Abstract
The myxomycete Physarum polycephalum expresses a calcium-independent nitric oxide (NO) synthase (NOS) resembling the inducible NOS isoenzyme in mammals. We have now cloned and sequenced this, the first nonanimal NOS to be identified, showing that it shares < 39% amino acid identity with known NOSs but contains conserved binding motifs for all NOS cofactors. It lacks the sequence insert responsible for calcium dependence in the calcium-dependent NOS isoenzymes. NOS expression was strongly up-regulated in Physarum macroplasmodia during the 5-day starvation period needed to induce sporulation competence. Induction of both NOS and sporulation competence were inhibited by glucose, a growth signal and known repressor of sporulation, and by l-N6–(1-iminoethyl)-lysine (NIL), an inhibitor of inducible NOS. Sporulation, which is triggered after the starvation period by light exposure, was also prevented by 1H-[1,2,4]oxadiazolo-[4,3-a]quinoxalin-1-one (ODQ), an inhibitor of NO-sensitive guanylate cyclase. In addition, also expression of lig1, a sporulation-specific gene, was strongly attenuated by NIL or ODQ. 8-Bromo-cGMP, added 2 h before the light exposure, restored the capacity of NIL-treated macroplasmodia to express lig1 and to sporulate. This indicates that the second messenger used for NO signaling in sporulation of Physarum is cGMP and links this signaling pathway to expression of lig1.
Keywords: nitric oxide synthase, cGMP, glucose, sporulation, Physarum polycephalum
Physarum polycephalum is a plasmodial slime mold or myxomycete and is also classified as Mycetozoa (Baldauf and Doolittle 1997). It has a life cycle involving both mobile and stationary stages (Raub and Aldrich 1982; Wick and Sauer 1982). Macroplasmodia are giant, single multinuclear cells. They live in the dark on moist organic matter, feed by phagocytosis, and move by creeping. The nuclei undergo naturally synchronous mitosis without cell division. On maturation or when food becomes limited, macroplasmodia differentiate into sporangia, a process that depends on migration toward light. After meiosis, the sporangia release haploid spores, which germinate into flagellated myxamoebae. Myxamoebae—usually from different plasmodia, thus allowing for genetic recombination—can fuse into a zygote, which grows into a new plasmodium. Alternatively, in response to adverse environmental conditions, Physarum plasmodia can encyst into dormant multinucleated macrocysts (or sclerotia). These remain viable for several years. In vitro, plasmodia grown in shaken liquid culture instead of on solid surfaces form so-called microplasmodia, which differentiate into microsclerotia or spherules, analogous structures to the naturally occurring sclerotia. Plasmodia can also reversibly fragment within 5 h when challenged with low temperatures (Kakiuchi and Ueda 1999).
Because nuclear division in the plasmodium is uncoupled from cell division and the unicellular state cannot be overcome, Physarum is considered a ‘lower eukaryote’, although it is clearly more complex than protists. Molecular phylogenetic studies place Physarum together with the cellular slime mold Dictyostelium discoideum and other members of the Mycetozoans among the multicellular eukaryotes, representing a group more closely related to the animal-fungal clade than green plants (Baldauf and Doolittle 1997).
In the developmental cycle of Physarum, the different stages clearly involve differential expression of genes, some of which have been characterized (Bailey 1995; Kroneder 1999). In higher animals, one signaling molecule that has been found to have roles in proliferation, apoptosis, differentiation, and development is nitric oxide (NO; Peunova and Enikolopov 1995; Peunova et al. 1996; Beck et al. 1999; Kim et al. 1999). In previous work, we detected NO synthase (NOS) activity in Physarum and showed that this organism has biosynthetic activities for the production of folic acid as well as of 5,6,7,8-tetrahydrobiopterin (H4-biopterin; Werner-Felmayer et al. 1994), an essential cofactor of all known NOS isoenzymes (Mayer and Hemmens 1997). Biochemically, purified Physarum NOS shares a number of features with inducible NOS (NOS 2) from mammals, the most prominent one being independence from exogenous calcium (Werner-Felmayer et al. 1994). In the present study, we aimed to clarify whether the observed biochemical similarity of the Physarum NOS to the inducible NOS isoenzyme from mammals reflected a high degree of molecular conservation during evolution of this protein. We also wished to define conditions inducing Physarum NOS and, if possible, to identify a biological role for endogenous NO formation.
Results
Analysis of Physarum NOS sequences
Scaling up the purification protocol for Physarum NOS (Werner-Felmayer et al. 1994) allowed the internal peptide sequences to be determined, and primers were designed to recognize either nucleotide sequences deduced from these peptide sequences or consensus regions of other available NOS sequences from mammals, birds, insects, and a mollusc. A 600-bp probe for Physarum NOS was generated by polymerase chain reaction (PCR) and was used for screening a Physarum cDNA library (detailed in Materials and Methods). Two NOS cDNAs, designated physnosa and physnosb, were isolated. Both clones contained complete reading frames for NOS: The 3571-bp physnosa clone (GenBank accession no. AF145041) encoded a 1055 amino acid protein with a calculated molecular mass of 118,180.89 Da; the 3316-bp physnosb clone (GenBank accession no. AF145040) encoded a 1046 amino acid protein of 117,566.52 Da. This agrees with results obtained by denaturing-gel electrophoresis of Physarum NOS purified according to the optimized protocol, which ran at about 120 kD (see Materials and Methods). Physnosb lacked amino acids at positions 726, 1040–1043, and 1050–1055 of physnosa but otherwise shared 82% amino acid identity with physnosa. Northern blot analysis showed that both mRNA species were similarly expressed during cell cycle, spherulation, and sporulation of Physarum (data not shown).
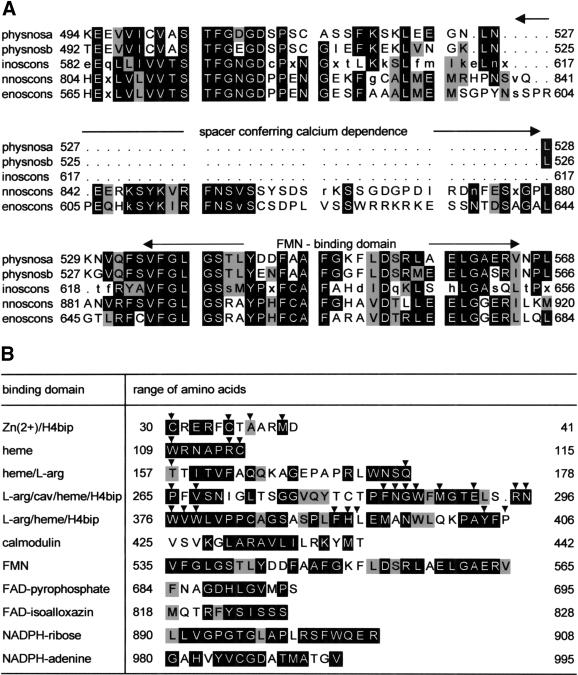
A PILEUP of the deduced protein sequences with consensus sequences of the three NOS isoenzymes (Fig. 1A) shows that like mammalian inducible NOS, both Physarum NOS proteins lacked the spacer sequence responsible for the calcium dependence of neuronal and endothelial NOS from mammals (Salerno et al. 1997). The established binding motifs of mammalian NOSs for FMN, FAD, NADPH, calmodulin (Bredt et al. 1991), heme (Chen et al. 1994), H4-biopterin (Cho et al. 1995; Crane et al. 1998), zinc (Raman et al. 1998; Fishmann et al. 1999), and caveolin (Garcia-Cardena et al. 1997; Crane et al. 1998) were all conserved in both Physarum NOS proteins. The conserved binding domains for physnosa are shown in Figure 1B. Of the 27 residues reported to be in contact with heme, H4-biopterin, and arginine (closer than 3.6 Å; Fischmann et al. 1999), 24 (89%) were conserved in both Physarum NOS cDNAs. One of the replacements, T157, was conservative, leaving only two nonconservative replacements (L393 and P406; Fig. 1B).
