Abstract
The accurate perception of sound frequency by vertebrates relies upon the tuning of hair cells, which are arranged along auditory organs according to frequency. This arrangement, which is termed a tonotopic gradient, results from the coordination of many cellular and extracellular features. Seeking the mechanisms that orchestrate those features and govern the tonotopic gradient, we used expression microarrays to identify genes differentially expressed between the high- and low-frequency cochlear regions of the chick (Gallus gallus). Of the three signaling systems that were represented extensively in the results, we focused on the notch pathway and particularly on DNER, a putative notch ligand, and PTPζ, a receptor phosphatase that controls DNER trafficking. Immunohistochemistry confirmed that both proteins are expressed more strongly in hair cells at the cochlear apex than in those at the base. At the apical surface of each hair cell, the proteins display polarized, mutually exclusive localization patterns. Using morpholinos to decrease the expression of DNER or PTPζ as well as a retroviral vector to overexpress DNER, we observed disturbances of hair-bundle morphology and orientation. Our results suggest a role for DNER and PTPζ in hair-cell development and possibly in the specification of tonotopy.
Keywords: auditory system, hair bundle, planar cell polarity, signaling
The mechanosensory hair cells of the cochlea specialize in detecting particular frequencies of sound. Like the keys on a piano, cells tuned to similar frequencies lie adjacent to one another. Between the cochlear apex and base, hair cells span the entire range of audition, from the lowest to the highest frequencies. This arrangement is termed the tonotopic gradient.
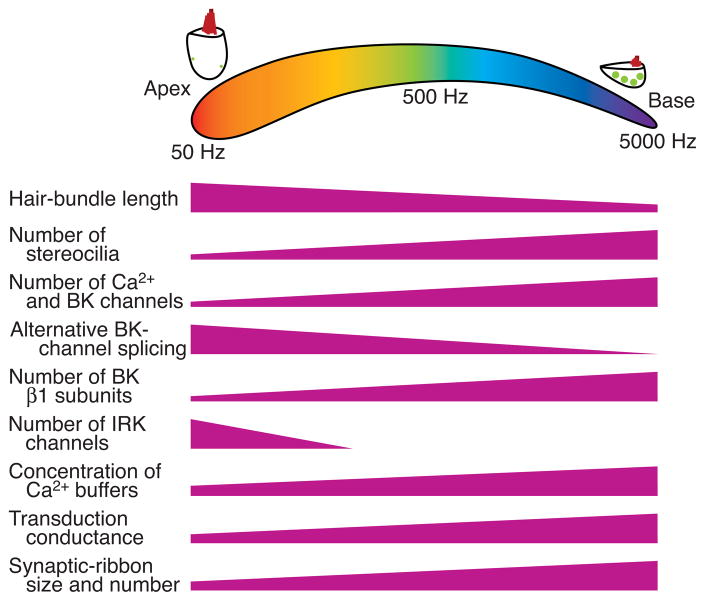
Many cellular and extracellular characteristics collude to produce precise tuning (Fig. 1). In mammals, the stiffness of the basilar membrane on which the hair cells rest increases from the cochlear apex, which is tuned to low frequencies, to the base, which responds to high frequencies (von Békésy, 1960). The length of the mechanoreceptive hair bundles generally declines from the apex to the base; in the chicken, for example, this gradient extends from 5.5 μm to 1.5 μm. The number of stereocilia in each hair bundle conversely rises from the apex to the base, from near 50 to more than 300 (Tilney and Saunders, 1983).
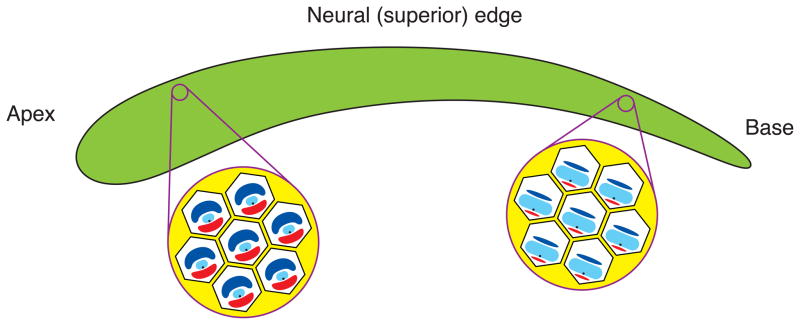
Figure 1.
Manifestations of tonotopy. The schematic diagram at the top portrays the basilar papilla or sensory epithelium of the chicken’s cochlea, which contains some 10,000 hair cells. The color scale represents the range of characteristic frequencies in a chromatic spectrum. Representative low- and high-frequency hair cells are shown above; the dots represent synaptic ribbons. The wedges delineate some of the gradients in morphology and gene expression along the cochlea. In addition to references cited in the text, data were taken from: Ramanathan et al., 1999; Hackney et al., 2003; Ricci et al., 2003; and Carninci et al., 2007.
Electrical resonance, a tuning mechanism found in the hair cells of many vertebrates, also exhibits a tonotopic gradient (Fettiplace and Fuchs, 1999). The gradient in electrical tuning results from differences along the tonotopic axis in the number and properties of Ca2+-sensitive, high-conductance K+ (BK) channels (Hudspeth and Lewis, 1988; Rosenblatt et al., 1997; Samaranayake et al., 2004; Beisel et al., 2007; Miranda-Rottmann et al., 2010). Electrical tuning is additionally shaped by other channels, particularly the inwardly rectifying K+ channel Kir2.1 (IRK1) in the low-frequency region of the cochlea (Navaratnam et al., 1995). The size and number of synaptic ribbons, specializations involved in synaptic-vesicle trafficking and release, increase with the characteristic frequency in many species (Martinez-Dunst et al., 1997; Schnee et al., 2005).
To yield correct tuning, all of these features must be orchestrated over hundreds of cell diameters. The tonotopic variations occur gradually over the cochlea’s length, suggesting that tonotopic identity is encoded in a continuous manner rather than by specification of discrete cellular subtypes. Few if any other biological systems possess this quality. The mechanism by which the tonotopic gradient is established and maintained remains unknown.
Because tonotopic identity encompasses so many components, it is important to understand the appearance of the tonotopic gradient in relation to other developmental processes shaping the cochlea. For example, how is tonotopic identity assigned to the hair cells and surrounding supporting cells as they differentiate after being specified by lateral inhibtion through the notch pathway (Daudet and Lewis, 2005)? As hair bundles form, how are they instructed about the appropriate morphology to assume? The radial pattern of the cochlea, governed by the planar-cell-polarity pathway, results in the alignment of hair bundles’ axes of sensitivity along the direction orthogonal to the tonotopic gradient (Montcouquiol et al., 2003). Are the radial and longitudinal arrangements be coupled? Answering these and similar questions would bring us closer to understanding the mechanism by which tonotopy develops.
METHODS
RNA extraction and labeling
For each of five microarray analyses we dissected 80–100 cochleas from 14–16-day-old White Leghorn chickens (Gallus gallus) of both sexes. After removing the lagena and tegmentum vasculosum, we isolated the basal and apical quarters of each cochlea. The tissue samples therefore contained connective tissue and some neurons as well as hair and supporting cells. We pooled the samples into apical and basal samples, isolated total RNA from each (Trizol, Invitrogen), and reverse-transcribed it to biotin-labeled cDNA (MessageAmp II-Biotin Enhanced Single Round aRNA Amplification kit, Ambion). Labeled cDNA was then hybridized to Affymetrix arrays that were washed and scanned by standard protocols.
For qPCR analysis of apical and basal segments, we performed one round of RNA amplification (Arcturus RiboAmp mRNA amplification kit).
