Abstract
Communication between different brain regions, and between local circuits in the same brain region, is an important area of study for basic and translational neuroscience research. Selective and chronic manipulation of one of the components in a given neural pathway is frequently required for development and plasticity studies. We designed an in vitro platform that captures some of the complexity of mammalian brain pathways but permits easy experimental manipulation of their constituent parts. Organotypic cultures of brain slices were carried out in compartments interconnected by microchannels. We show that cocultures from cortex and hippocampus formed functional connections by extending axons through the microchannels. We report synchronization of neural activity in cocultures, and demonstrate selective pharmacological manipulation of activity in the constituent slices. Our platform enables chronic, spatially-restricted experimental manipulation of pre- and post-synaptic neurons in organotypic cultures, and will be useful to investigators seeking to understand development, plasticity, and pathologies of neural pathways.
Introduction
The information processing circuits of the mammalian brain are formed by synaptic connections between neurons. Understanding the processes that guide the development of these synaptic circuits may provide new insights into the pathophysiology of important disorders of the central nervous system. Examples include limbic system pathways between prefrontal cortex, hippocampus, amygdala, and hypothalamus, important in the research into causes of psychiatric disorders1, 2, and more local pathways linking sub-regions of the hippocampus, which are of importance for understanding mechanisms of learning and memory3 and pathophysiology of epilepsy4, 5.
A number of signaling cascades are known to be important in axon guidance and targeting6–8. Activation of axonal pathways also plays an important role in the development of brain circuitry9–12, and abnormal patterns of activity have been hypothesized to play a role in the pathogenesis of neurological disorders2, 13–15. Understanding the relationships between neural activity and synaptogenesis is thus important for gaining insight into the normal development of the brain and its pathologies.
Research into circuit-to-circuit communication frequently requires chronic manipulation of one or more components in a given neural pathway. A variety of in vivo approaches 16–24 have generated a wealth of information. However, there are methodological limitations to in vivo experiments, primarily concerning selective and controlled manipulation of well-defined portions of the overall network. This limitation can be overcome through the use of in vitro culture systems, in which exquisite chronic pharmacological control of the activity of networks of neurons is possible. We have therefore focused on the design of an in vitro platform that captures some of the complexity of mammalian brain pathways but permits easy experimental manipulation of the pathway components. Our platform will complement in vivo studies of circuit-to-circuit axonal pathways, their activity, and their role in development.
We modified and combined a number of techniques – microfabrication, organotypic tissue culture, and microfluidics – to create a platform that permits selective and chronic pharmacological manipulation of interconnected local circuits. We have elected to use brain slice cultures (organotypic cultures) because these organotypic cultures retain circuit architecture and physiology specific for their source region, and come as close to the complexity of the brain as is possible in an in vitro culture system 25–27. Extensive axon sprouting and synaptogenesis occur in organotypic cultures of brain slices. This property of slice cultures has been exploited to create co-culture models of thalamocortical28–30, entorhinal cortex-hippocampal31, 32, and septo-hippocampal projections33. Unfortunately, the pharmacological manipulations in these experiments were limited to bath applications of drugs to the entire network34, 35. To achieve pharmacological selectivity, we adopted the method of creating compartmented cultures, or ‘Campenot’ chambers36, to organotypic brain slice cultures. In the original ‘Campenot’ chambers, neurons were placed in plastic compartments, which were sealed against the culture substrate. Axons could extend from one compartment to another while neural soma were restricted to their original compartment; in this manner, the influence of growth factors and other compounds on axon growth has been investigated. This method has been updated recently for use with central nervous system (CNS) neurons with a soft lithography37, 38 based microfabrication technique, and applied to the study of axonal injury and regeneration39.
Here, we applied a method to maintain organotypic slices in soft-lithography defined chambers to create an in vitro platform where brain slices are cultured in compartments interconnected by microchannels. Slices formed functional connections with each other by extending axons through the microchannels, while pharmacological isolation was maintained by varying the levels of fluid in the two compartments. We cultured pairs of hippocampal slices to create a model of axon sprouting in a sub-region of the hippocampal formation, CA1. Extensive sprouting of axon collaterals has been repeatedly observed in area CA1 in epileptic tissue, and this sprouting is hypothesized to cause hyperexcitability in epilepsy40–42. We also demonstrate that our platform can be used to study pathways between other brain regions, by creating connected co-cultures of hippocampus and entorhinal cortex. In principle, slices from any brain region that extend axons in vitro (including thalamus and many cortical regions in addition to entorhinal cortex and the hippocampus) can be used as building blocks to create interconnected, but pharmacologically isolated co-cultures. We demonstrate selective, acute and chronic pharmacological manipulation of activity in the hippocampal slices linked by sprouted CA1 axons. Our platform enables independent experimental manipulation of axonally-linked circuits, and will be useful to investigators seeking to understand development, function, and pathologies of axonal pathways in the brain.