Figure 1.
Physarum nitric oxide synthase (NOS) sequences do not contain the spacer sequence conferring calcium dependence and show high conservation of various binding domains. (A) Alignments of Physarum NOS forms a [AF145041] and b [AF145040] with inoscons (consensus sequence of inducible NOSs from man [L09210], rat [L12562], mouse [M87039], guinea pig [AF027180], and chicken [U56540]), with nnoscons (consensus sequence of neuronal NOSs from man [L02881], mouse [D14552], rat [X59949], and rabbit [U91584]), and with enoscons (consensus sequence of endothelial NOSs from man [M95296], mouse [ U53142], bovine [M89952], and pig [U59924]) have been created using the PILEUP program. Only the region containing the spacer conferring calcium dependence and the FMN-binding domain is shown. A postscript file of the complete alignment of physnosa and physnosb with the three isoenzyme consensus sequences is available on request (gabriele.werner-felmayer@uibk.ac.at). Amino acids in inverse mode are conserved in more than three of the compared sequences. Similar amino acids are painted gray. Similarity is assumed for all pairs with positive elements in the amino acid substitution matrix (Henikoff and Henikoff 1992). Capital letters in consensus sequences denote conserved amino acids; small letters, amino acids conserved in a majority of the sequences; and x, a nonconserved amino acid. (B) Parts of the amino acid sequence of physnosa are shown, and conserved residues are displayed as detailed for A. The motifs were assigned according to Bredt et al. (1991) for the reductase domain (downstream position 425). Motifs in the oxygenase domain (upstream position 425) were assigned on the basis of NOS oxygenase domain crystal structures (Crane et al. 1998; Raman et al. 1998; Fischmann et al. 1999). The arrows indicate the 27 residues, which in the NOS oxygenase domain crystal structures are closer than 3.6 Å to the functional groups zinc, heme, H4-biopterin (H4bip), and/or L-arginine (Fischmann et al. 1999). cav denotes the caveolin binding region (Garcia-Cardena et al. 1997).
The overall identity to NOS protein sequences from other species, however, was less than 39%. In accordance with this low degree of identity, antisera raised versus mammalian endothelial, neuronal, or inducible NOS did not stain purified Physarum NOS in Western blots (data not shown). Transient transfection of Sf9 cells by baculovirus carrying the physnosa cDNA, modified at the Kozak motif (see Materials and Methods), yielded a specific activity (measured as formation of 3H-citrulline) of 517 pmol/(mg•min) versus 28 pmol/(mg•min) found in mock-transfected cells. This verifies that the physnosa cDNA encoded a functional NOS.
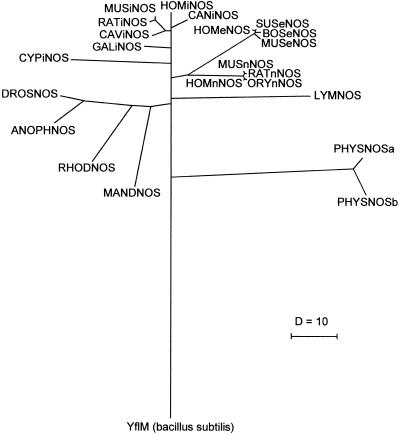
Despite the lack of the spacer sequence responsible for calcium dependence, Physarum NOSs were similarly remote from all three NOS isoenzymes of higher eukaryotes. Phylogenetic tree analysis of a 295-amino-acid, highly conserved region of the oxygenase domain of NOS protein sequences by the DISTANCES program placed Physarum NOS on an individual branch evolving from a low position of the tree (Fig. 2). The distance of Physarum NOSs to NOSs from other species was comparable to the one observed for the not further characterized reading frame designated YflM from Bacillus subtilis, which is homologous to the NOS oxygenase domain (Yamamoto et al. 1997).
Figure 2.
Phylogenetic tree of nitric oxide synthases (NOSs). A 295– amino acid– long, highly conserved region, including the heme-binding domain, was aligned using PILEUP, and this alignment was used to calculate phylogenetic distances using Kimura's method (Kimura 1983) with the DISTANCE program. From the calculated distances, a tree was assembled by GROWTREE. The following sequences (species, accession number) have been included (ordered alphabetically by abbreviation; i is for inducible; n, neuronal; and e, endothelial): ANOPHNOS [Anopheles stephensi, AF053344] CANiNOS [Canis familiaris, AF077821], CAViNOS [Cavia porcellus, AF027190], CYPiNOS [Cyprinos carpio, AJ242906], DROSNOS [Drosophila melanogaster, U25117], GALiNOS [Gallus gallus, U46504], HOMeNOS [Homo sapiens, M95296], HOMiNOS [Homo sapiens, L09210], HOMnNOS [Homo sapiens, L02881], LYMNOS [Lymnaea stagnalis, AF012531], MANDNOS [Manduca sexta, AF062749], MUSeNOS [Mus musculus, U53412], MUSiNOS [Mus musculus, M87039], MUSnNOS [Mus musculus, D14552], ORYnNOS [Oryctolagus cuniculus, U91584], PHYSNOSa [Physarum polycephalum, AF145041], PHYSNOSb [Physarum polycephalum, AF145040], RATiNOS [Rattus rattus, L12562], RATnNOS [Rattus rattus, X59949], RHODNOS [Rhodnius prolixus, U59389], SUSeNOS [Sus scrofa, U59924], YflM [Bacillus subtilis, D86417]. The inserted scale shows the distance of 10 Kimura units.
NOS expression is induced during achievement of sporulation competence
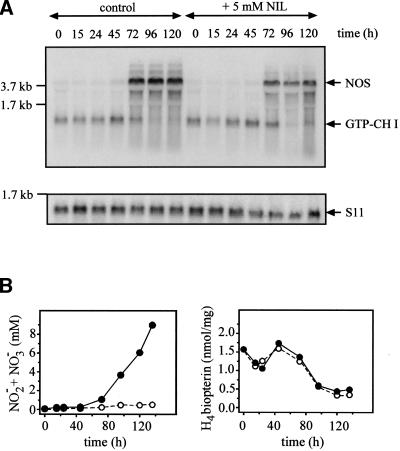
To make them competent for sporulation, Physarum macroplasmodia were starved for 5 d in the dark. During this period, NOS mRNA levels (Fig. 3A) and activity, as determined by accumulation of nitrite plus nitrate (Fig. 3B) were strongly induced from 0.1 mM to 9 mM at the end of the starvation phase (120 h), the time at which the light pulse was set, which then triggered formation of sporangia within the following 14 h. Thus, Physarum NOS is an inducible isoenzyme.
Figure 3.