Microarray analysis
We analyzed microarray data with GeneSpring GX 10 (Agilent Technologies). The results were normalized by robust multi-array analysis (Irizzary et al., 2003). Probes were then filtered on the basis of expression above the twentieth percentile in at least one of the arrays, yielding 33,319 entities, and a Student’s t-test was performed to compare the apical and basal samples. Using a p-value criterion of 0.05, we pared the list to 598 differentially expressed transcripts, then filtered the list on the basis of expression ratio.
Partial annotation of the probes was provided by Affymetrix. When the annotation was not available, we used the retrieved probe sequences (http://www.affymetrix.com/analysis/index.affx) to perform a search using the nblast protocol of the NCBI nr/nt database (http://blast.ncbi.nlm.nih.gov/Blast) and sought annotated genes with high homology that we then used in annotation.
Using the database for annotation, visualization and integrated discovery (DAVID; http://david.abcc.ncifcrf.gov/; Dennis et al., 2003; Huang et al., 2009), we performed gene ontology analysis for genes with expression ratios exceeding 1.5.
qRT-PCR
To perform qRT-PCR on embryonic inner-ear samples, we excised left and right E8 otocysts with surrounding tissue and extracted RNA with Trizol (Invitrogen). We synthesized cDNA with random hexamers (SuperScript III, Invitrogen) and performed qRT-PCR (ABI Prism 7900HT, Applied Biosystems) with melting a temperature of 59° C. To calibrate the efficiency of the amplification reactions, each reaction was run in triplicate using cDNA at two different concentrations per primer. The values were normalized to the amplification of β-actin transcript as an internal control. The transcript accession numbers and all primer sequences are provided in Supplementary Table 2.
RCAS construct preparation
The full-length cDNA encoding DNER was cloned from a chick cochlear cDNA library, placed in a pSlax13 shuttle vector, and cloned into the ClaI site of the RCASBP(A) plasmid. The orientation of the construct was verified by PCR and sequencing.
Embryo care and procedures
Fertilized White Leghorn chicken eggs (Charles River Laboratories) were incubated at 37.5° C with a humidity of 60% or greater. Eggs were windowed at E4, then returned to the incubator until microinjection at E5 or E6.
Using injection-ready solution provided by the supplier at a concentration of 0.5 mM and with methylene blue added for contrast, we injected vivo-morpholinos into the developing inner ear at E6. We injected the right ear, leaving the left one as an internal control. Viral plasmids were similarly injected at E5 at a concentration of 1 μg/ml.
Microelectroporation of the plasmids was performed by passing two trains of three 14-V, 25-ms pulses (ECM830, Harvard Apparatus) through a gold-coated anode, 5 mm in length and 0.5 mm in diameter, placed outside the electroporated tissue. A sharp titanium microelectrode, 3 mm in length and 0.125 mm in diameter, acted as a cathode situated in the tissue near the vesicular lumen. Following microelectroporation, a few drops of sterile phosphate-buffered saline solution (PBS) at pH 7.2 were applied at the site of manipulation. Embryos were sealed and returned to the incubator until the desired age.
Injection mildly affected certain features of the inner ear. Otoconia, normally present as small, crystalline particles (Li et al., 2006), were fused to form several large otoliths. In addition, the tegmentum vasculosum, a tissue that develops on the cochlear duct opposite the basilar papilla, was less distinctly segmented than in uninjected cochleas. These changes, unlike those described in the Results, did not depend on the sequence of the injected vivo-morpholino; they were induced by nonsense control oligonucleotides and could be discounted as non-specific artifacts of the technique.
Specimen collection and fixation
When the appropriate stage was reached, each embryo was removed from the egg. For ages E10 and below, we washed the embryo in PBS, decapitated it, perforated the mesencephalon for easier fixative access, bisected the head, and immersed the specimens overnight at 4° C in 4% formaldehyde in PBS. For an embryo of age E11 or greater, we bisected the head before fixation, bathed it in ice-cold artificial chick perilymph solution, dissected the temporal bones, removed the columellae, perforated the oval windows, and immersed the preparations overnight in fixative at 4°C. The fixative for light microscopy was 4% formaldehyde in PBS. For electron microscopy we used 1.2% formaldehyde in 120 mM sodium cacodylate, 25 mM sucrose, and 1 mM CaCl2. For young adult specimens, chickens were sacrificed and cochleas were dissected and fixed in 4% formaldehyde in PBS.
Immunolabeling
For the production of cryosections, embryos were cryoprotected in 30% (weight to volume) sucrose in PBS, frozen, and cut at thicknesses of 20–30 μm. For wholemount cochlear preparations, fixed cochleas were isolated from temporal bones and the tegmenta vasculosa were removed. Specimens were permeabilized for 8 min in acetone at −20° C, washed in PBS, blocked for one hour in 5%–10% heat-inactivated horse serum in PBS containing 0.1% Tween-20 (PBST), and incubated with primary antibodies overnight at 4° C. After several washes with PBST at room temperature, specimens were incubated with secondary antibodies and phalloidin (1:40) overnight at 4° C for wholemount preparations or 90 min at room temperature for tissue sections. They were then rinsed and mounted in Vectashield or ProLong Gold mounting medium and imaged on an Olympus FluoView FV1000 confocal microscope.
The polyclonal antiserum against DNER (AF2254, 1 μg/μl, R and D Systems) was used at a dilution of 1:200. The polyclonal antiserum against PTPζ (610180, 0.25 μg/μl, BD Bioscience) was used at a dilution of 1:40. The polyclonal antiserum against parvalbumin 3 was used as described (Heller et al., 2002). Monoclonal antibodies against supporting-cell antigen (D37; Kruger et al., 1999) were used at a dilution for the supernatant of 1:10. Monoclonal antibodies against gag protein (AMV-3C2, Developmental Studies Hybridoma Bank, University of Iowa), originally developed by Dr. D. Boettiger, were used at a dilution for the supernatant concentrate of 1:100.
Scanning electron microscopy
After fixation, specimens were rinsed in a solution of 120 mM sodium cacodylate, 25 mM sucrose, and 1 mM CaCl2. To loosen the tectorial membranes, the tegmenta vasculosa were opened and the samples were incubated in a solution containing 200 μg/ml of protease type XXIV (Sigma). Following a rinse, the specimens were immersed for 1 hr at 4° C in a solution of 50 mM osmium tetroxide, 120 mM sodium cacodylate, 25 mM sucrose, and 1 mM CaCl2, then rinsed in distilled water. After the tectorial membranes and surrounding tissue had been dissected, samples were dehydrated by immersion in a graded series of ethanol concentrations. The specimens were subjected to critical-point drying through liquid CO2 (Tousimis Autosamdri-815A), mounted on sample stubs with colloidal-silver glue, and dried in vacuum for 20 min. They were then sputter-coated with gold-palladium (Desk IV, Denton Vacuum) and examined in a LEO 1550 scanning electron microscope at an accelerating voltage of 5 kV.
Morpholino sequences
Because the high GC content in the DNER sequence and the uncertain translation-initiation site in the celsr1 mRNA prevented the design of effective translation-blocking morpholinos, we used morpholino oligonucleotides to block the splicing of DNER and celsr1 transcripts. The sequences were 5′-AAAGTCATCTTGTTACTCACCAATA-3′ for DNER (ENSGALG00000002958), targeting the boundary between the third exon and the following intron, and 5′-ATATTCTGAGAAGCAAAGGACAAGT-3′ for celsr1 (ENSGALG00000013399), targeting the boundary between exon 13 of the longer transcript and its preceding intron. The PTPζ morpholino, with the sequence 5′-AGCCTTGGTAAAGGACATCCTGCAT-3′, targeted the translation-initiation site. Finally, the sequence of the nonsense control morpholino was 5′-CCTCTTACCTCAGTTACAATTTATA-3′. Each of the molecules was synthesized with an octaguanidine dendrimer attached to its 3′ terminus (Gene Tools).