Experimental
A. PDMS mini-wells
Cross-linked polydimethylsiloxane (PDMS, sold as Sylgard 184 by Dow Corning) was used to fabricate slice culture wells patterned with microchannels via modified soft lithography. As the first step in the fabrication sequence, silicon mold masters were prepared by defining a negative relief of microchannel pattern via SU-8 (5) (Microchem) photolithography on a 4″ diameter silicon wafer. SU-8 patterns of 3 μm height, 50 μm width, and 100 μm center-to-center spacing (corresponding to microchannel depth, width, and spacing, respectively) were fabricated. Liquid PDMS was spin-coated onto the silicon master at 500 rpm, and cured (cross-linked) at 70°C for at least 4 hours. The resulting flexible 120 μm Sylgard membrane bearing imprinted microchannels was removed from the silicon wafer. Two mini-wells with dimensions of 1.8 × 1.6 mm, separated by 200–600 μm PDMS spacer containing connecting microchannels were cut in the same PDMS membrane. The center region of 35 mm tissue-culture grade Petri dishes was coated with 200 μl of 1 mg/ml sterile solution of poly-D-lysine (PDL, Sigma) in borate buffer at pH 8.5. The Petri dishes were incubated in PDL in humidified atmosphere at 37 °C overnight, washed in three changes of sterile deionized water, and dried. Pre-cut Sylgard mini-wells were sterilized in 100% ethanol, dried, and sealed against the PDL-coated center region of 35 mm Petri dishes. The watertight seal between Sylgard and polystyrene dish surface forms spontaneously on contact, provided that both surfaces are thoroughly cleaned, and stays stable in aqueous solutions for several weeks. The dishes were then filled with 1.5 ml of serum-free medium (see below), and incubated at 37 °C overnight to allow medium to fill the microchannels and remove any soluble impurities from PDMS.
B. Organotypic cultures
Slice cultures of rat hippocampus and entorhinal cortex were prepared as described in our previous work43. Briefly, we cut 350 μm hippocampal or cortical slices using McIlwain tissue chopper under sterile conditions, and placed the slices onto poly-D-lysine coated 35 mm Petri dishes with polydimethylsiloxane (PDMS) mini-wells and microchannels. Cultures were kept in medium containing 25% horse serum, Basal Eagle Medium, and Hank’s Balanced Salt Solution in a 1:2:1 proportion with 1mM glutaMAX (glutamine substitute, Invitrogen) and 30 μg/ml gentamicin for 24 hours, and then swithched to serum-free, chemically defined NeurobasalA/B27 medium (Invitrogen) containing 0.5 mM glutaMAX and 30 μg/ml gentamicin for the rest of the culture period. Medium changes were carried out every three days.
C. Chronic drug application
We included 2 mM kynurenic acid in the culture medium, and placed 100 μl of kynurenate-containing medium per well on one side of the dish, and 150 μl of regular medium per well on the other side, to maintain a pressure gradient-dependent flow for pharmacological isolation. Kynurenate-containing and regular media were changed every 2 days.
D. Electrophysiology
Petri dishes with organotypic cultures were removed from the tissue culture incubator and transferred to a recording chamber kept at 36°C, with a humidified atmosphere of 95% air/5% CO2. Recordings were carried out in an interface configuration, with two separate perfusion channels supplying ACSF and drugs to each slice of co-culture independently. We used two tungsten microelectrodes placed into CA1 areas of respective slices in co-cultures. Signals were detected with pre-amplifiers and a multi-channel amplifer (Dagan Corporation), digitized, and recorded with custom-designed software dClamp (© Staley Laboratory).
E. Immunohistochemistry
We placed a crystal of FluoroRuby (Invitrogen) on CA1 of one of the slices in the co-culture, and placed the dishes back in the incubator overnight for retrograde transport of the dye. We then washed cultures in ACSF, fixed in 4% paraformaldehyde for 2 hours at room temperature, and permeabilized for 4 hours in 0.3% Triton X-100. The slices were blocked in 10% goat serum, and primary antibodies to NeuN (rabbit Alexa Fluor 488 conjugated IgG, Microchem), and tetramethylrhodamine (mouse IgG, Microchem) were applied at 1:100 dilutions for 3 days at 4°C. Slices were then washed, and secondary goat anti-mouse Alexa Fluor 546 conjugated antibody (Invitrogen) was applied at 1:200 dilution for 3 days at 4°C. Slices were then washed and mounted. We recorded epifluorescent images with Nikon microscope at 4x magnification and confocal images at 20x magnification with Zeiss LSM 5 Pascal laser confocal microscope. Images were processed with ImageJ (NIH) software.