Nitric oxide synthase (NOS) and GTP cyclohydrolase I expression during starvation and sporulation of Physarum in absence or presence of NIL. (A) Northern blots for NOS, GTP cyclohydrolase I (GTP-CH I), and the ribosomal protein S11 at different times in absence or presence of 5 mM NIL. (B) Data on nitrite plus nitrate accumulation in supernatants and intracellular H4-biopterin levels at different times. Closed symbols with a full line indicate untreated controls; open symbols with a dashed line, results in presence of 5 mM NIL.
We also checked the mRNA expression of GTP cyclohydrolase I, the enzyme of pteridine biosynthesis that can limit the supply of the H4-biopterin cofactor for NOS. In mammalian cells, GTP cyclohydrolase I is induced in parallel with NOS (for review, see Werner et al. 1998). In contrast, mRNA levels of GTP cyclohydrolase I mRNA decreased during starvation of Physarum (Fig. 3A). Intracellular H4-biopterin amounts also declined, from 1.7 nmol/mg of protein to about 500 pmol/mg (Fig. 3B). However, even this final amount is higher than the maximal level reported for induced cytokine-activated mammalian cells (Werner et al. 1998) and, thus, is more than sufficient for NO synthesis.
L-N6–(1-iminoethyl)-lysine (NIL), a selective inhibitor of inducible NOS (Moore et al. 1994), efficiently inhibited NOS activity (Fig. 3B) without affecting NOS mRNA expression or pteridine biosynthesis (Fig. 3A,B). The concentration causing half-maximal inhibition was 200 μM both for NOS activity in homogenates and accumulation of nitrite plus nitrate in supernatants from intact macroplasmodia (data not shown).
Glucose is a repressor of NOS induction and of sporulation
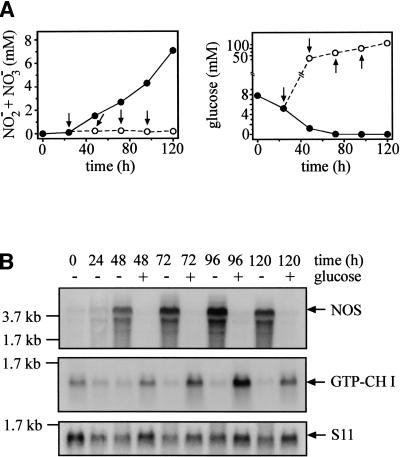
For the Physarum isolate used in this study, starting starvation to obtain sporulation competence in presence of reduced nutrients (see Materials and Methods), instead of completely removing nutrients (Daniel and Rusch 1962a), turned out to yield optimal sporulation ratios. Nitrite plus nitrate levels in supernatants started to increase ∼30 h after beginning starvation and reached 7 mM at 120 h in the experiment shown in Figure 4. The 7.8-mM glucose present at the beginning of sporulation experiments was completely consumed within 48 to 72 h (Fig. 4A). Maintaining glucose levels at 62.4 mM, the usual concentration for promoting growth, effectively inhibited NOS activity (Fig. 4A) by suppressing induction of NOS mRNA expression (Fig. 4B). In contrast, GTP cyclohydrolase I mRNA expression was strongly induced by glucose (Fig. 4B).
Figure 4.
Effects of glucose on nitric oxide synthase (NOS) and GTP cyclohydrolase I expression. (A) Accumulation of nitrite plus nitrate in cultures supplemented with glucose (left panel) and glucose levels (right panel) during the starvation period inducing sporulation competence. Macroplasmodia were inoculated in sporulation medium containing 7.8 mM glucose. At the time points labeled by arrows (24, 48, 72, 96 h), glucose was added to half of the cultures to 62.4 mM, the concentration used in growth medium. At 0 and 24 h as well as 24 h after each glucose addition (48, 72, 96, 120 h), nitrite plus nitrate (left panel) and glucose (right panel) were measured in supernatants from both control and glucose-supplemented cultures. Data from control cultures are shown in filled circles with solid lines; data from glucose-treated cultures, in open circles with dotted lines. Values are means of three independent cultures. (B) NOS and GTP cyclohydrolase I mRNA expression in starved or glucose-treated cultures. Macroplasmodia were harvested for RNA extraction at the times indicated. In the case of glucose treatment, RNA was extracted 24 h after each glucose addition (compare with A). Northern blots were hybridized with probes for NOS, GTP cyclohydrolase I (GTP-CHI), and the ribosomal protein S11.
cGMP is the second messenger used for NO signaling in lig 1 mRNA expression and in sporulation of Physarum
We then were interested to see whether the strong induction of NOS during development of sporulation competence was functionally linked to sporulation and whether cGMP was used as a second messenger. We therefore studied the effect of different drugs interfering with NO/cGMP signaling on sporulation rates of macroplasmodia, as well as on lig1 mRNA expression. Lig1 is a recently discovered early gene expressed during phytochrome-controlled sporulation. Its expression level correlates positively with the probability to sporulate (Kroneder et al. 1999). In addition, we tested the effect of glucose, a physiological repressor of sporulation (Daniel and Rusch 1962a).
Macroplasmodia treated with NIL were no longer able to sporulate (Fig. 5). Also, lig1 mRNA expression was strongly reduced in NIL-treated plasmodia compared with untreated control plasmodia (Fig. 5). Macroscopically, NIL-treated cultures looked identical to untreated cultures, and activity of glucose-6-phosphate dehydrogenase remained unaffected by NIL, indicating that the lack of sporulation is not caused by possible toxic side effects of NIL (data not shown). Inhibition of sporulation was only observed if NIL was added at the beginning of starvation. Addition of the inhibitor at 45 h and 118 h (i.e., 2 h before exposing the cultures to the sporangia-inducing light stimulus) could not prevent sporulation (data not shown). Complete (100%) inhibition of sporulation was caused by 250 μM NIL. With 100 μM of NIL, three of 15 cultures (20%) sporulated. Aminoguanidine (1 mM), another NOS inhibitor preferentially affecting inducible NOS (Misko et al. 1993), also inhibited sporulation (two of 19 cultures [10%] sporulated).
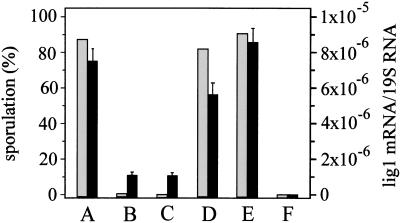
Figure 5.
Manipulation of sporulation and lig1 mRNA levels in Physarum polycephalum by drugs affecting nitric oxide (NO)/cGMP signaling and by glucose. Macroplasmodia were inoculated in sporulation medium without further supplements (A) or were supplemented with NIL (an inhibitor of iNOS, 250 μM) (B) or ODQ (an inhibitor of soluble, NO-sensitive guanylate cyclase, 150 μM) (C). 8-Bromo-cGMP (a cGMP substitute, 500 μM) was added to NIL-treated cultures (D) or to control cultures (E) at 118 h, which is 2 h before the light pulse triggering formation of mature sporangia. Some cultures were supplemented with glucose to 62.4 mM at 24, 48, 72, and 120 h (F). Sporulation (gray bars) is shown as percentage of the cultures sporulating (left y-axis), number of cultures tested were 72 (A), 36 (B), 35 (C), 16 (D), 21 (E), and 24 (F). Lig1 mRNA levels (black bars, right y-axis) were quantified using real-time PCR (Taqman technology). Values show lig1 mRNA levels at 8 h after setting the light pulse and are means from triplicate PCR reactions of one of two similar experiments (for details, see Materials and Methods).