Phenotypic analysis of hair bundles
Hair bundles from the basal halves of basilar papillae were visualized by phalloidin staining. Confocal images were processed using ImageJ. A line was drawn along the base of each bundle and its angle with respect to an external reference was recorded. Angular distributions obtained from different images were superimposed according to their mean values. Their von Mises character, corresponding to a normal distribution on a circle, was confirmed. A parametric test for the concentration parameter with the significance level of 0.01 was performed to assert statistical differences between the distributions (Batschelet, 1981).
RESULTS
Microarray analysis
In an effort to identify proteins that underlie the specification of tonotopic position, we used expression microarrays to compare gene expression between the apex and base of the chicken’s cochlea. This approach was predicated upon the observation that regenerating hair cells express normal tonotopic features (Woolley and Rubel, 2002), which implies that tonotopic information persists in the adult cochlea. This approach yielded 429 positive probe sets, corresponding to 385 transcripts, with expression ratios exceeding 1.5; 215 of these probe sets, reflecting 186 transcripts, displayed expression differing by more than twofold (Supplementary Table 1). In validation of the experimental approach, some of the transcripts previously known to be tonotopically expressed along the cochlea occurred among the results. These transcripts encoded the Kir2.1 channel and brain-derived neurotrophic factor, both of which are more highly expressed at the apex, as well as calretinin, more highly expressed at the base.
To classify the significant transcripts, we used gene ontology to describe gene products with respect to biological processes, cellular components, and molecular functions (Ashburner et al., 2000). Although the annotation of the chicken genome is incomplete, an analysis of term enrichment revealed three groups of terms overrepresented in descriptions of differentially expressed transcripts. The first group includes terms such as “synaptic transmission,” “vesicle,” and “neurotransmitter secretion.” The second group embraces “gated channel activity,” “ion transport,” and “calcium.” The final group consists of descriptions such as “developmental process,” “mechanoreceptor differentiation,” and “cell proliferation.” Closer inspection shows that the list contains genes linked to human sensorineural defects (Petit and Richardson, 2009) such as deafness (ESPN, MYO3a, MYO6, OTOF, and TMHS), Usher syndrome (GPR98 and USH2A), blindness (DGKI), and retinitis pigmentosa (CERKL). In addition to the products of several of those genes, the transcripts include those encoding known structural components of hair cells such as β-spectrin and the protein tyrosine phosphatase receptor Q found in the basal tapers of stereocilia (Goodyear et al., 2003). We also found several transcription factors, including isl1, tbx19, id4, etv4, and emx2. The last of these is known to play a critical role in hair-cell differentiation (Holley et al., 2010).
To better understand the molecular signaling that underlies the tonotopic gradient, we investigated proteins with known functions in signaling pathways as well as those with predicted signaling activity. Among the differentially expressed genes we found three pathways represented by at least four members apiece. The notch pathway showed the greatest number of differentially expressed transcripts. The proteins included delta/notch-like EGF-related (DNER), lunatic fringe (LFNG), hairy and enhancer of split 5 (HES5), hairy/enhancer-of-split related with YRPW motif 1 (HEY1), achaete-scute complex homologue 1 (ASCL1), receptor type protein tyrosine phosphatase ζ (PTPζ), and pleiotrophin (PTN). The wnt pathway was represented by differential expression of frizzled-8 (FZD8), r-spondins 2 and 3 (RSPO2 and RSPO3), dapper, antagonist of β-catenin, homolog 2 (DACT2), and the novel low-density-lipoprotein receptor-related protein LRP11. Finally, four transcripts were identified as encoding elements of the fibroblast-growth-factor signaling cascade: fibroblast growth factor 3 (FGF3), fibroblast-growth-factor receptor-like 1 (FGFRL1), fibroblast growth factor-binding protein 1 (FGFBP1), and iroquois homeobox 2 (IRX2).
Validation of array results
We verified the differential expression of these and many other transcripts by quantitative real-time polymerase chain reactions (qPCRs; Table 1). In general, the qPCR results accorded with the array data. Because the dynamic range of detection of transcripts by qPCRs exceeds that of an array, however, the differences revealed by the qPCR method were more striking. This is evident upon comparison of the first and third columns of Table 1: the transcript encoding FGF3, for example, was shown by the array data to be expressed 11 times more strongly at the apex than at the base, whereas qPCR results pointed to 190-fold difference.
Table 1.
Results of expression analyses
| qPCR ratio | Error | Array ratio | Gene symbol | Description |
|---|---|---|---|---|
| 19 | TMC2 | transmembrane channel-like 2 | ||
| 190 | ±50 | 11 | FGF3 | fibroblast growth factor 3 |
| 51 | ±9 | 6.7 | finished cDNA, clone ChEST905o6 | |
| 18 | ±2 | 6.2 | BDNF | brain-derived neurotrophic factor |
| 52 | ±2 | 5.6 | ALPHA 10 nAChR | alpha 10 subunit of nicotinic acetylcholine receptor |
| 15 | ±2 | 4.9 | OTOF | otoferlin |
| 4.8 | TMCC2 | transmembrane and coiled-coil domain family 2 | ||
| 4.8 | SPTBN5 | spectrin, beta, non-erythrocytic 5 | ||
| 19 | ±3 | 4.7 | A2M | alpha-2-macroglobulin |
| 4.6 | finished cDNA, clone ChEST117a22 | |||
| — | — | — | — | — |
| 4.3 | LRP11 | low-density-lipoprotein receptor-related protein 11 | ||
| 4.7 | ±0.5 | 3.2 | KCNJ2 | potassium inwardly-rectifying channel, subfamily J, member 2 |
| 7.2 | ±0.5 | 3.2 | FGFRL1 | fibroblast growth factor receptor-like 1 |
| 29 | ±4 | 3.1 | HES5 | hairy and enhancer of split 5 (Drosophila) |
| 6.9 | ±0.6 | 3.0 | PTPRZ1 | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| 5.7 | ±1.3 | 2.3 | DACT2 | dapper, antagonist of beta-catenin, homolog 2 |
| 7.6 | ±0.4 | 2.2 | DNER | delta/notch-like EGF repeat containing |
| 1.9 | ±0.2 | 1.8 | LFNG | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| 3.2 | ±0.8 | 1.7 | FZD8 | frizzled homolog 8 (Drosophila) |
| 1.6 | PTN | pleiotrophin (heparin-binding growth factor 8, neurite growth-promoting factor 1) | ||
| 13 | ±4 | 1.6 | FGFBP1 | fibroblast growth factor binding protein 1 |
| 4.1 | ±1 | 1.6 | HEY1 | hairy/enhancer-of-split related with YRPW motif 1 |
| — | — | — | — | |
| 0.57 | RSPO3 | R-spondin 3 homolog | ||
| 0.42 | ±0.05 | 0.54 | CALB2 | calbindin 2, 29 kDa (calretinin) |
| 0.2 | ±0.03 | 0.46 | RSPO2 | R-spondin 2 homolog |
| 0.18 | ±0.01 | 0.39 | ASCL1 | achaete-scute complex homolog 1 |
| — | — | — | — | — |
| 0.54 | ±0.17 | 0.36 | GPR22 | G-protein-coupled receptor 22 |
| 0.15 | ±0.02 | 0.36 | IRX2 | iroquois homeobox 2 |
| 0.5 | ±0.1 | 0.33 | AKR1D1 | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) |
| 0.19 | ±0.03 | 0.32 | GREM2 | gremlin-2 |
| 0.29 | LOC776531 | similar to placenta growth factor | ||
| 0.16 | ±0.01 | 0.24 | CHODL | chondrolectin |
| 0.21 | SPP1 | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I) | ||
| 0.13 | MMP-13 | matrix metalloproteinase-13 (collagenase-3) | ||
| 0.086 | finished cDNA, clone ChEST275i11 | |||
| 0.076 | OSTN | osteocrin |
The table summarizes the notable results of the expression analysis. For each of the indicated genes, the numerical values represent the ratio of the expression level at the apex to that at the base as determined by normalized qPCR (with the error value for three measurements) and by microarray analysis. Blank spaces indicate that qPCR reactions were not conducted. The first ten entries represent the transcripts with the largest ratios of apical to basal expression and the final ten entries correspond to the lowest ratios. The middle segments of the table include transcripts mentioned in the text and others related to known hair-cell functions or of potential interest in signaling.
