominates), and also “Post Conversion” (overlay of green and red channels, but after UV conversion red fluorescence predominates). Following netrin-1 addition, the green fluorescence increases after 30 min in Zbp1+/+ growth cones, but not those from Zbp1−/− embryos (last two images in the series are the green channel only and shown in false color; see Figure 6A legend for explanation of false color scale). C, Application of netrin-1 results in an increase in green fluorescence in Zbp1+/+ growth cones transduced with PalX2-Dendra2-β-actin 3′UTR, as compared to Zbp1−/− growth cones transduced with the same construct. Neurons that received a vehicle control instead of netrin-1 or were transduced with a construct without the β-actin 3′UTR did not show an increase in fluorescence over basal translation rates. Data are represented as ΔF/F0 and then multiplied by 100. *p ≤ 0.01, Repeated-measures ANOVA with Bonferroni post hoc and correction (α = 0.01). D, Data shown in C, but only at the 30 min time point. E, Q-FISH to dendra2 mRNA demonstrates that the average intensity of dendra2 mRNA in Zbp1−/− growth cones was significantly reduced as compared to wild-type neurons. Both neuronal types were transduced with the PalX2-Dendra2-β-actin 3′UTR construct. *p ≤ 0.05, Mann–Whitney. F, Preincubation with a protein synthesis inhibitor, anisomycin, before stimulation with netrin-1 abolishes the increase in β-actin reporter translation. All groups were treated with netrin-1 in this experiment, and “+Vehicle” in the figure legend refers to the vehicle for anisomycin. Data are represented as ΔF/F0 and then multiplied by 100. *p ≤ 0.025, Repeated-measures ANOVA with Bonferroni post hoc and correction (α = 0.025).
To further examine whether the deficit in Zbp1−/− neurons was due to a reduction in mRNA localization or translation or both, we transduced the PalX2-Dendra2-β-actin 3′UTR construct into Zbp1+/+ and Zbp1−/− neurons and then performed quantitative FISH to dendra2 mRNA. We found that the average intensity of dendra2 mRNA in Zbp1−/− growth cones was significantly reduced as compared to wild-type neurons (Fig. 8E). However, we were still able to visualize dendra2 reporter mRNA granules in Zbp1−/− axons and growth cones, and as such, these data suggest that there is most likely a deficit in both 3′UTR-dependent mRNA localization and translation in Zbp1−/− neurons. Finally, preincubation with anisomycin before stimulation with netrin-1 significantly reduced the increase in green fluorescence after photoconversion (Fig. 8F). Together, these data suggest that ZBP1 is required for netrin-1-induced local translation of β-actin mRNA in the growth cone.
Discussion
Posttranscriptional regulation in the axon
It has been shown that the local translation of mRNA within the axon regulates processes such as outgrowth, survival, and regeneration (Zheng et al., 2001; Verma et al., 2005; Cox et al., 2008; Hengst et al., 2009; Vogelaar et al., 2009). Few studies have demonstrated a role of local axonal translation in growth cone steering and collapse in vitro. Netrin-1- and BDNF-induced attractive turning in Xenopus required the asymmetric local synthesis of β-actin (Leung et al., 2006; Yao et al., 2006). Another study demonstrated that local translation of RhoA within the axon is necessary for Sema3A-induced growth cone collapse of rat DRGs (Wu et al., 2005). Protein synthesis is also required for Slit2-induced collapse of Xenopus retinal growth cones (Piper et al., 2006). However, it is important to note that not all cell types have been shown to respond to guidance cues in a protein synthesis-dependent manner. A recent paper suggests that chick neurons do not require protein synthesis to respond to a number of different guidance cues, including Sema3A and nerve growth factor (Roche et al., 2009). Thus, protein synthesis-dependent responses to guidance cues may be cell type specific, concentration dependent, or developmentally regulated. In vivo, one study suggests that a guidance receptor, EphA2, is locally translated in the distal axon during and after midline crossing, which implies a role in axon guidance (Brittis et al., 2002). However, the molecular mechanisms and trans-acting factors involved in the above examples are unclear.