Complete (100%) inhibition of sporulation and a strong decrease of lig1 mRNA levels were also caused by 150 μM ODQ, a selective inhibitor of NO-sensitive guanylate cyclase (Fig. 5; Garthwaite et al. 1995). In the presence of 100 μM ODQ, nine of 28 cultures (32%) sporulated.
8-bromo-cGMP (500 μM), a nonhydrolysable cGMP analog (Francis et al. 1988), added to NIL-treated macroplasmodia at 118 h of starvation (2 h before the light pulse) effectively reversed inhibition of sporulation by 250 μM NIL (Fig. 5). Also lig1 mRNA levels were clearly restored (Fig. 5). Adding only 250 μM of 8-bromo-cGMP to cultures treated with 250 μM NIL resulted in sporulation of four of 11 macroplasmodia (36%). 8-Bromo-cGMP could not trigger NIL-treated macroplasmodia to sporulate when added 12 h before or immediately at the beginning of the light pulse. Also, addition of 8-bromo-cGMP at the end of the 4 h of light treatment, or 6 h or 12 h thereafter, could not reverse inhibition of sporulation by NIL (data not shown). Furthermore, 8-bromo-cGMP had no effect on sporulation or lig1 mRNA expression of otherwise untreated macroplasmodia (Fig. 5) and could not accelerate sporulation when added on day 2 or 4 of the starvation period (data not shown).
Macroplasmodia were unable to sporulate and to express lig1 mRNA in presence of glucose (Fig. 5). The effects of 250 μM NIL, 150 μM ODQ, and 500 μM 8-bromo-cGMP in presence of 250 μM NIL and of glucose on sporulation (shown in Fig. 5) are highly significant as determined by Kruskal-Wallis analysis (calculated using SPSS for Windows 9.0.1): P is <1.0 × 10−14 for NIL versus controls and for ODQ versus controls, 4.43 × 10−9 for 8-bromo-cGMP plus NIL versus NIL alone, and 2.25 × 10−14 for glucose-treated versus starved plasmodia. Also, the attenuation of lig1 mRNA expression by NIL, ODQ, or glucose (P<0.0001), as well as the reversal of the NIL effect by 8-bromo-cGMP on lig1 mRNA expression (P<0.001) shown in Figure 5, were highly significant (Student's t-test).
Discussion
For mammals, the biological significance of endogenously formed NO as a signal molecule in vasodilation (Moncada and Higgs 1995), neurotransmission (Bredt 1996), gene regulation (Hentze and Kuhn 1996; Beck et al. 1999), and development (Peunova et al. 1996; Kim et al. 1999), as well as its janus-faced role as a cytotoxic/cytoprotective agent in host defense, is well established (Nathan 1997; Kim et al. 1999). In recent years, NOS has been discovered in a variety of nonmammalian animals, including birds (Lin et al. 1996), fish (Laing et al. 1996, 1999; Saeij et al. 2000), insects (for review, see Müller 1997), and molluscs (Korneev et al. 1998). There is also evidence that NOS is not restricted to the animal kingdom; NOS or NOS-like activity has been detected in slime molds (Werner-Felmayer et al. 1994; Ninnemann and Maier 1996; Tao et al. 1997), fungi (Ninnemann and Maier 1996), plants (Ninnemann and Maier 1996; Delledonne et al. 1998; Durner et al. 1998), and parasitic protists (Ghigo et al. 1995; Paveto et al. 1995; Basu et al. 1997). NOS also seems to occur in bacteria (Chen and Rosazza 1995; Morita et al. 1997; Sari et al. 1998). However, until now, full-length NOS sequences have only been reported for animals.
Here, two distinct cDNA clones encoding calcium-independent NOS with similar size and characteristics sharing 82% amino acid identity were isolated from Physarum; this is a similar degree of difference as exists between human and canine inducible NOSs. Parallel occurrence of two types of the same NOS isoenzyme has not been described for other species. Interestingly, the plasmodium-specific mRNA hapP from Physarum, which encodes for a protein of unknown function, occurs in two forms differing by 9.6% (predicted amino acid sequence), which were traced to two alleles of the same gene. Both heterozygous plasmodia expressing both forms and homozygous plasmodia were identified (Lépine et al. 1995). Although it remains to be determined whether the two NOS sequences observed in the diploid Physarum isolate used in this work are products from two differing alleles of the same gene, such a hypothesis is supported by the observation that the two NOS forms of Physarum are not differentially regulated during cell cycle, spherulation, and sporulation. Also, protein sequencing data indicated that NOS purified from Physarum macroplasmodia was a mixture of physnosa and physnosb. Neither cDNA analysis (this study) nor enzyme purification (Werner-Felmayer et al. 1994) yielded any hints of additional NOS isoenzymes, that is, calcium-dependent NOSs in Physarum.
The N terminus of the proteins encoded by the two Physarum mRNAs corresponded to position 75 of mammalian inducible NOSs. Still, it extended 30 amino acids upstream of the fully conserved zinc binding motif (Raman et al. 1998; Fischmann et al. 1999). As can be concluded from investigations on the oxygenase domain of mammalian inducible NOS, where catalytic properties of deletion mutants lacking amino acids 1–65 were equivalent to the wild-type protein (Ghosh et al. 1997), Physarum NOS contains all sequence sections essential for function.
Phylogenetic tree analysis places Physarum NOSs on an individual and low branch distant from mollusc, insect, bird, and mammalian NOSs. In fact, B. subtilis YflM, a characterized sequence homologous to the oxygenase domain of mammalian NOSs (Yamamoto et al. 1997), and the same region of Physarum NOSs are comparably distant from NOSs of other species. However, in line with our finding that the biochemistry of Physarum NOS is strikingly similar to that of mammalian inducible NOS (Werner-Felmayer et al. 1994), motifs and residues essential for NOS function are highly conserved in Physarum NOS despite a comparatively small overall homology to NOS from other species. Data from molecular phylogenetic analysis group Physarum and other Mycetozoa, including D. discoideum among the eukaryote crown taxa between fungi and green plants (Baldauf and Doolittle 1997). Thus, the Physarum NOS sequences might provide a link to a still hypothetical NOS from green plants, in which endogenous NO has been identified as a signal in host defense (Delledonne et al. 1998; Durner et al. 1998; Hausladen and Stamler 1998).
In D. discoideum (Tao et al. 1997), as well as in Neurospora crassa (Ninnemann and Maier 1996), endogenous NO and NO gas in Dictyostelium or sodium nitroprusside in Neurospora inhibited differentiation. In contrast, NO is a differentiation-promoting signal in nerve growth factor–induced differentiation of PC12 cells into neurons (Peunova and Enikolopov 1995). In Physarum, we found that NO is formed during differentiation of macroplasmodia into the sporulation competent state, a process that includes growth arrest, condensation of cellular material, a mitosis, migration, and development of light-sensitivity. The light pulse triggering sporulation is processed by a phytochrome-type photoreceptor (Starostzik and Marwan 1995). Stimulation of this photoreceptor induces expression of several genes, of which one, lig1, has been characterized recently (Kroneder et al. 1999). Thus, we performed experiments to see if the NO was playing a signaling role and checked for a possible link to lig1 expression. We manipulated NOS activity and endogenous cGMP levels during this differentiation process using NIL, an inhibitor of inducible NOS, by ODQ, a selective inhibitor of NO-sensitive soluble guanylate cyclase; by 8-bromo-cGMP, a nonhydrolysable analog of cGMP; and by glucose, a physiological repressor of Physarum sporulation. A schematic summary of our results and conclusions is shown in Figure 6.