Immunolabeling of DNER and associated proteins
Of the pathways mentioned, the notch system was represented by the most members and accordingly elicited the closest examination. We directed our attention toward the proteins exhibiting the greatest differences in expression and displaying differences in the most probe sets as well as those whose function in hair cells had not previously been described. Immunolabeling of one of these, the transmembrane putative notch ligand DNER, confirmed its tonotopic distribution along the cochlea: the protein occurs exclusively in hair cells and at a high concentration at the cochlear apex but a low level at the base. DNER exhibits a distinctive distribution pattern, concentrating in intracellular punctae near the apical surface of each hair cell and adjacent to the kinociliary edge, at the front of the hair bundle (Fig. 2A–I). The extracellular domain of DNER is thought to interact with that of its receptor, notch, in adjacent cells. Consistent with this interaction, notch is expressed in supporting cells of the chick cochlea (Daudet and Lewis, 2005). notch signaling downstream of DNER binding is dependent on deltex proteins (Eiraku et al., 2005). We confirmed by RT-PCR that at least one member of that family, deltex-2, occurs in the chicken’s cochlea (data not shown).
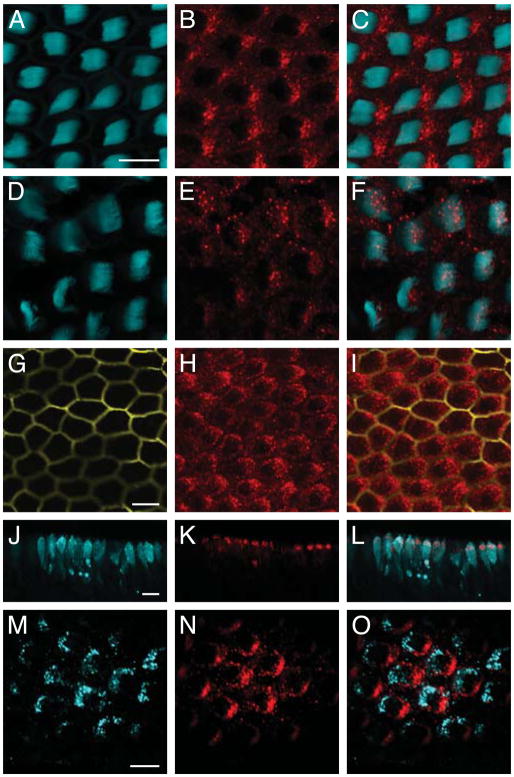
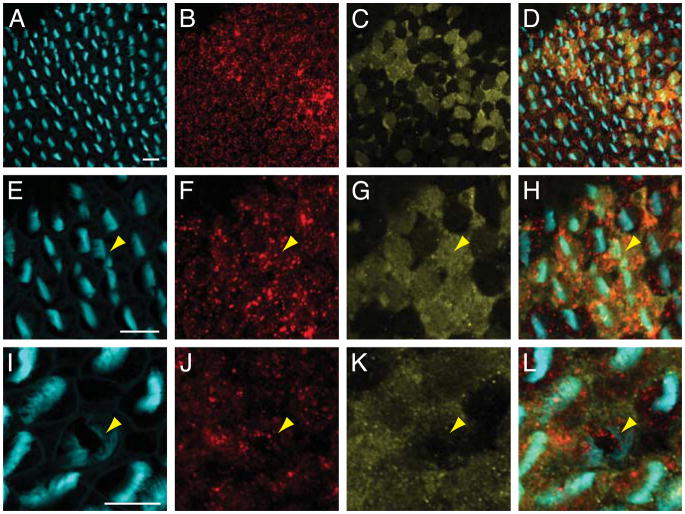
Figure 2.
DNER and PTPζ immunolabeling of the chick’s cochlea. A–C: In a wholemount preparation of the cochlear apex, DNER (red) is expressed in a punctate pattern at the apical surfaces of hair cells on the edges adjacent to the kinocilia. Phalloidin (cyan) labels the hair bundles. D–F: DNER expression (red) is strikingly lower in basal hair cells. G–I: DNER (red) is excluded from supporting cells (yellow), whose narrow apical surfaces are marked by the expression of supporting-cell antigen. J–L: The expression of DNER (red) is apparent in a section through an E8 chick vestibule. Nascent hair cells are marked by the expression by parvalbumin 3 (cyan). M–O: The expression pattern of PTPζ (cyan) is complementary to that of DNER (red). In these and subsequent illustrations of wholemounts, kinocilia are oriented toward the right. The scale bars represent 10 μm.
We determined that the onset of DNER expression coincides with hair-cell differentiation, as defined by appearance of the Ca2+ buffer parvalbumin 3 (Heller et al., 2002) (Fig. 2J–L). The characteristic intracellular foci of expression develop on embryonic days 16–17, following the establishment of the pattern of planar polarity.
Another protein that exhibited an apical-to-basal difference in expression, PTPζ, has recently been shown to dephosphorylate DNER and regulate its intracellular trafficking (Fukazawa et al., 2008). Immunolabeling showed that PTPζ is confined to the apical part of the hair cell and that its pattern of expression is mutually exclusive to that of DNER. In the plane of the epithelium, PTPζ occurs behind the hair bundle, opposite its kinociliary edge (Fig. 2M–O).
Vivo-morpholino knockdown experiments
To assess the role of the DNER and PTPζ in cochlear development, we established a method of transiently reducing the expression of selected genes in the developing inner ear. At embryonic day 6 (E6), the onset of hair-cell differentiation in the cochlear duct of the chick (Daudet and Lewis, 2005), we injected the inner ears of embryos with a solution containing antisense morpholino oligonucleotides linked to octaguanidine. These modified oligonucleotide analogs, known as vivo-morpholinos, are transported across cellular membranes owing to the highly charged octaguanidine dendrimer, which mimics the polyarginine sequence in the HIV tat peptide (Morcos et al., 2008). The modification allows gene expression to be reduced in vivo without the need for electroporation, which we found ineffective for targeting hair cells in late-stage embryos (data not shown).
We used vivo-morpholinos to reduce the expression of DNER and PTPζ. As a positive control, we also diminished the expression of celsr1 (c-flamingo-1), a protein with a readily assayed phenotype that functions in the specification of planar cell polarity in the cochlear hair cells (Curtin et al., 2003; Davies et al., 2005).
To confirm the reduced expression of DNER by immunohistochemistry, we sectioned morpholino-injected embryos at E8, when DNER was already robustly expressed in nascent vestibular hair cells. In an injected ear we observed a marked decrease in immunofluorescence by comparison to the same embryo’s uninjected ear (Fig. 3A–F). Because the morpholino targeting DNER was designed to block splicing, we sought a decrease in the correctly spliced form by performing semiquantitative RT-PCRs with primers that spanned the blocked intron-exon boundary on RNA samples obtained from embryonic fragments containing the inner ear. Although we observed on the morpholino-injected side a marked decrease in the intensity of the signal for DNER transcripts, we did not observe another band of a different length. The misspliced product may therefore have been degraded. This result was confirmed by qPCR analysis, which showed a decrease in the amount of DNER transcript to one-third to one-half its control value. The PTPζ transcript displayed a still greater reduction, to less than one-seventh its control value, raising the possibility that the downstream effects of DNER include transcriptional regulation of PTPζ. The amount of transcript for a hair-cell marker, parvalbumin 3, did not change appreciably, assuring us that the number of hair cells remained constant.