Little is known about whether a defect in axon guidance may result from the loss of an RNA binding protein. One example is Musashi1, which is an RNA binding protein that interacts with Robo3 mRNA either directly or indirectly (Kuwako et al., 2010). This study demonstrated that in Musashi1-deficient mice, axon midline crossing of precerebellar neurons is impaired, and Robo3 protein levels are reduced. However, it is unknown whether Musashi1 or Robo3 mRNA are present in the axon, and as such, whether this process involves local translation is unclear. In Drosophila, loss of the mRNA binding protein, ELAV, results in defects in commissural axon midline crossing (Simionato et al., 2007), and loss of function of the Drosophila homolog of FMRP results in guidance defects (Morales et al., 2002; Michel et al., 2004). However, not all studies conducted thus far have shown that loss of an RNA binding protein results in axon guidance defects. The loss of cytoplasmic polyadenylation element binding protein 1 (CPEB1), either by antisense morpholino in Xenopus or use of the CPEB1 knock-out mouse, does not result in retinal axon pathfinding defects (Lin et al., 2009). Our study reveals that the RNA binding protein, ZBP1, regulates axon guidance in vitro, and its loss results in the local reduction of β-actin mRNA and protein levels. Furthermore, we find that ZBP1 is responsible not only for β-actin localization but also its translational regulation leading to enrichment of β-actin in the growth cone. Continuing work, although outside the scope of the current study, is examining the role of ZBP1 in axon guidance in vivo and will provide important insights into the role of local mRNA translation in the formation of proper neuronal circuitry in the developing nervous system.
ZBP1 regulates the spatiotemporal expression of β-actin under stimulated conditions
Here we demonstrate that although axon outgrowth is not affected in Zbp1-deficient neurons, both filopodial dynamics and axon guidance are impaired. It is likely that Zbp1-deficient neurons are still able to transport β-actin protein to growth cones by protein transport mechanisms and so absence of a locally synthesized pool of β-actin may have only a very modest impact on the growth cone under basal conditions. Indeed, the only noticeable impairment in Zbp1-deficient neurons under basal conditions was a modest reduction in filopodial length. However, the phenotypes become more noticeable when the growth cone must react to a cue; here even a modest defect in filopodial length, coupled with loss of new β-actin synthesis, could lead to loss of axon guidance. Thus, it is likely that local protein synthesis allows the axon and growth cone to respond quickly and in a compartmentalized fashion to stimulation with growth and/or guidance factors.
Our findings of defects in growth cone turning in ZBP1 knock-out neurons, but not axon outgrowth, imply a specific role for ZBP1 to regulate actin dynamics. Axon outgrowth is largely a microtubule-based process, whereas filopodial dynamics is largely actin based (Suter and Forscher, 2000). Because β-actin protein is highly enriched in growth cones, unlike γ-actin (Bassell et al., 1998), this enrichment may be controlled by ZBP1-mediated local β-actin protein synthesis. Deficiency of ZBP1 may have only a modest impact on β-actin at steady state, yet could compromise the ability of a growth cone to enrich β-actin in filopodia and lamellipodia in response to external stimuli. Because ZBP1 does bind other mRNAs in addition to β-actin, such as Arp proteins involved in actin polymerization (Jønson et al., 2007; Gu et al., 2009), it will be interesting to further assess how loss of ZBP1 could affect actin dynamics needed for growth cone steering.
While our data do reveal a role of ZBP1 (IMP1) as a regulator of stimulus-induced changes in β-actin mRNA, further work is needed to assess redundant roles of other ZBP1 paralogs. ZBP1 does not seem to be necessary for steady-state maintenance of β-actin mRNA in the growth cone (Fig. 6E). Additionally, our translation reporter demonstrated that basal levels of β-actin mRNA translation in the growth cone are not affected by the loss of ZBP1 (Fig. 8C,D). Because a previous study demonstrated Vg1RBP granules moving within growth cones and filopodia of Xenopus neurons (Leung et al., 2006), it will be important to assess respective roles for ZBP1 and IMP3/Vg1RBP.
In terms of stimulation paradigms affecting β-actin protein, previous studies have demonstrated that a netrin-1 or BDNF gradient results in an asymmetric distribution of β-actin across the growth cone (Leung et al., 2006; Yao et al., 2006). This is thought to provide a pool of newly synthesized β-actin at the location where it is needed to affect actin dynamics and initiate the turn. In the current study, we found that stimulus-induced β-actin protein expression was not uniformly compromised by loss of ZBP1 in the growth cone (Fig. 7E–I). Together, these data suggest that the function of β-actin mRNA localization by VICKZ/IMP proteins may not be to target β-actin protein, which presumably can get to the growth cone by protein transport mechanisms as well, but rather to enrich β-actin in a spatiotemporal manner needed for cue-induced turning. It may be that there are microdomains of β-actin mRNA translation in the growth cone, as suggested by our immunofluorescence data, which allow for β-actin protein enrichment in microdomains following stimulation. Thus, our data provide a unifying picture as to how slow axonal transport mechanisms may be combined with local translation to perform varying functions as needed during axonal growth and guidance.