F. Data Analysis and Statistics
Burst initiation delays were determined manually in dClamp, by measuring the time difference between points at which the signal exceeded two times the standard deviation of baseline noise. For cross-correlation analysis, data were imported into Matlab, filtered with a 4th order high-pass Butterworth filter (cut-off frequency = 4 Hz), re-sampled at 1 kHz, and cross-correlated using phase delays of +/− 1 second. The resulting coefficients were normalized by dividing with the square root of the product of each channels’ auto-correlation coefficient at delay of 0 to derive r values. Mean values and standard deviations of maximum r values of recorded co-cultures were then calculated for each experimental group (number of days in vitro). Significance of the differences between cross-correlation coefficients was analyzed with Kruskal-Wallis non-parametric test, with Dunn’s post-hoc analysis. Variance of burst timing was calculated with F-test, and statistical significance of the burst timing differences was determined by t-test for equal variance.
Results and discussion
A. Compartmented co-cultures of organotypic slices
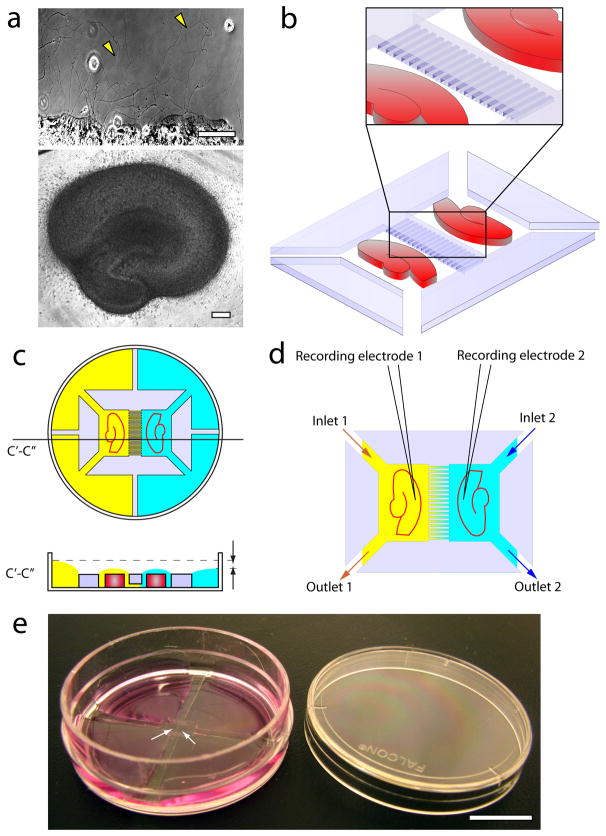
Organotypic co-cultures of entorhinal cortex and hippocampus form functional slice-to-slice synapses via extended axons (Supplementary Fig. 1, Supplementary Methods). Neurons in both hippocampal and entorhinal cortex slices, cultured in polydimethylsiloxane (PDMS) mini-wells, readily extend axons on growth-permissive substrate such as polylysine-coated tissue culture plastic within first few days in vitro (DIV) (Fig. 1a, Supplementary Fig. 2a, b), sprouting 1 mm or more from the slice border within the first week of culture. Microfabrication using “soft lithography” enabled us to fabricate slice compartments with a fluid barrier containing microchannels for axon growth (Fig. 1b, Fig. 2a). The PDMS compartments were then placed in a standard tissue culture dish that was pre-coated with polylysine (Fig. 1e). PDMS, a very hydrophobic polymer, repels water-based culture medium and causes it to bead on the hydrophilic polylysine-coated polystyrene. In our design, hydrophobic medium-free PDMS surrounds the slice compartment, creating a droplet of culture medium in which the slice is maintained at air-liquid interface. The slice compartment is connected by inlet and outlet channels (cut through the PDMS, and 300 – 400 μm wide where they join the slice compartment) to larger medium reservoirs as shown in Fig. 1c. Fluid isolation is maintained by adjusting fluid levels in the larger reservoirs. The resulting pressure difference between two sides of the dish causes a slow fluid flow through axon-containing microchannels. This counteracts diffusion, and permits us to confine drugs or other molecules to the compartment with the lower fluid level for the duration of culture (Fig. 1c). We kept the channel height under 5 μm to increase fluid resistance and maintain a fluid level difference between compartments over at least 3 days. We found that adding 150 μl of medium per reservoir on one side of the dish, and 100 μl of medium per reservoir on the other side, for a total difference of 100 μl between left and right side, provides sufficient pressure differential to counter diffusion through microchannels. For electrical recordings of activity, we superfused the slices separately using two inlet and outlet channels (Fig. 1d). Fluid level adjustment was not necessary during recording, since the fluid flow rate was significantly faster than the diffusion rate through microchannels.