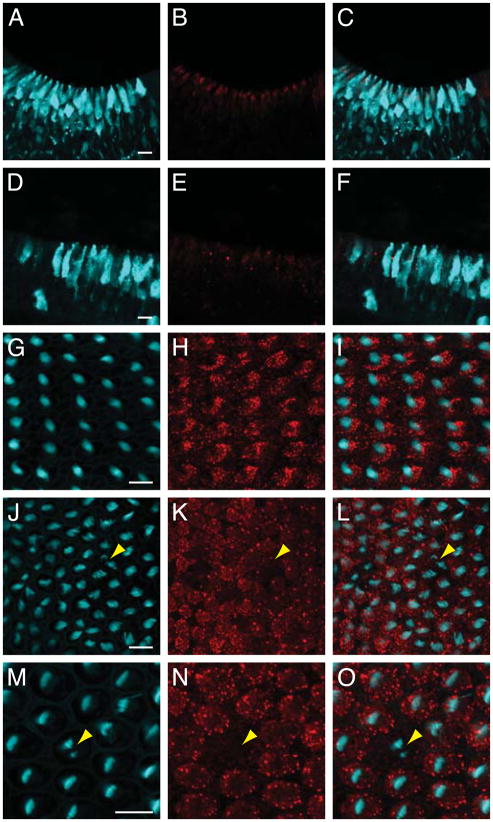
Figure 3.
Reducing DNER expression with a splice-blocking morpholino. A–C: DNER (red) is apparent at E8 in a cryosection of nascent vestibular hair cells from an uninjected ear. Parvalbumin 3 (cyan) marks the hair-cell somata. D–F: Morpholino injection reduces the amount of DNER (red). G–I: An ear injected with control morpholino retains a regular arrangement of phalloidin-labeled hair bundles (cyan). J–L: An ear injected previously with DNER morpholino displays a patchy reduction in the concentration of DNER (red) and misorientation of hair bundles (arrowhead) labeled with phalloidin (cyan). M–O: A higher-magnification micrograph shows a central hair cell with little DNER (red) and a split hair bundle (arrowhead) labeled with phalloidin (cyan). The micrographs in G–O are surface views of E16 wholemount preparations. The scale bars represent 10 μm.
To assess the morphant phenotype at the level of hair bundles, we performed immunohistochemistry and scanning electron microscopy on cochleas at E16–E17. In 20 cochleas injected with DNER morpholino, 12% of the hair cells exhibited reduced DNER expression, presumably because only some cells took up morpholino in sufficient quantities to display an effect lasting ten or more days. Diminished labeling sometimes occurred in clone-like patches (Fig. 3G–L). Thirty-five percent of the hair cells with reduced DNER expression had misshapen hair bundles. We also observed hair bundles of unusual length and morphology; in some the stereocilia lay in two or three discrete tufts distant from the kinocilium (Fig. 4).
Figure 4.
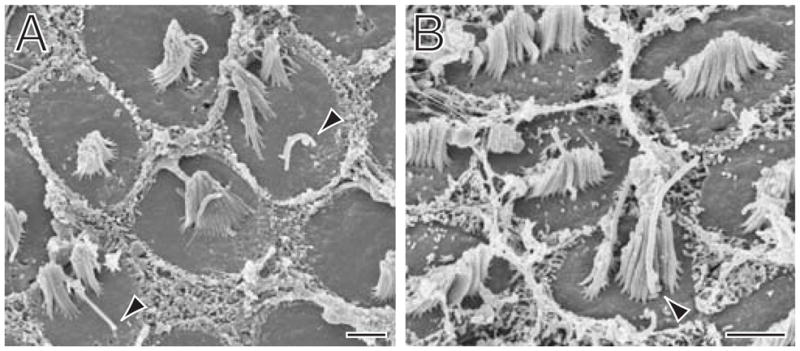
The effect of DNER morpholino on hair bundles. A: A scanning electron micrograph portrays two hair bundles with split clusters of stereocilia mislocalized to the edges of the apical surfaces opposite their kinocilia (arrowheads). B: A micrograph of hair bundles from near the cochlear base includes a single bundle (arrowhead) that is conspicuously taller and narrower than its neighbors. The aberrant bundle resembles those from a more apical position in the basilar papilla. The scale bars represent 2 μm.
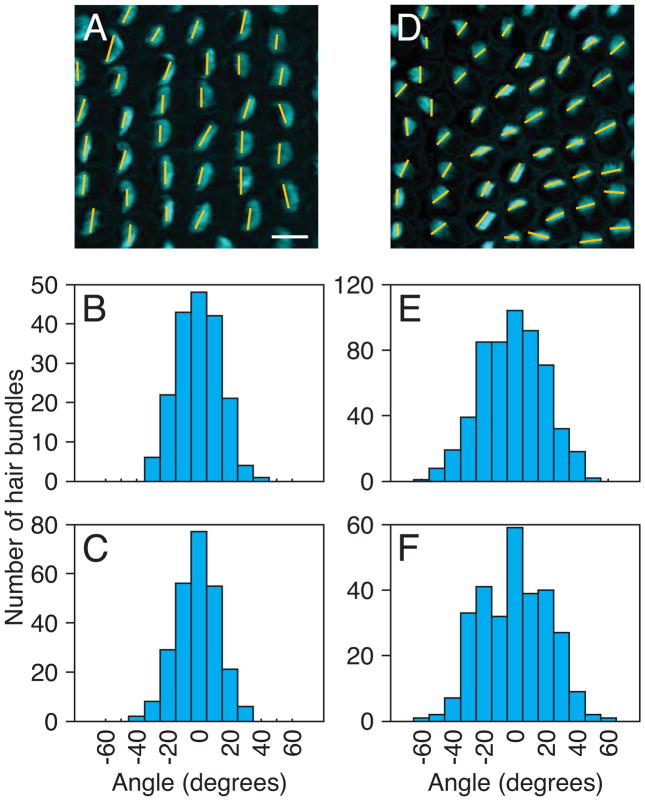
The largest effect, however, involved the orientation of hair bundles. Wild-type hair bundles are uniformly oriented in the developing cochlea; we measured an angular dispersion of 13° (Fig. 5A–B) for hair cells in the basal halves of the cochleas, which showed the most regular arrangement. The seven cochleas injected with a control, nonsense vivo-morpholino displayed wild-type hair-bundle morphology and orientation (Fig. 5C). The dispersion in the angle of bundle orientation was statistically identical to that of uninjected cochleas. In contrast, the hair-bundle orientations in the same region of morpholino-injected cochleas were strongly deranged, with a dispersion of 20° (Fig. 5D–E).
Figure 5.
Hair-bundle misorientation in DNER- and celsr1-depleted cochleas. A: In a normal E17 cochlea, a line drawn across each phalloidin-stained hair bundle (cyan) indicates its orientation. The deviation in orientations is relatively small. B: A histogram of the angular distribution of 254 hair-bundle orientations from uninjected cochleas shows a narrow peak. C: The histogram for 187 hair bundles from cochleas injected with a control morpholino is also narrow. D: In a cochlea treated with DNER morpholino, the orientations of hair bundles are disrupted. E: The angular-distribution histogram for 556 hair bundles from cochleas injected with DNER morpholino shows a broadened peak. F: Cochlear injection of a morpholino against celsr1 has a similar effect on 293 hair bundles. The scale bar represents 10 μm in panels A and D.