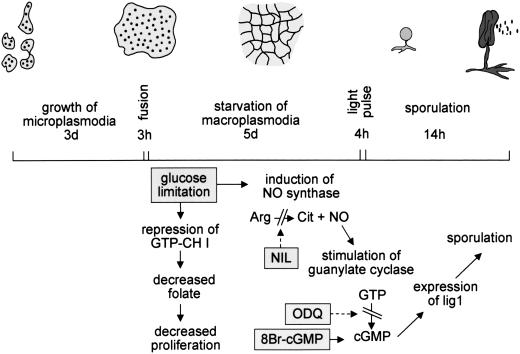
Figure 6.
Schematic presentation of morphological and biochemical changes during sporulation of Physarum polycephalum. Starvation of macroplasmodia in the dark for 5 d induces sporulation competence. During this phase, the plasmodium develops into a net of condensed cellular matrix. Biochemically, this phase is associated with glucose limitation which caused induction of nitric oxide synthase (NOS) and repression of GTP cyclohydrolase I (GTP-CH I). Although intracellular H4-biopterin levels were still high enough to support NOS activity, this repression of GTP cyclohydrolase I might affect proliferation via decreasing folate availability. Having attained sporulation competence at the end of the 5 d starvation period, a light pulse then induces formation of sporangia, a process paralleled by phytochrome-dependent induction of lig1. NIL and ODQ interfere with NO/cGMP signaling by inhibiting NOS and NO-sensitive guanylate cyclase, respectively. NIL and ODQ as well as glucose, which prevented NOS induction, suppressed both sporulation and lig1 expression. 8-Bromo-cGMP counteracted the inhibitory effect of NIL on sporulation and lig1 expression.
Macroplasmodia are obtained by fusion of microplasmodia grown for 3 d in the dark and under optimal growth-promoting conditions. Under limiting glucose in the dark, these macroplasmodia develop from a giant cell into a characteristic plasmodial net within 5 d. Sporangia formation is accomplished 14 to 16 h after setting the essential light pulse. Refeeding with glucose, but not with other nutrients such as tryptone or tryptone plus yeast extract has been shown to inhibit sporulation (Daniel and Rusch 1962a). As we found here, NOS mRNA expression and activity were inversely linked, and GTP cyclohydrolase I mRNA expression directly linked, to the available glucose concentration. Consumption of glucose during the 5-d starvation period that induces sporulation competence resulted in strongly up-regulated NOS and down-regulated GTP cyclohydrolase I. In Physarum, GTP cyclohydrolase I is essential for biosynthesis not only of H4-biopterin but also of folic acid (Werner-Felmayer et al. 1994). Growth arrest and physical condensation of macroplasmodia are prerequisites for obtaining sporulation competence (Daniel and Rusch 1962a). It is therefore conceivable that GTP cyclohydrolase I is down-regulated to limit the supply of a cofactor essential for cell proliferation (i.e., folic acid), while still providing enough H4-biopterin for NOS to function (see Results). Only starved but not glucose-fed plasmodia expressed lig1 mRNA and sporulated in response to the light pulse. Inhibition of endogenous NO or cGMP formation throughout the starvation period impaired light-induced lig1 mRNA expression and prevented sporulation. When added 2 h before the light pulse, 8-bromo-cGMP restored the capacity of NIL-treated macroplasmodia to express lig1 mRNA and to sporulate. These findings support the hypothesis that NO-mediated cGMP formation is functionally linked to processing the light signal, which then triggers phytochrome-induced lig1 expression and sporulation.
In higher plants, cGMP mediates phototransduction by modulating gene expression of phytochromes (Bowler et al. 1994), prominent partners in a complicated network of photoreceptors (for review, see Mustilli and Bowler 1997). In a not yet understood way, sugar-dependent signaling pathways interact with phytochrome-regulated pathways (Mustilli and Bowler 1997). Future work will analyze how NO/cGMP contribute to phytochrome-dependent phototransduction in sporulation of Physarum, a process that involves NOS induction by glucose limitation and phytochrome-dependent expression of lig1.
Materials and methods
Conditions for growing Physarum vegetative states and for inducing sporulation
Microplasmodia of P. polycephalum strain M3b, a Wis 1 isolate, were cultured in submersed shake-culture in semi-defined medium (Daniel and Baldwin 1964), supplemented with 0.013% (w/v) hemoglobin instead of hematin. Macroplasmodia were prepared by coalescence of exponentially growing microplasmodia and were cultured in the same medium as microplasmodia but grown in plastic dishes on filter paper supported by glass beads. Both micro- and macroplasmodia were grown in the dark at 25°C.
Sporulation is achieved by reduction of nutrients at low pH and strictly depends on addition of niacin and a daylight pulse (Daniel and Rusch 1962a,b). As a modification, macroplasmodia prepared by coalescence (3 h) from microplasmodia grown for 72 h (Affolter et al. 1979) were directly put into sporulation medium, a salt solution of pH 4.6 supplemented with peptone from meat (10 g/L), yeast extract (1.5 g/L), and glucose (7.8 mM), as well as 0.01% (w/v) nicotinamide, 0.1% (w/v) CaCO3, and 0.14 mM CuCl2. In some experiments, glucose was added to 62.4 mM, the growth medium concentration, at 24, 48, 72 and 96 h of the starvation period. After 120 h in the dark at 25°C, cultures were exposed to daylight (OSRAM L8W/12) for 4 h. About 12 to 14 h after the light treatment, sporangia were formed and differentiation was completed when the spores had developed their characteristic dark melanin color.
Treatment of macroplasmodia with NOS inhibitors, ODQ and 8-bromo-cGMP
NIL (0.1 to 5 mM), aminoguanidine (1 mM), and ODQ (100–250 μM), all obtained from Alexis Corporation, were added at time 0 of the starvation phase necessary for inducing sporulation competence. 8-bromo-cGMP (10 to 500 μM) (Sigma) was added 2 h before the light pulse, that is, 118 h after starting starvation and addition of NIL. In some experiments, 8-bromo-cGMP was added 12 h before, at the beginning of, and at the end (4 h) of the light treatment, and 6 and 12 h afterwards.