Fig. 1.
Compartmented organotypic slice culture platform. (a) Organotypic hippocampal culture at 5 DIV (bottom), scale bar is 200 μm; axons and dendrites are extended by neurons within the slice beyond the slice border (top), arrows point to axons, scale bar is 100 μm, (b) Schematic of the hippocampus-hippocampus co-culture in compartments cut in a thick (150 μm) PDMS film. Microchannels (5 μm high, 50 μm wide) are imprinted into PDMS barrier separating the two slices via soft lithography. (c) PDMS compartments in a 35 mm Petri dish. The culture dish is separated into four quadrants with PDMS barriers, isolating solutions on the left and the right sides of the dish. During incubation, higher fluid level on the left side of the dish creates a pressure-driven flow from left to right, preventing diffusion of pharmaceuticals in the opposite direction. (d) During recording, extracellular electrodes are placed into both slices, which are superfused through separate fluid inlets and outlets. (e) Photograph of the culture platform (without the slices), consisting of microchannel-linked PDMS compartments and medium reservoirs built into a 35 mm Petri dish. The dish is filled with culture medium. Arrows point to the slice compartments. Scale bar is 10 mm.
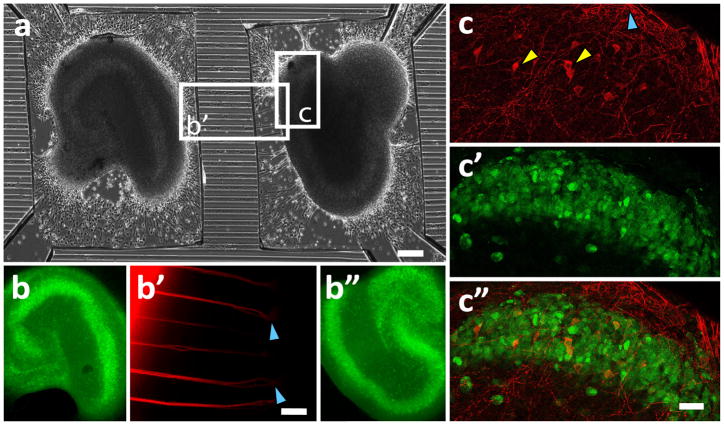
Fig. 2.
Morphology of organotypic slices and axonal connections. (a) Phase contrast micrograph of two hippocampus slices in PDMS compartments at 18 DIV. Sidewalls of microchannels imprinted into the PDMS appear as bright horizontal lines. The channels are 50 μm wide with 50 μm channel-to-channel intervals, scale bar is 200 μm. (b, b″) The morphology of slices maintained in this preparation is typical of hippocampal organotypic cultures, including intact CA1, CA3, and DG, readily observed with anti-NeuN immunohistochemistry. (b′) The axons in microchannels, poorly visible with phase contrast optics, are brightly stained with DiI (red), crystals of DiI were placed on the left slice. Blue arrows show the point where axons split and enter the right slice. Scale bar is 50 μm. (c) FluoroRuby (red), a retrograde tracer dye, was applied to CA1 of one of the slices in the co-culture. The dye was transported in the retrograde directions through axons in microchannels, into the neurons (yellow arrows) in the other slice. Blue arrow shows axons exiting the slice toward the microchannels. (c′) The slice was counterstained with anti-NeuN (green) to reveal the position of the CA1 pyramidal layer. (c″) FluoroRuby-positive neurons were located in the CA1 pyramidal layer. Scale bar is 50 μm.
Slices of rat hippocampus and entorhinal cortex were maintained in the compartmented dishes for up to 4 weeks. The slices maintained the layered organization of their source regions. We carried out anti-NeuN (specific for neural nuclei and soma) immunohistochemistry on hippocampal slices shown in Fig. 2a, and found well-defined areas CA1, CA3, and dentate gyrus (Fig. 2b). Axon outgrowth could be observed after three days in vitro (Fig. 1a, Supplementary Fig. 2a,b), with multiple axons extending through the channels and to the co-cultured slice by 10–14 DIV. To aid with the visualization of axons in microchannels, we applied crystals of membrane tracer dye, DiI, to one of the slices (left slice in Fig. 2a, axons are shown in Fig. 2b′). We have also used a retrograde tracer, FluoroRuby, applied as a crystal to one of the slices in co-culture, to reveal the neurons with slice-to-slice axons. Most of these neurons were found in the CA1 area (Fig. 2c-c″, FluoroRuby (red) and anti-NeuN (green)) (n = 3 co-cultures).