Signaling mechanisms underlying regulation of cue-induced axon guidance by ZBP1
The phosphorylation state of ZBP1 is an essential component to understanding how ZBP1 regulates its mRNA cargo. ZBP1 is phosphorylated by Src kinase at its tyrosine 396 residue; this phosphorylation acts as a switch to cause the release and subsequent translation of its β-actin mRNA cargo (Hüttelmaier et al., 2005). A recent study from our laboratory examined the importance of the phosphorylation state of ZBP1, using the same nonphosphorylatable mutant ZBP1 construct used in the present study (Sasaki et al., 2010). However, overexpression of this construct does not demonstrate a physiologic role for ZBP1 because the nonphosphorylatable mutant ZBP1 construct likely sequesters most of the β-actin mRNA in the cell and does not release it. Thus, the current study, which employs a null organism, directly demonstrates a genetic requirement for ZBP1 in axon guidance and local translation of β-actin mRNA. Although we do not currently completely understand the cascade through which netrin-1/DCC or BDNF/TrkB activates Src, which then leads to the phosphorylation of ZBP1, a recent study has shed some light on the subject. In axons, DCC has been shown to colocalize with translational machinery (Tcherkezian et al., 2010).
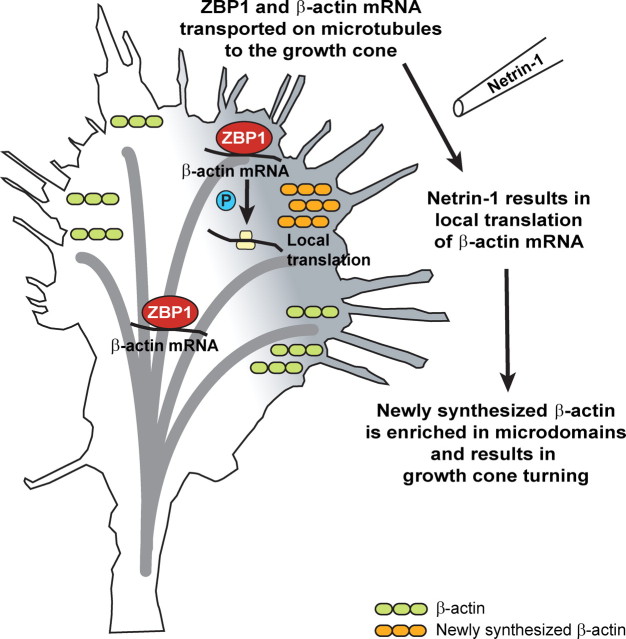
Here, we demonstrate that ZBP1 and the phosphorylation of ZBP1 are required for attractive guidance of cortical neurons, and appropriate filopodial dynamics. Thus, ZBP1 may orchestrate a series of events that are necessary for responses to attractive guidance cues: (1) ZBP1 transports β-actin mRNA from the soma to the growth cone; (2) in response to a growth or guidance cue in the growth cone, Src kinase phosphorylates ZBP1, which results in the local translation of β-actin; and (3) this newly synthesized β-actin is then locally integrated into the growth cone, resulting in a change in growth cone dynamics and/or guidance (Fig. 9). As we have demonstrated, the loss of ZBP1 results in the perturbation of this cascade, and thus could disrupt the formation of proper neuronal circuitry in the developing nervous system. Further studies should reveal the diverse roles played by ZBP1 and other paralogs in protein synthesis-dependent axon guidance.
Figure 9.
ZBP1 regulates local β-actin synthesis and growth cone guidance. ZBP1 orchestrates a series of events that are necessary for responses to attractive guidance cues. First, ZBP1 binds to β-actin mRNA in the soma and transports it to the growth cone on microtubules. Then, in response to a growth or guidance cue, Src kinase phosphorylates ZBP1, which results in the local translation and enrichment of β-actin. This newly synthesized β-actin is locally integrated into the growth cone, resulting in a change in growth cone dynamics and/or guidance. As we have demonstrated, the loss of ZBP1 results in the perturbation of this cascade, and thus can disrupt normal cell motility.
Footnotes
This research was supported by awards from NINDS (F32NS064727) to K.W. and NICHD (R01HD046368) to G.J.B. This research project was also supported in part by the Viral Vector Core and the Microscopy Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We thank Michael Davidson for the original Dendra2 plasmid. We thank all members of the Bassell laboratory for helpful comments during the development of this manuscript, but specifically acknowledge Wilfried Rossoll and Sharon Swanger for their critical reading of the manuscript. We also specifically thank Yukio Sasaki for technical assistance and Wilfried Rossoll for obtaining the Zbp1 mutant mice. Finally, we would like to acknowledge Andrew Swanson for technical support with imaging and analysis.