Purification of NOS
To obtain enough purified Physarum NOS for sequencing, the purification protocol detailed in Werner-Felmayer et al. (1994) was scaled up and modified as follows: Microplasmodia were disintegrated by means of a French press (16,000 psi). A 50,000 g supernatant was prepared by centrifugation and precipitated by 50% (w/v) ammonium sulfate. The precipitate was dissolved in 50 mM Tris.HCl (pH 8.0), containing 10% (v/v) glycerol, 1 mM phenylmethanesulphonyl fluoride, 5 mM 1,4-dithioerythritol, 10 μM H4-biopterin, 50 μM L-arginine, and 10 mM 2-mercaptoethanol, subjected to affinity chromatography with 2‘-5′-ADP-Sepharose 4B (Amersham Pharmacia Biotech) and eluted with 10 mM NADPH. This was followed by gelfiltration with Sephacryl S-300 HR (28 × 540 mm, Amersham Pharmacia Biotech) and ion exchange chromatography with Q-Sepharose (fast flow, 10 × 100 mm, Amersham Pharmacia Biotech). The protein was eluted with a gradient of 0 to 0.5 M NaCl. Active fractions were concentrated with Amicon 100 filters (Amicon Inc.), supplemented with 25% (v/v) glycerol and stored at −80°C. The purification factor achieved by this modified protocol was about 16,000-fold. In one run, about 15 μg (∼125 pmol) of NOS could be isolated from 1200 mL of microplasmodia, corresponding to ∼10 g of total protein. As observed before (Werner-Felmayer et al. 1994), the protein ran as a double band in 7.5% denaturing polyacrylamide gels. The size of the more abundant larger band was estimated to be approximately 120 kD from comparison with a 116-kD protein marker (Sigma C3312, 29 to 205 kD). The smaller band was most likely a degradation product of the 120-kD band, because it occured in varying amounts in different preparations and because two internal peptide sequences were identical to those identified for the 120-kD band (WITA sequencing service). Furthermore, the two bands showed an identical isoelectric point of 6.6, as determined by isoelectric focusing with a pH gradient of 3 to 10. A similar double band, in which the smaller band turned out to be a proteolytic degradation fragment from the intact enzyme, was also observed for inducible NOS from murine macrophages (Vodovotz et al. 1995). From the 120-kD band, eight internal peptide sequences were obtained from custom sequencing at the Harvard Microchemistry Facility (Harvard University). Four of these could be unambigously aligned to NOS consensus sequences from other species and were therefore used for primer design. The N terminus of Physarum NOS is blocked, and therefore, no N-terminal sequence could be identified. The purified protein was subjected to Western blot analysis, according to standard procedures using the ECL Plus detection system (Amersham Pharmacia Biotech). Antisera raised to mouse inducible NOS (a kind gift from Q. Xie and C. Nathan, Cornell University), recombinant neuronal NOS from rat, and recombinant endothelial NOS from man (kindly provided by B. Mayer, University of Graz) were tested, and cell extracts from interferon-γ/lipopolysaccharide-treated RAW264.7 mouse macrophages, recombinant neuronal NOS from rat, or recombinant human endothelial NOS were used as positive controls.
Cloning of Physarum NOS
To obtain poly-A+ RNA, S-phase macroplasmodia were ground in liquid nitrogen, total RNA was isolated using the RNeasy Plant Mini kit from Qiagen, and mRNA was isolated using the Oligotex mRNA kit from Qiagen. A library was produced by ligation of oligo-(dT)-primed cDNA into the Lambda ZapII vector (digested with Eco RI and dephosphorylated) using the Gigapack II packaging kit from Stratagene.
The amplified library had a titer of 2.7 × 109 plaque-forming units/mL. For screening, a NOS-specific probe was generated by PCR using degenerate primers. The primers that succeeded were directed to the nucleotide sequence deduced from one of the internal peptide sequences identified from purified NOS (sense primer termed pnos30B) and to a consensus sequence from all available NOS sequences (antisense primer termed pnos31D). Primer pnos30B was designed for the nucleotide sequence of peptide P6 (TGTYFQTIEEL) in the sense direction and had the sequence 5′-ACIGGIACNTAYTTCCAAAC-3′. Primer pnos31D bound to the nucleotide sequence corresponding to the consensus amino acid sequence GWYMGT in the antisense direction and had the sequence 5′-GTGTCCATR WACCATCC-3′. PCR conditions were 40 cycles of denaturation at 94°C (30 sec), annealing at 50°C (1 min), and extension at 72°C (1 min), with an initial step at 95°C for 10 min to activate AmpliTaq Gold DNA polymerase (Perkin Elmer) and a final step of 7 min at 72°C using a GeneAmp9600 PCR instrument from Perkin Elmer. Reverse transcribed (SuperScript RNase H− reverse transcriptase, Life Technologies) poly-A+ RNA (500 ng in a final volume of 50 μL) was used for PCR, which was performed in presence of 5% (v/v) of dimethylsulfoxide. A 600-bp fragment was amplified, and sequence analysis clearly showed that it was a partial NOS sequence. Using this probe, two types of Physarum NOS were isolated and termed physnosa and physnosb (see Results). The abundance of Physarum NOS in the cDNA library was about 1 in 2 × 105 clones, as estimated from various rounds of screening. As it turned out, the 600-bp fragment was part of the physnosb type that cross-reacted with physnosa. Nucleotide sequencing and primer synthesis were performed by custom service (Microsynth). Analysis showed that the NOS probe bound to a region from nucleotides 587 to 1188, corresponding to amino acids 91 to 291 of the physnosa clone.
Sequence analysis
Sequences were analyzed using the Wisconsin Sequence Analysis Package version 9.1 from the Genetics Computer Group.
Functional expression of Physarum NOS
The physnosa cDNA was cloned into the pFastBac1 vector (GIBCO BRL) using the SalI and XbaI restriction sites. An optimized Kozak sequence (GCCATGG) was introduced by PCR-based mutagenesis (QuickChange, Stratagene). The plasmid was then transformed into baculovirus using the Bac-to-Bac expression system (GIBCO BRL), and the transposition was confirmed by PCR. Sf9 cells grown in TC100 (Sigma) were transfected with baculovirus, according to the manufacturer’s instructions. After 72 h, cells were harvested and assayed for NOS activity by the 3H-citrulline assay, essentially as outlined in Werner-Felmayer et al. (1994).
Northern blot analysis
Total RNA was isolated from macroplasmodia at different times during sporulation experiments and on different treatments. Samples frozen in liquid nitrogen were ground and homogenized using the RNeasy Plant Mini kit from Qiagen. Ten or 20 μg of total RNA were resolved in 1% agarose/6% formaldehyde gels, vacuum-blotted onto Duralon-UV nylon membranes (Stratagene), and cross-linked using ultraviolet irradiation (Stratalinker, Stratagene). Blots were hybridized overnight with 106 cpm/mL of 32P-dCTP-labeled probes at 65°C, according to standard protocols, and exposed to PhosphoImager Screens.
For detection of NOS mRNA, blots were hybridized with the 600-bp PCR fragment generated as detailed above. To discriminate between physnosa and physnosb, PCR fragments from the untranslated 3′-ends of the two types of Physarum clones, which differed considerably, were generated. Primers physnosa1 (5′-CCATCCAAGAAAGCCGATGC-3′, sense) and physnosa2 (5′-CAGGAATTCCGGTGGTACAG-3′, antisense) amplified a 157-bp fragment of physnosa. Primers physnosb1 (5′-CCTAAA TAGGTTCGGCTAGGC-3′, sense) and physnosb2 (5′-CTTTG CAAGGTCTGGAGTGG-3′, antisense) amplified 168 bp of physnosb. Two nanograms of plasmid DNA from physnosa and physnosb, respectively, were used in a final volume of 50 μL. PCR conditions were: 2 min at 94°C, 30 cycles of 30 sec at 94°C, 1 min at 57°C, 1 min at 72°C, and a final step of 7 min at 72°C.