B. Selective pharmacological control of neural activity
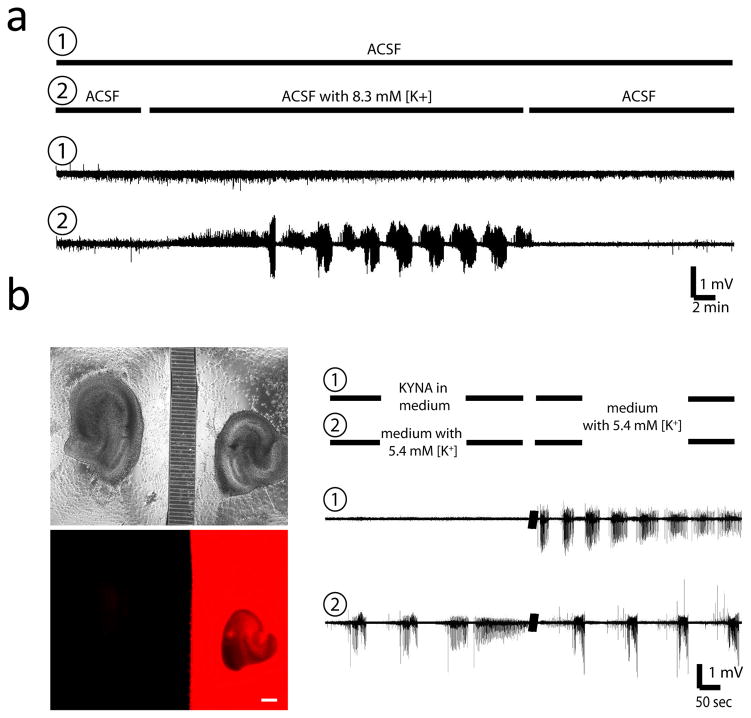
We tested whether we could control the neural activity in the co-cultured slices independently, by varying the composition of recording or culture medium in fluidically isolated compartments. We transiently superfused one of the slices in the co-culture with artificial cerebrospinal fluid (ACSF) containing a high K+ concentration, while the other slice in the co-culture was continuously superfused with regular ACSF (Fig. 3a). High K+ concentration in the recording solution depolarized neurons and caused epileptiform discharges in slice 2. The slice activity returned to normal, including single unit activity and small-magnitude bursts following a wash-out period with regular ACSF. The activity in slice 1 stayed constant throughout the recording. This experiment demonstrated that the fluidic isolation imposed by the microchannel-containing barrier between the two slice compartments was sufficient to isolate the slices pharmacologically during two-channel perfusion. To test chronic fluid isolation (under static culture conditions), we added either rhodamine (red fluorescent dye) or kynurenic acid (KYNA, a glutamate receptor antagonist that prevents most excitatory neurotransmission) to the culture medium of one of the slices in the co-cultured slice pairs. A higher volume (+ 100 μl) of unmodified culture medium was added to the other slice to set up a pressure-driven flow to counteract the diffusion of rhodamine or kynurenate. After two days, we verified fluidic isolation by checking rhodamine diffusion between compartments, and finding no rhodamine fluorescence in the compartment with higher medium volume (Fig. 3b). We recorded the levels of spontaneous activity in the co-cultures to check for diffusion of kynurenic acid. The slice with KYNA in the culture medium had no spontaneous activity requiring glutamatergic synapses, as expected, while the slice in regular medium exhibited high levels of spontaneous activity (Fig. 3c) characteristic of organotypic hippocampal cultures recorded in slightly elevated potassium44, 45 (our culture medium, NeurobasalA/B27, contains 5.4 mM [K+]). We then recorded spontaneous activity in two slices after replacing the original medium with fresh culture medium. Spontaneous epileptiform discharges appeared in slice 1, while the activity level in slice 2 was not significantly changed, signifying lack of kynurenate diffusion from compartment 1 to compartment 2 under static culture conditions (n = 6 co-cultures).
Fig. 3.
Acute and chronic fluidic isolation. (a) Slices in co-culture at 12 DIV were superfused through separate fluid channels as indicated. Slice 1 activity level stayed constant, while slice 2 developed epileptiform discharges when superfused with high [K+] containing ACSF. Activity in slice 2 returned to normal after [K+] washout. Co-cultures at 12 DIV are not yet functionally connected. (b) Phase contrast (top) and fluorescent (bottom) micrographs of the co-culture 2 days after rhodamine application to the right compartment. Rhodamine fluorescence (red) is contained in the right compartment, with no observed diffusion through the microchannels. Scale bar is 200 μm, (c) Co-culture with regular culture medium (including 5.4 mM [K+]) in compartment 2, and medium containing 2mM kynurenic acid in compartment 1, recorded at 10 DIV. No spontaneous activity is observed in slice 1, while slice 2 exhibits characteristic burst activity, indicating no diffusion of kynurenic acid from compartment 1 to compartment 2. When fresh medium, with no kynurenic acid, is placed on both slices, both exhibit spontaneous bursting.