Eight cochleas injected with a morpholino targeting celsr1, a protein essential for the specification of planar cell polarity, showed normal hair-bundle morphology, but the orientations of the bundles deviated severely from the regular pattern observed in controls (Fig. 6D–F). The angular dispersion was 21° (Fig. 5F), a result in accord with the disturbed bundle orientation in murine celsr1 mutants (Curtin et al., 2003). This control confirms that vivo-morpholino microinjection is an effective method for the selective reduction of gene expression in the chick’s embryo. Despite abnormal bundle orientations, immunohistochemistry revealed no decrease in DNER levels. As the focal distribution of DNER became established, labeling followed the same pattern. Even if a hair bundle was severely misoriented, DNER was always concentrated adjacent to the kinocilium. Immunohistochemistry confirmed that no cells in uninjected or control morpholino-injected cochleas exhibited reduced DNER expression.
Figure 6.
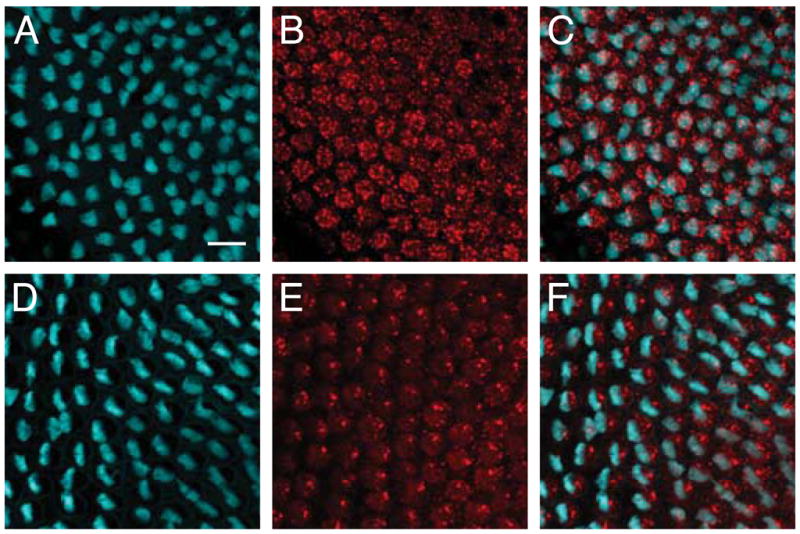
Misorientation of hair bundles in morpholino-injected cochleas. A–C: Injection of a morpholino against PTPζ causes disruption in the orientation pattern of hair bundles labeled with phalloidin (cyan). The localization of DNER (red) is also perturbed. D–F: A positive-control morpholino against celsr1 produces misorientation of phalloidin-labeled hair bundles (cyan) and mislocalization of DNER labeling (red). The scale bar represents 10 μm.
To test the hypothesis that DNER and PTPζ act through a common pathway, we injected five cochleas with a vivo-morpholino against the PTPζ transcript. The procedure yielded an orientation phenotype similar to that following use of the DNER vivo-morpholino, with an angular dispersion of 21.5° (Fig. 6A–C).
Retrovirus-mediated gene overexpression
To test the effect of DNER overexpression in the developing inner ear, we used the retroviral vector RCASBP(A) to drive exogenous expression (Hughes et al., 1987). When otic vesicles were electroporated with the DNER expression construct at E5, cells took up the plasmid and produced additional virus that subsequently spread throughout the ear. We confirmed that the overexpression of DNER overlapped with production of the viral gag protein (Fig. 7A–D). For eight cochleas examined at E16 the extent of the spread of infection varied between specimens. As assessed by immunohistochemistry, however, both hair cells and supporting cells were infected and expressed DNER.
Figure 7.
The effect of DNER overexpression on hair-bundle morphology. A–D: E16 cochleas subjected to the RCAS-DNER virus at E5 display viral gag protein (yellow) both in circular hair cells and in hourglass-shaped supporting cells. In areas of viral infection, DNER (red) is overexpressed and phalloidin-labeled hair bundles (cyan) are misoriented. E–L: Higher-power micrographs with the same color coding demonstrate abnormalities of selected hair bundles marked by arrowheads. The scale bars represent 10 μm.
As in cochleas injected with DNER morpholino, many cellular patches expressing the virus contained hair cells with hair bundles of abnormal, split morphology (Fig. 7E–H). Some bundles lost their ordinary polarization and assumed a circular shape (Fig. 7I–L). This held true both for hair cells expressing exogenous DNER and for some hair cells apparently free of infection but adjoined by infected supporting cells. When an RCAS-GFP construct was electroporated as a control, none of the GFP-expressing cochleas displayed abnormal hair-bundle morphology.
DISCUSSION
Despite the many manifestations of cochlear tonotopy, there is a dearth of information regarding possible mechanisms underlying the tonotopic gradient. To our knowledge, no mutants have been demonstrated to display abnormal tonotopy, nor have there been reports of congenital disease attributable to tonotopic defects. The tonotopic gradient is an emergent phenomenon with potentially complex developmental regulation. It is unclear whether its different components are regulated by a common pathway or by several coordinated cascades. Even more puzzling is the high precision of frequency tuning, which is established across thousands of cells.
Several mechanisms could give rise to the tonotopic arrangement. It is possible that a gradient in the concentration of a morphogen underlies the process. It is unclear, however, whether such a gradient could yield the fine frequency resolution found in the cochlea, where thousands of distinct phenotypes are specified over a few millimeters. Another possibility is that an intercellular-relay mechanism similar to that of the planar-cell-polarity pathway governs the regular arrangement of hair cells (Montcouquiol et al., 2003; Lopez-Schier and Hudspeth, 2006). An obvious difference between the planar-cell-polarity pathway and a putative system for specifying the tonotopic gradient is that the former needs to convey only the direction of polarization whereas the latter must also specify frequency information. Other conceivable mechanisms could depend on the timing of cell division or differentiation, resulting in different properties of hair cells produced at different times.
To infer some of the mechanisms involved in establishing tonotopy, we analyzed the transcriptional differences between high- and low-frequency regions of the cochlea. The results of the microarray analysis encompass several classes of genes. One comprises genes encoding likely downstream manifestations of tonotopy. This class includes components of the hair bundle: espin, PTPRQ, β-spectrin, G protein-coupled receptor 98, and myosins VI and IIIa. Because hair-bundle morphology and the number of stereocilia change dramatically along the tonotopic gradient, the expression of those proteins would be expected to differ between the high- and low-frequency regions of the cochlea. Other transcripts that are important for hair-cell function exhibited expression differences as well, including the putative Ca2+ sensor otoferlin, the Ca2+-binding proteins calbindin and CABP2, and the IRK-1 channel that participates in electrical resonance. Proteins involved in the formation and functioning of synapses were also present in the results of our analysis, including synaptojanin 2, SNAP-91, and neuronal pentraxin II. Finally, we encountered several transcription factors. Because many cellular characteristics vary along the cochlea, one would expect the transcription factors that coordinate the expression of the proteins involved to be differentially expressed between its apical and basal ends.
The basilar papilla is narrower at the base than at the apex of the cochlea, so a greater proportion of basal transcripts might have arisen from cells in the fibrocartilaginous plates. This could account for several connective-tissue markers, such as osteopontin and osteocrin (Oldberg et al., 1986; Thomas et al., 2003), that registered as upregulated in the high-frequency region of the cochlea. Some of these proteins do, however, occur as well in cell types other than cartilage (Takarada and Yoneda, 2009).