References
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Mattson MP. NT-3 and BDNF protect CNS neurons against metabolic/excitotoxic insults. Brain Res. 1994;640:56–67. doi: 10.1016/0006-8993(94)91857-0. [DOI] [PubMed] [Google Scholar]
- Chien CB, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JR, Höpker VH, Ming GL, Poo MM, Tessier-Lavigne M, Hemmati-Brivanlou A, Holt CE. Turning of retinal growth cones in a netrin-1 gradient mediated by the netrin receptor DCC. Neuron. 1997;19:1211–1224. doi: 10.1016/s0896-6273(00)80413-4. [DOI] [PubMed] [Google Scholar]
- Fivaz M, Meyer T. Specific localization and timing in neuronal signal transduction mediated by protein-lipid interactions. Neuron. 2003;40:319–330. doi: 10.1016/s0896-6273(03)00634-2. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini R, Choi-Lundberg D, Thompson MJ, Mitchell CB, Foa L. Homer regulates calcium signalling in growth cone turning. Neural Dev. 2009;4:29. doi: 10.1186/1749-8104-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, Winkler C, Sendtner M. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19:1951–1966. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Pan F, Singer RH. Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci. 2009;122:1895–1905. doi: 10.1242/jcs.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kater SB, Rehder V. The sensory-motor role of growth cone filopodia. Curr Opin Neurobiol. 1995;5:68–74. doi: 10.1016/0959-4388(95)80089-1. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kuwako K, Kakumoto K, Imai T, Igarashi M, Hamakubo T, Sakakibara S, Tessier-Lavigne M, Okano HJ, Okano H. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010;67:407–421. doi: 10.1016/j.neuron.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jia YC, Cui K, Li N, Zheng ZY, Wang YZ, Yuan XB. Essential role of TRPC channels in the guidance of nerve growth cones by brain-derived neurotrophic factor. Nature. 2005;434:894–898. doi: 10.1038/nature03477. [DOI] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Tan CL, Lin CL, Strochlic L, Huang YS, Richter JD, Holt CE. Cytoplasmic polyadenylation and cytoplasmic polyadenylation element-dependent mRNA regulation are involved in Xenopus retinal axon development. Neural Dev. 2009;4:8. doi: 10.1186/1749-8104-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohof AM, Quillan M, Dan Y, Poo MM. Asymmetric modulation of cytosolic cAMP activity induces growth cone turning. J Neurosci. 1992;12:1253–1261. doi: 10.1523/JNEUROSCI.12-04-01253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Wong ST, Henley J, Yuan XB, Song HJ, Spitzer NC, Poo MM. Adaptation in the chemotactic guidance of nerve growth cones. Nature. 2002;417:411–418. doi: 10.1038/nature745. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Murray AJ, Peace AG, Shewan DA. cGMP promotes neurite outgrowth and growth cone turning and improves axon regeneration on spinal cord tissue in combination with cAMP. Brain Res. 2009;1294:12–21. doi: 10.1016/j.brainres.2009.07.071. [DOI] [PubMed] [Google Scholar]
- O'Connor TP, Duerr JS, Bentley D. Pioneer growth cone steering decisions mediated by single filopodial contacts in situ. J Neurosci. 1990;10:3935–3946. doi: 10.1523/JNEUROSCI.10-12-03935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XR, Ming GL, Xie Y, Hong Y, Sun DM, Zhao ZQ, Feng Z, Wang Q, Shim S, Chen ZF, Song HJ, Mei L, Xiong WC. Focal adhesion kinase in netrin-1 signaling. Nat Neurosci. 2004;7:1204–1212. doi: 10.1038/nn1330. [DOI] [PubMed] [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J Neurosci. 2010;30:9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Barrios N, Duloquin L, Boissonneau E, Lecorre P, Agnès F. The Drosophila RNA-binding protein ELAV is required for commissural axon midline crossing via control of commissureless mRNA expression in neurons. Dev Biol. 2007;301:166–177. doi: 10.1016/j.ydbio.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Spooner BS, Holladay CR. Distribution of tubulin and actin in neurites and growth cones of differentiating nerve cells. Cell Motil. 1981;1:167–178. doi: 10.1002/cm.970010202. [DOI] [PubMed] [Google Scholar]
- Suter DM, Forscher P. Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J Neurobiol. 2000;44:97–113. [PubMed] [Google Scholar]
- Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–115. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Li Y, Zhang Z, Cui K, Wang S, Yuan XB, Wu CP, Poo MM, Duan S. Nerve growth cone guidance mediated by G protein-coupled receptors. Nat Neurosci. 2002;5:843–848. doi: 10.1038/nn899. [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zheng JQ, Wan JJ, Poo MM. Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient. J Neurosci. 1996;16:1140–1149. doi: 10.1523/JNEUROSCI.16-03-01140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]