C. Synchronization of activity in co-cultures
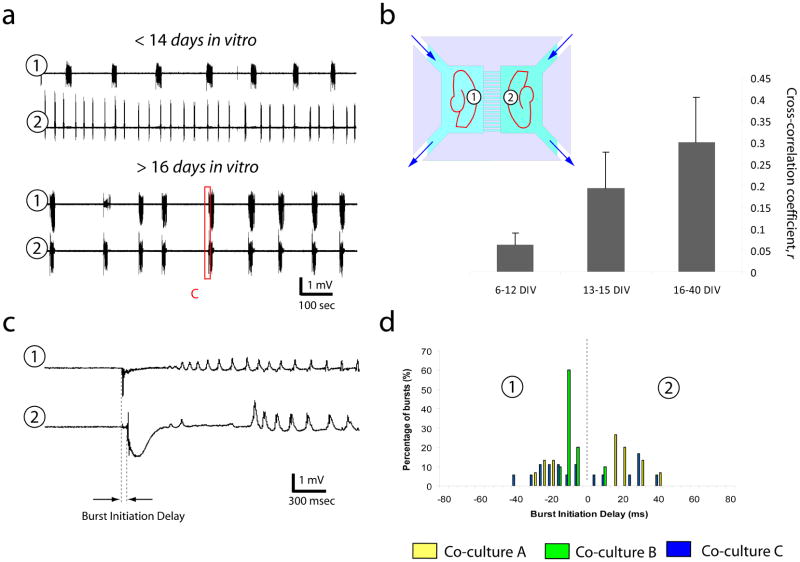
We then examined whether the axons connecting the slices through a microchannel-perforated barrier could synchronize the two-slice network. We recorded the cultures in the ACSF containing GABAA and GABAB antagonists, picrotoxin (Sigma) and CGP 55845 (Tocris Bioscience) (Fig. 4b, inset schematic), and observed no synchronization prior to 14 DIV (Fig. 4a, upper trace, n = 7 co-cultures). However, after 16 DIV, bursts in two connected slices became highly synchronized (Fig. 4a, lower trace, n = 7 hippocampal-hippocampal co-cultures, also Supplementary Figure 2c, n = 2 entorhinal-hippocampal co-cultures). Slices recorded between 14 and 16 DIV exhibited a mixture of synchronized and unsynchronized activity (n = 5 co-cultures). We calculated cross-correlation coefficients for activity in slice pairs at different culture ages, and plotted the results in Fig. 4b. We also measured delays in burst onset between co-cultured slices (Fig. 4c) to determine whether one, or both, of the slices were the sites of burst initiation. The results for three of the co-cultures are plotted in Fig. 4d. The presence of positive and negative delays in each co-culture signifies that the initiating site alternated between the two slices during the recording.
Fig. 4.
Synchronization of activity in co-cultures. (a) Activity is recorded with extracellular electrodes in positions indicated by (1) and (2) while both slices are perfused with ACSF containing GABAA and GABAB antagonists (b, inset). Top trace shows a typical example of activity recorded in co-cultures at less than 14 DIV, with no synchronization of bursts between the two slices. Bottom trace shows typical recording from co-cultures older than 16 DIV. Burst timing is synchronized in slice 1 and slice 2. (b) Mean values of the cross-correlation coefficients between slice 1 and slice 2 are plotted for cultures at 6–12 DIV (n = 7 co-cultures), 13–15 DIV (n = 5 co-cultures), and 16–40 DIV (n = 7 co-cultures). Difference between r is very significant between age groups (p = 0.0011, Kruskal-Wallis test, values of r are very significantly different between 6–12 DIV and 16–40 DIV, p = 0.0006, Dunn’s post-hoc analysis). Error bars correspond to standard deviation from the mean cross-correlation coefficient. (c) Burst initiation delay was measured as the difference between the burst onset in slice 1 and slice 2. (d) Histogram of the burst initiation delays in 3 co-cultures. Negative delays indicate initiation in slice 1, while positive delays indicate initiation in slice 2.
During the past two decades, co-cultures of organotypic slices have been used to create realistic in vitro models of the development of thalamocortical and perforant path axonal projections28–32. However, these models do not allow separate experimental manipulation of the individual slices in co-cultures, something that would be of great use in addressing a number of important questions in the study of normal and pathological neural development.