GTP cyclohydrolase I mRNA was detected by a PCR-generated 282-bp probe that was obtained using consensus primers 3 (sense) and 8 (antisense) for GTP cyclohydrolase I from different species (Maier et al. 1995). PCR was done by 30 cycles of 30 sec at 94°C, 1 min at 55°C, and 1 min at 72°C, with an initial denaturation step at 94°C for 2 min. Having verified that the PCR fragment is homologous to GTP cyclohydrolase I from other species, this probe was also used for cloning the full-length Physarum GTP cyclohydrolase I sequence from the cDNA library that was functionally active as was verified by recombinant expression in baculovirus-transfected Sf9 cells (Golderer et al. 2001). The sequence was deposited to the GenBank (accession no. AF177165).
For use as a control, a 500-bp cDNA fragment with high homology to ribosomal protein S11 (63% amino acid sequence identity with S11 from Xenopus laevis [GenBank accession no. X78805]) was amplified. The putative Physarum S11 sequence is available from GenBank (accession no. AF177283).
Quantification of lig1 mRNA levels using real-time PCR
Total RNA was isolated from macroplasmodia at the beginning and at the end of the 4-h light treatment and 4, 8, and 12 h thereafter using the RNeasy Plant Mini kit from Qiagen (see above). Because of condensation of the plasmodium, cellular material is very limited at this stage of the experiment. It therefore was pooled from 5 plasmodia treated in parallel. Further, 5 to 10 parallel cultures were used to determine sporulation rates. Random hexamer-primed cDNAs were prepared from 500 ng of total RNA using Superscript II RNase H− reverse transcriptase. Real-time PCR analysis was performed using the Taqman technology (AbiPrism 7700 sequence detector, Applied Biosystems) and the Brilliant Quantitative PCR Core Reagent kit from Stratagene. Probes (5′-FAM/3′-TAMRA label) and primers, both synthesized by Microsynth, were selected using the Primer Express software (Applied Biosystems). Sequences (5′-3′ direction) of probes and primers were as follows: lig1 probe, TGGCGCA AGCGTGAATCTATTATTCGA; lig1–136F (sense), GATGGC ATGCAGGTTTGGA; lig1–212R (antisense), CCATTGTTCA AACTCTCTATTCTGTACTC; 19 S probe, CCCGTGTTGAG TCAAATTAAGCCGCA; 19S-1273F (sense), ACGGAAGGG CACACAAAGCG; 19S-1350R (antisense), CGTGTATCCGG ACCTGGTG. Lig1 mRNA levels were related to 19 S ribosomal RNA levels. In accordance with previous work (Kroneder et al. 1999), lig1 mRNA levels increased up to 50-fold at 8 and 12 h after setting the light pulse as compared to the levels found at the beginning of the light treatment.
Determination of intracellular H4-biopterin and of nitrite plus nitrate in supernatants
Intracellular H4-biopterin was quantified as detailed before (Werner-Felmayer et al. 1994; Werner et al. 1997) by high performance liquid chromatography (HPLC) after oxidation with iodine in acidic (total biopterin) or alkaline (7,8-dihydrobiopterin plus biopterin) media. The amount of H4-biopterin is given in pmol per mg of protein. Nitrite plus nitrate was determined in supernatants as outlined previously (Werner-Felmayer et al. 1994). Briefly, samples were applied to reversed phase HPLC and eluted with 5% NH4Cl (pH 7.0). Nitrate was reduced to nitrite using a cadmium reactor. Nitrite was quantified by measuring ultraviolet absorption at 546 nm after post-column mixing with the Griess-Ilosvay reagent from Merck.
Determination of glucose
Glucose in supernatants at different times of sporulation experiments was determined using the glucose/GOD-Perid kit from Boehringer Mannheim (Roche Diagnostics).
Acknowledgments
We are indebted to Christine Heufler-Tiefenthaler, Department of Dermatology, University of Innsbruck, for her help with preparation of the Physarum cDNA library. Thanks are also owed to Emanuela Felley-Bosco, Institut de Pharmacologie et de Toxicologie, Université de Lausanne, for discussions on evolutionary tree analysis; to Bernd Mayer, Institut für Pharmakogie und Toxikologie, University of Graz, for his generous help on various aspects of this work; and to Ben Hemmens, Graz, for critical reading of the manuscript. The excellent technical assistance of Renate Kaus, Petra Höfler, Anabella Weisskopf, and Bettina Fritz is gratefully acknowledged. This work was supported by the Austrian Funds “Zur Förderung der wissenschaftlichen Forschung”, project P13580-MOB.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gabriele.werner-felmayer@uibk.ac.at; FAX 43-512-507-2865.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.890501.
References
- Affolter HU, Behrens K, Seebeck T, Braun R. Large scale isolation of ribosomal DNA from giant surface cultures of Physarum polycephalum. FEBS Lett. 1979;107:340–342. doi: 10.1016/0014-5793(79)80403-2. [DOI] [PubMed] [Google Scholar]
- Bailey J. Plasmodium development in the myxomycete Physarum polycephalum: Genetic control and cellular events. Microbiology. 1995;141:2355–2365. doi: 10.1099/13500872-141-10-2355. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Doolittle WF. Origin and evolution of the slime molds (Mycetozoa) Proc Natl Acad Sci. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu NK, Kole L, Ghosh A, Das PK. Isolation of a nitric oxide synthase from the protozoan parasite, Leishmania donovani. FEMS Microbiol Lett. 1997;156:43–47. doi: 10.1111/j.1574-6968.1997.tb12703.x. [DOI] [PubMed] [Google Scholar]
- Beck KF, Eberhardt W, Frank S, Huviler A, Messmer UK, Mühl H, Pfeilschifter J. Inducible NO synthase: Role in cellular signalling. J Exp Biol. 1999;202:645–653. doi: 10.1242/jeb.202.6.645. [DOI] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua NH. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Bredt DS. Targeting nitric oxide to its targets. Proc Soc Exp Biol Med. 1996;211:41–48. doi: 10.3181/00379727-211-43950f. [DOI] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Chen PF, Tsai AL, Wu KK. Cystein 184 of endothelial nitric oxide synthase is involved in heme coordination and catalytic activity. J Biol Chem. 1994;269:25062–25066. [PubMed] [Google Scholar]
- Chen Y, Rosazza JPN. Purification and characterization of nitric oxide synthase (NOSNoc) from a Nocardia species. J Bacteriol. 1995;177:5122–5128. doi: 10.1128/jb.177.17.5122-5128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Martin E, Xie QW, Sassa S, Nathan C. Inducible nitric oxide synthase: Identification of amino acid residues essential for dimerization and binding of tetrahydrobiopterin. Proc Natl Acad Sci. 1995;92:11514–11518. doi: 10.1073/pnas.92.25.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- Daniel JW, Rusch HP. Method for inducing sporulation of pure cultures of the myxomycete Physarum polycephalum. J Bacteriol. 1962a;83:234–240. doi: 10.1128/jb.83.2.234-240.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Niacin requirement for sporulation of Physarum polycephalum. J Bacteriol. 1962b;83:1244–1250. doi: 10.1128/jb.83.6.1244-1250.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature. 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci. 1998;95:10328–10333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, et al. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nature Struct Biol. 1999;6:233–242. doi: 10.1038/6675. [DOI] [PubMed] [Google Scholar]