The proteins of greatest interest to us are members of signaling pathways that could play a role in establishing the tonotopic gradient, rather than in serving as its downstream products. Several of the pathways identified in our array analysis could play roles in the establishment or maintenance of tonotopy. FGF3 in particular stands out, for it is expressed 190-fold as much in the apical as in the basal segment of the cochlea. The poorly characterized r-spondins 2 and 3, together with their receptor frizzled 8 (Nam et al., 2006) and potential co-receptor LRP11, might form another cascade regulating tonotopy. We chose to investigate in greatest detail the most extensively represented pathway, the cascade potentially involving notch, DNER, PTPζ, and pleiotrophin. Canonical notch signaling participates at least twice in the development of the chick’s inner ear, first in the specification of prosensory patches and later in the determination of hair-cell and supporting-cell fates through lateral inhibition (Daudet and Lewis, 2005). Proteins such as notch are thus already known to occur in the ear’s sensory epithelia. DNER is also known to be expressed in murine hair cells and spiral-ganglion neurons (Hartman et al., 2010).
DNER-related signaling has been described only in the cerebellum (Eiraku et al., 2005; Fukazawa et al., 2008). DNER occurs in the dendrites of Purkinje cells, where it promotes differentiation of Bergmann glial cells through activation of non-canonical—RBP-J-independent and deltex-dependent—notch signaling. In the Purkinje cells themselves, DNER’s presence promotes dendrite extension. PTPζ, which can be inhibited by pleiotrophin, dephosphorylates DNER, leading to its endocytosis from the membrane and thus inhibiting neuritogenesis. It is noteworthy that murine pleiotrophin mutants suffer moderate hearing impairment (Zou et al., 2006).
Tonotopic information could be transmitted between hair cells through the notch pathway of intervening supporting cells, a potentially robust relay mechanism for establishing a finely resolved gradient. The mutually exclusive expression patterns of DNER and PTPζ suggest that the proteins are part of a molecular circuit in which PTPζ restricts the expression of DNER. Control of DNER endocytosis through PTPζ-dependent dephosphorylation could help to elaborate the gradient, particularly if DNER also controls PTPζ expression in a feedback loop. Expression of pleiotrophin, the ligand of PTPζ, also varies between high- and low-frequency regions, potentially adding another layer of regulation. The presence of deltex proteins in the cochlea suggests that non-canonical notch signaling is active during the formation of the tonotopic gradient.
When assessing the effects of misexpression of components of the putative signaling cascade, we used hair bundles as a convenient index of tonotopic identity. The phenotype of cochleas with reduced or elevated DNER expression levels suggests that DNER is involved in the regulation of hair-bundle development. Many bundles in these specimens displayed abnormalities of orientation and some of stereociliary length. Moreover, the stereocilia of an affected bundle often formed several clusters at a distance from the kinocilium, which is thought to orchestrate the bundle’s development (Tilney et al., 1992). This result suggests that DNER signaling has a more general effect on cellular morphology, making it a candidate molecule for orchestrating the development of tonotopy.
In cochleas infected with a DNER-expressing retrovirus, we observed defects not only in virus-containing cells but also in neighboring cells, an indication that DNER-mediated signaling affects hair bundles in a non-cell autonomous manner. Such non-autonomy is one of the principal characteristics expected of a molecule involved in intercellular signaling. In the morphant cochleas the number of hair cells that displayed abnormal bundle phenotypes exceeded the number manifesting decreased DNER expression. This finding could stem from non-cell autonomous signaling, but might instead reflect the ejection, degradation, or dilution of the morpholino. Although the misexpression phenotypes do not correspond directly to switching between extreme apical and basal fates, the results do not preclude the possibility that this pathway directly regulates the tonotopic gradient. The protein concentrations achieved with manipulations may lie outside the physiological range, driving the system to extreme phenotypes.
That both DNER and PTPζ morphants show disrupted orientation of hair cell bundles raises the possibility that the signaling pathways for the specification of planar cell polarity and the tonotopic gradient are linked. DNER concentrates near the kinocilium, which plays a role in the interpretation of planar-cell-polarity information (Jones et al., 2008). Consistent with this idea, the localization of DNER in celsr1 morphant cochleas follows the misplaced cilium.
The orientations of hair bundles deviate systematically from a radial pattern in the apical portion of the avian basilar papilla (Manley, 1990). Because DNER and PTPζ are more heavily expressed at the apex, signaling by these proteins might help to establish this deviation from the canonical orientation pattern.
The distribution of DNER within each hair cell is asymmetric and polarized in the direction of hair-bundle polarity, orthogonally to the tonotopic axis and the gradient in DNER expression (Fig. 8). If DNER signaling resembles the planar-cell-polarity pathway, this arrangement suggests that DNER and PTPζ mediate communication between cells, not of different characteristic frequencies, but rather of the same frequency. It is possible that this signaling functions as a spatial averaging mechanism between cells on an isofrequency line. Each cell might make a noisy measurement of its tonotopic position, based on a yet-unspecified signal, translate the result into DNER and PTPζ activity, and convey that information to cells along the same isofrequency line. The signaling noise would therefore be averaged so that all cells along the signaling path would arrive at a common value of characteristic frequency. A similar mechanism has been proposed for development of the hunchback gradient in the Drosophila embryo (Gregor et al., 2007).
Figure 8.
Schematic summary of the results. Hair cells in the chicken’s cochlea display a tonotopic gradient in the sizes of their hair bundles (light blue), which encompass as few as 50 stereocilia at the apex and more than 300 at the base. The putative notch ligand DNER (red), which accumulates at the cellular surface adjacent to the kinocilium (black dot), occurs at a high concentration near the cochlear apex and in progressively lower amounts toward the base. DNER may be dephosphorylated by PTPζ, which is also concentrated toward the cochlear apex but occurs in a complementary pattern within in each hair cell, accumulating at the cellular surface on the edge opposite the kinocilium. Notch (yellow), the receptor for DNER and related ligands, is known to occur in the supporting cells whose narrow apical processes separate all hair cells.
Our study establishes molecular differences between high- and low-frequency regions of the chick’s cochlea and demonstrates a role for signaling by DNER and PTPζ in the control of hair-bundle morphology. We propose that this signaling cascade plays a role in establishment of the tonotopic gradient and is interlinked with the mechanism governing cellular polarity in the orthogonal direction.
Supplementary Material
RESEARCH HIGHLIGHTS.
Hair cells in the cochlea are arranged by frequency, forming the tonotopic gradient.
We identified genes differentially expressed in high- and low-frequency regions.
Three signaling pathways were represented in results, including the notch pathway.
DNER, a notch ligand, and PTPζ, a receptor phosphatase, localize in a polarized way.
Interference with DNER expression causes hair-bundle and orientation defects.