We have found that we did not need to place slices in physical contact with each other in order to establish axonal interconnections. This enabled us to construct, via microfabrication of fluidically isolated compartments, a co-culture platform where slices could be individually manipulated. The number of long (> 1 mm) axons that extended through microchannels in the fluid barrier was high enough to synchronize network activity in two slices, when recorded in the presence of GABA receptor antagonists. We observed a delay of 4 to 6 days between the time that axons have extended from one slice to another, and the onset of synchronization. This was likely the time required for axon elaboration within the slice and the formation of synapses. Axonal connections in our cultures were bidirectional; each slice was capable of initiating synchronized bursts as evidenced by positive and negative delays in Fig. 4d.
The level of activity synchronization between the two slices, measured with extracellular electrodes, represents a feasible assay of the physiological significance of sprouted axons and newly formed synapses. Our platform enables a number of experimental manipulations, including long-term incubation of one of the slices with various pharmaceuticals to determine the effects on formation of new connections, and acute applications of drugs to one or both slices to test the effect on activity synchronization. We have demonstrated an example of long term application of kynurenic acid (Fig. 3b), effectively achieving suppression of spontaneous activity in one of the slices, but permitting spontaneous bursting in the other, throughout the time of culture. This makes possible experiments that test the role of spontaneous activity and formation of connections between neurons during normal development.
In our platform, neurons that extend axons and their post-synaptic targets are pharmacologically isolated from each other; therefore, the activity of pre- and post- synaptic neurons can be modulated independently, which is of great interest to investigation of various forms of long-term synaptic plasticity46. Finally, pathological axonal sprouting and formation of excessive connections in epilepsy are hypothesized to be influenced by epileptiform activity47. Chronic pharmacological isolation of a portion of connected network, achieved in our platform, will permit studies of the relative role of synaptic activity in inducing sprouting versus strengthening and stabilizing excessive connections.
Supplementary Material
Acknowledgments
We thank Helen Sabolek for helpful discussion and for proof-reading the manuscript. This work was supported in part by NIH Grant P41EB002503. Y.B. was supported by NIH F32MH079662.
References
- 1.de Kloet ER, Joels M, Holsboer F. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 2.Herman JP, Cullinan WE. Trends Neurosci. 1997;20:78–84. doi: 10.1016/s0166-2236(96)10069-2. [DOI] [PubMed] [Google Scholar]
- 3.Bliss T, Collingridge G, Morris R. In: The Hippocampus Book. Andersen P, Morris R, Amaral D, Bliss T, O’Keefe J, editors. Oxford University Press; New York: 2007. pp. 343–474. [Google Scholar]
- 4.Scheibel ME, Crandall PH, Scheibel AB. Epilepsia. 1974;15:55–80. doi: 10.1111/j.1528-1157.1974.tb03997.x. [DOI] [PubMed] [Google Scholar]
- 5.Sutula T, Cascino G, Cavazos J, Parada I, Ramirez L. Ann Neurol. 1989;26:321–330. doi: 10.1002/ana.410260303. [DOI] [PubMed] [Google Scholar]
- 6.Tessier-Lavigne M, Goodman CS. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 7.Dickson BJ. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto N, Tamada A, Murakami F. Prog Neurobiol. 2002;68:393–407. doi: 10.1016/s0301-0082(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 9.Innocenti GM, Price DJ. Nat Rev Neurosci. 2005;6:955–965. doi: 10.1038/nrn1790. [DOI] [PubMed] [Google Scholar]
- 10.Katz LC, Shatz CJ. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 11.Zhang LI, Poo MM. Nat Neurosci. 2001;4(Suppl):1207–1214. doi: 10.1038/nn753. [DOI] [PubMed] [Google Scholar]
- 12.Kerschensteiner D, Morgan JL, Parker ED, Lewis RM, Wong RO. Nature. 2009;460:1016–1020. doi: 10.1038/nature08236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikegaya Y. J Neurosci. 1999;19:802–812. doi: 10.1523/JNEUROSCI.19-02-00802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swann JW, Smith KL, Lee CL. Int Rev Neurobiol. 2001;45:89–118. doi: 10.1016/s0074-7742(01)45007-0. [DOI] [PubMed] [Google Scholar]
- 15.Jankord R, Herman JP. Ann N Y Acad Sci. 2008;1148:64–73. doi: 10.1196/annals.1410.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesel TN, Hubel DH. J Neurophysiol. 1963;26:1003–1017. doi: 10.1152/jn.1963.26.6.1003. [DOI] [PubMed] [Google Scholar]
- 17.Wiesel TN, Hubel DH. J Neurophysiol. 1965;28:1029–1040. doi: 10.1152/jn.1965.28.6.1029. [DOI] [PubMed] [Google Scholar]
- 18.Holtmaat A, Wilbrecht L, Knott GW, Welker E, Svoboda K. Nature. 2006;441:979–983. doi: 10.1038/nature04783. [DOI] [PubMed] [Google Scholar]
- 19.Horton JC, Hocking DR. J Neurosci. 1997;17:3684–3709. doi: 10.1523/JNEUROSCI.17-10-03684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomber SG, Malhotra S. Nat Neurosci. 2008;11:609–616. doi: 10.1038/nn.2108. [DOI] [PubMed] [Google Scholar]
- 21.Shatz CJ, Stryker MP. Science. 1988;242:87–89. doi: 10.1126/science.3175636. [DOI] [PubMed] [Google Scholar]
- 22.Martin JH, Ghez C. J Neurosci Methods. 1999;86:145–159. doi: 10.1016/s0165-0270(98)00163-0. [DOI] [PubMed] [Google Scholar]
- 23.Lodge DJ, Grace AA. J Neurosci. 2008;28:7876–7882. doi: 10.1523/JNEUROSCI.1582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Echegoyen J, Neu A, Graber KD, Soltesz I. PLoS One. 2007;2:e700. doi: 10.1371/journal.pone.0000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gahwiler BH. J Neurosci Methods. 1981;4:329–342. doi: 10.1016/0165-0270(81)90003-0. [DOI] [PubMed] [Google Scholar]
- 26.Stoppini L, Buchs PA, Muller D. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 27.Gahwiler BH, Thompson E, Audinat E, Robertson RT. In: Culturing Nerve Cells. Banker G, Goslin K, editors. MIT Press; Cambridge: 1998. pp. 379–411. [Google Scholar]
- 28.Yamamoto N, Kurotani T, Toyama K. Science. 1989;245:192–194. doi: 10.1126/science.2749258. [DOI] [PubMed] [Google Scholar]
- 29.Bolz J, Novak N, Gotz M, Bonhoeffer T. Nature. 1990;346:359–362. doi: 10.1038/346359a0. [DOI] [PubMed] [Google Scholar]
- 30.Molnar Z, Blakemore C. Exp Neurol. 1999;156:363–393. doi: 10.1006/exnr.1999.7032. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Field PM, Raisman G. Exp Neurol. 1996;142:151–160. doi: 10.1006/exnr.1996.0186. [DOI] [PubMed] [Google Scholar]
- 32.Del Turco D, Deller T. Methods Mol Biol. 2007;399:55–66. doi: 10.1007/978-1-59745-504-6_5. [DOI] [PubMed] [Google Scholar]
- 33.Fischer Y, Gahwiler BH, Thompson SM. J Physiol. 1999;519(Pt 2):405–413. doi: 10.1111/j.1469-7793.1999.0405m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uesaka N, Hirai S, Maruyama T, Ruthazer ES, Yamamoto N. J Neurosci. 2005;25:1–9. doi: 10.1523/JNEUROSCI.3855-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uesaka N, Hayano Y, Yamada A, Yamamoto N. J Neurosci. 2007;27:5215–5223. doi: 10.1523/JNEUROSCI.4685-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campenot RB. Proc Natl Acad Sci U S A. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 38.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 39.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esclapez M, Hirsch JC, Ben-Ari Y, Bernard C. J Comp Neurol. 1999;408:449–460. doi: 10.1002/(sici)1096-9861(19990614)408:4<449::aid-cne1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 41.Smith BN, Dudek FE. J Neurophysiol. 2001;85:1–9. doi: 10.1152/jn.2001.85.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Shao LR, Dudek FE. J Neurophysiol. 2004;92:1366–1373. doi: 10.1152/jn.00131.2004. [DOI] [PubMed] [Google Scholar]
- 43.Berdichevsky Y, Sabolek H, Levine JB, Staley KJ, Yarmush ML. J Neurosci Methods. 2009;178:59–64. doi: 10.1016/j.jneumeth.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBain CJ, Boden P, Hill RG. J Neurosci Methods. 1989;27:35–49. doi: 10.1016/0165-0270(89)90051-4. [DOI] [PubMed] [Google Scholar]
- 45.Dyhrfjeld-Johnsen J, Berdichevsky Y, Swiercz W, Sabolek HR, Staley KJ. Journal of Clinical Neurophysiology. doi: 10.1097/WNP.0b013e3181fe0709. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson SB, Turrigiano GG. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staley KJ, Dudek FE. Epilepsy Curr. 2006;6:199–202. doi: 10.1111/j.1535-7511.2006.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.