Acknowledgments
The authors thank Ms. J. McCarthy, Dr. S. Miranda-Rottman, Mr. S. Pylawka, and Dr. M. Vologodskaia for their contributions to the array studies and cloning. We are grateful to Dr. C. Cepko for the RCAS-GFP vector, Dr. S. Hughes for the RCASBP(A) vector, and Dr. G. Richardson for monoclonal antibody D37. The members of our research group, and especially Dr. J. A. N. Fisher, Dr. M. Gleason, Mr. S. Patel, and Dr. J. Schwarz, as well as Dr. H. Lopez-Schier provided helpful comments on the manuscript. This research was supported by grant DC000241 from the National Institutes of Health. L. K. was the recipient of a fellowship from Boehringer Ingelheim Fonds; A. J. H. is an Investigator of Howard Hughes Medical Institute
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschelet E. Circular Statistics in Biology. London: Academic Press; 1981. [Google Scholar]
- Beisel KW, Rocha-Sanchez SM, Ziegenbein SJ, Morris KA, Kai C, Kawai J, Carninci P, Hayashizaki Y, Davis RL. Diversity of Ca2+-activated K+ channel transcripts in inner ear hair cells. Gene. 2007;386:11–23. doi: 10.1016/j.gene.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM, Nolan PM, Steel KP, Brown SD, Gray IC, Murdoch JN. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol. 2003;13:1129–1133. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–551. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- Davies A, Formstone C, Mason I, Lewis J. Planar polarity of hair cells in the chick inner ear is correlated with polarized distribution of c-flamingo-1 protein. Dev Dyn. 2005;233:998–1005. doi: 10.1002/dvdy.20376. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Fettiplace R, Fuchs PA. Mechanisms of hair cell tuning. Annu Rev Physiol. 1999;61:809–834. doi: 10.1146/annurev.physiol.61.1.809. [DOI] [PubMed] [Google Scholar]
- Fukazawa N, Yokoyama S, Eiraku M, Kengaku M, Maeda N. Receptor type protein tyrosine phosphatase zeta-pleiotrophin signaling controls endocytic trafficking of DNER that regulates neuritogenesis. Mol Cell Biol. 2008;28:4494–4506. doi: 10.1128/MCB.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear RJ, Legan PK, Wright MB, Marcotti W, Oganesian A, Coats SA, Booth CJ, Kros CJ, Seifert RA, Bowen-Pope DF, Richardson GP. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci. 2003;23:9208–9219. doi: 10.1523/JNEUROSCI.23-27-09208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney CM, Mahendrasingam S, Jones EM, Fettiplace R. The distribution of calcium buffering proteins in the turtle cochlea. J Neurosci. 2003;23:4577–4589. doi: 10.1523/JNEUROSCI.23-11-04577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Nelson BR, Reh TA, Bermingham-McDonogh O. Delta/Notch-Like EGF-Related Receptor (DNER) is expressed in hair cells and neurons in the developing and adult mouse inner ear. J Assoc Res Otolaryngol. 2010;11:187–201. doi: 10.1007/s10162-009-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller S, Bell AM, Denis CS, Choe Y, Hudspeth AJ. Parvalbumin 3 is an abundant Ca2+ buffer in hair cells. J Assoc Res Otolaryngol. 2002;3:488–498. doi: 10.1007/s10162-002-2050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley M, Rhodes C, Kneebone A, Herde MK, Fleming M, Steel KP. Emx2 and early hair cell development in the mouse inner ear. Dev Biol. 2010;340:547–556. doi: 10.1016/j.ydbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudspeth AJ, Lewis RS. A model for electrical resonance and frequency tuning in saccular hair cells of the bull-frog, Rana catesbeiana. J Physiol. 1988;400:275–297. doi: 10.1113/jphysiol.1988.sp017120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- Kruger RP, Goodyear RJ, Legan PK, Warchol ME, Raphael Y, Cotanche DA, Richardson GP. The supporting-cell antigen: a receptor-like protein tyrosine phosphatase expressed in the sensory epithelia of the avian inner ear. J Neurosci. 1999;19:4815–4827. doi: 10.1523/JNEUROSCI.19-12-04815.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Gong M, Huang YN, Tang ZQ, Chen L. Morphometry of otoliths in chicken macula lagena. Neurosci Lett. 2006;404:83–86. doi: 10.1016/j.neulet.2006.05.046. [DOI] [PubMed] [Google Scholar]
- López-Schier H, Hudspeth AJ. A two-step mechanism underlies the planar polarization of regenerating sensory hair cells. Proc Natl Acad Sci USA. 2006;103:18615–18620. doi: 10.1073/pnas.0608536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley GA. Peripheral Hearing Mechanisms in Reptiles and Birds. Berlin: Springer; 1990. [Google Scholar]
- Martinez-Dunst C, Michaels RL, Fuchs PA. Release sites and calcium channels in hair cells of the chick’s cochlea. J Neurosci. 1997;17:9133–9144. doi: 10.1523/JNEUROSCI.17-23-09133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Rottmann S, Kozlov AS, Hudspeth AJ. Highly specific alternative splicing of transcripts encoding BK channels in the chicken’s cochlea is a minor determinant of the tonotopic gradient. Mol Cell Biol. 2010;30:3646–3660. doi: 10.1128/MCB.00073-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. BioTechniques. 2008;45:613–614. 616, 618. doi: 10.2144/000113005. passim. [DOI] [PubMed] [Google Scholar]
- Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J Biol Chem. 2006;281:13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- Navaratnam DS, Escobar L, Covarrubias M, Oberholtzer JC. Permeation properties and differential expression across the auditory receptor epithelium of an inward rectifier K+ channel cloned from the chick inner ear. J Biol Chem. 1995;270:19238–19245. doi: 10.1074/jbc.270.33.19238. [DOI] [PubMed] [Google Scholar]
- Oldberg A, Franzén A, Heinegård D. Cloning and sequence analysis of rat bone sialoprotein (osteopontin) cDNA reveals an Arg-Gly-Asp cell-binding sequence. Proc Natl Acad Sci USA. 1986;83:8819–8823. doi: 10.1073/pnas.83.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan K, Michael TH, Jiang GJ, Hiel H, Fuchs PA. A molecular mechanism for electrical tuning of cochlear hair cells. Science. 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- Ricci AJ, Crawford AC, Fettiplace R. Tonotopic variation in the conductance of the hair cell mechanotransducer channel. Neuron. 2003;40:983–990. doi: 10.1016/s0896-6273(03)00721-9. [DOI] [PubMed] [Google Scholar]
- Rosenblatt KP, Sun ZP, Heller S, Hudspeth AJ. Distribution of Ca2+-activated K+ channel isoforms along the tonotopic gradient of the chicken’s cochlea. Neuron. 1997;19:1061–1075. doi: 10.1016/s0896-6273(00)80397-9. [DOI] [PubMed] [Google Scholar]
- Samaranayake H, Saunders JC, Greene MI, Navaratnam DS. Ca2+ and K+ (BK). channels in chick hair cells are clustered and colocalized with apical-basal and tonotopic gradients. J Physiol. 2004;560:13–20. doi: 10.1113/jphysiol.2004.069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee ME, Lawton DM, Furness DN, Benke TA, Ricci AJ. Auditory hair cell-afferent fiber synapses are specialized to operate at their best frequencies. Neuron. 2005;47:243–254. doi: 10.1016/j.neuron.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Takarada T, Yoneda Y. Transactivation by Runt related factor-2 of matrix metalloproteinase-13 in astrocytes. Neurosci Lett. 2009;451:99–104. doi: 10.1016/j.neulet.2008.12.037. [DOI] [PubMed] [Google Scholar]
- Thomas G, Moffatt P, Salois P, Gaumond MH, Gingras R, Godin E, Miao D, Goltzman D, Lanctôt C. Osteocrin, a novel bone-specific secreted protein that modulates the osteoblast phenotype. J Biol Chem. 2003;12:50563–50571. doi: 10.1074/jbc.M307310200. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Saunders JC. Actin filaments, stereocilia, and hair cells of the bird cochlea. I. Length, number, width, and distribution of stereocilia of each hair cell are related to the position of the hair cell on the cochlea. J Cell Biol. 1983;96:807–821. doi: 10.1083/jcb.96.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Cotanche DA, Tilney MS. Actin filaments, stereocilia and hair cells of the bird cochlea. VI. How the number and arrangement of stereocilia are determined. Development. 1992;116:213–226. doi: 10.1242/dev.116.1.213. [DOI] [PubMed] [Google Scholar]
- von Békésy G. In: Experiments in Hearing. Wever EG, editor. New York: McGraw-Hill; 1960. [Google Scholar]
- Woolley SMN, Rubel EW. Vocal memory and learning in adult Bengalese Finches with regenerated hair cells. J Neurosci. 2002;22:7774–7787. doi: 10.1523/JNEUROSCI.22-17-07774.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Muramatsu H, Sone M, Hayashi H, Nakashima T, Muramatsu T. Mice doubly deficient in the midkine and pleiotrophin genes exhibit deficits in the expression of beta-tectorin gene and in auditory response. Lab Invest. 2006;86:645–653. doi: 10.1038/labinvest.3700428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.