- Francis SH, Noblett BD, Todd BW, Wells JN, Corbin JD. Relaxation of vascular and tracheal smooth muscle by cyclic nucleotide analogs that preferentially activate purified cGMP-dependent protein kinase. Mol Pharmacol. 1988;34:506–517. [PubMed] [Google Scholar]
- Garcia-Cardena G, Martasek P, Masters BS, Skidd PM, Couet J, Li S, Lisanti MP, Sessa WC. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin binding domain in vivo. J Biol Chem. 1997;272:25437–25440. doi: 10.1074/jbc.272.41.25437. [DOI] [PubMed] [Google Scholar]
- Garthwaite J, Southam E, Boulton CL, Nielsen EB, Schmidt K, Mayer B. Potent and selective inhibition of nitric oxide-sensitive guanylyl cyclase by 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one. Mol Pharmacol. 1995;48:184–188. [PubMed] [Google Scholar]
- Ghigo D, Todde R, Ginsburg H, Costamagna C, Gautret P, Bussolino F, Ulliers D, Giribaldi G, Deharo E, Gabrielli G, et al. Erythrocyte stages of Plasmodium falciparum show a high nitric oxide synthase (NOS) activity and release an NOS-inducing soluble factor. J Exp Med. 1995;182:677–688. doi: 10.1084/jem.182.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh DK, Wu C, Pitters E, Moloney M, Werner ER, Mayer B, Stuehr DJ. Characterization of the inducible nitric oxide synthase oxygenase domain identifies a 49 amino acid segment required for subunit dimerization and tetrahydrobiopterin interaction. Biochemistry. 1997;36:10609–10619. doi: 10.1021/bi9702290. [DOI] [PubMed] [Google Scholar]
- Golderer G, Werner ER, Heufler C, Strohmaier W, Gröbner P, Werner-Felmayer G. GT cyclohydrolase I mRNA: Novel splice variants in the slime mold Physarum polycephalum and in human monocytes (THP-1) indicate conservation of mRNA processing. Biochem J. 2001;355:499–507. doi: 10.1042/0264-6021:3550499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A, Stamler JS. Nitric oxide in plant immunity. Proc Natl Acad Sci. 1998;95:10345–10347. doi: 10.1073/pnas.95.18.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi Y, Ueda T. Fragmentation of the plasmodium into equally sized pieces by low temperatures in the true slime mold Physarum polycephalum: A new morphogenesis. Protoplasma. 1999;206:131–136. [Google Scholar]
- Kim YM, Bombeck CA, Billiar TR. Nitric oxide as a bifunctional regulator of apoptosis. Circ Res. 1999;84:253–256. doi: 10.1161/01.res.84.3.253. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press; 1983. [Google Scholar]
- Korneev SA, Piper MR, Picot J, Phillips R, Korneeva EI, O'Shea M. Molecular characterization of NOS in a mollusc: Expression in a giant modulatory neuron. J Neurobiol. 1998;35:65–76. doi: 10.1002/(sici)1097-4695(199804)35:1<65::aid-neu6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Kroneder R, Cashmore AR, Marwan W. Phytochrome-induced expression of lig1, a homologue of the fission yeast cell-cycle checkpoint gene hus1, is associated with the developmental switch in Physarum polycephalum plasmodia. Curr Genet. 1999;36:86–93. doi: 10.1007/s002940050476. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Grabowski PS, Belosevic M, Secombes CJ. A partial sequence for nitric oxide synthase from a goldfish (Carassius auratus) macrophage cell line. Immunol Cell Biol. 1996;74:374–379. doi: 10.1038/icb.1996.65. [DOI] [PubMed] [Google Scholar]
- Laing KJ, Hardie LJ, Aartsen W, Grabowski PS, Secombes CJ. Expression of an inducible nitric oxide synthase gene in rainbow trout Oncorhynchus mykiss. Dev Comp Immunol. 1999;23:71–85. doi: 10.1016/s0145-305x(98)00036-6. [DOI] [PubMed] [Google Scholar]
- Lépine G, Laroch A, Lemieux G, Pallotta D. The two alleles of the hapP gene in Physarum polycephalum code for different proteins. Biochim Biophys Acta. 1995;1264:271–274. doi: 10.1016/0167-4781(95)00191-3. [DOI] [PubMed] [Google Scholar]
- Lin AW, Chang CC, McCormick CC. Molecular cloning and expression of an avian macrophage nitric-oxide synthase cDNA and the analysis of the genomic 5′-flanking region. J Biol Chem. 1996;271:11911–11919. doi: 10.1074/jbc.271.20.11911. [DOI] [PubMed] [Google Scholar]
- Maier J, Witter K, Gütlich M, Ziegler I, Werner T, Ninnemann H. Homology cloning of GTP-cyclohydrolase I from various unrelated eukaryotes by reverse-transcription polymerase chain reaction using a general set of degenerate primers. Biochem Biophys Res Commun. 1995;212:705–711. doi: 10.1006/bbrc.1995.2026. [DOI] [PubMed] [Google Scholar]
- Mayer B, Hemmens B. Biosynthesis and action of nitric oxide in mammalian cells. Trends Biochem Sci. 1997;22:477–481. doi: 10.1016/s0968-0004(97)01147-x. [DOI] [PubMed] [Google Scholar]
- Misko TP, Moore WM, Kasten TP, Nickols GA, Corbett JA, Tilton RG, McDaniel ML, Williamson JR, Currie MG. Selective inhibition of the inducible nitric oxide synthase by aminoguanidine. Eur J Pharmacol. 1993;233:119–125. doi: 10.1016/0014-2999(93)90357-n. [DOI] [PubMed] [Google Scholar]
- Moncada S, Higgs EA. Molecular mechanisms and therapeutic strategies related to nitric oxide. FASEB J. 1995;9:1319–1330. [PubMed] [Google Scholar]
- Moore WM, Webber RK, Jerome GM, Tjoeng FS, Misko TP, Currie MG. L-N6–(1-iminoethyl)lysine: A selective inhibitor of inducible nitric oxide synthase. J Med Chem. 1994;37:3886–3888. doi: 10.1021/jm00049a007. [DOI] [PubMed] [Google Scholar]
- Morita H, Yoshikawa H, Sakata R, Nagata Y, Tanaka H. Synthesis of nitric oxide from the two equivalent guanidino nitrogens of L-arginine by Lactobacillus fermentum. J Bacteriol. 1997;179:7812–7815. doi: 10.1128/jb.179.24.7812-7815.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. The nitric oxide system in insects. Prog Neurobiol. 1997;51:363–381. doi: 10.1016/s0301-0082(96)00067-6. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Bowler C. Tuning in to the signals controlling photoregulated gene expression in plants. EMBO J. 1997;19:5801–5806. doi: 10.1038/sj.emboj.7590554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Inducible nitric oxide synthase: What difference does it make? J Clin Invest. 1997;100:2417–2423. doi: 10.1172/JCI119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninnemann H, Maier J. Indications of the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem Photobiol. 1996;64:393–398. doi: 10.1111/j.1751-1097.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- Paveto C, Pereira C, Espinosa J, Montagna AE, Farber M, Esteva M, Flawiá MM, Torres HN. The nitric oxide transduction pathway in Trypanosoma cruzi. J Biol Chem. 1995;270:16576–16579. doi: 10.1074/jbc.270.28.16576. [DOI] [PubMed] [Google Scholar]
- Peunova N, Enikolopov G. Nitric oxide triggers a switch to growth arrest during differentiation of neuronal cells. Nature. 1995;375:68–73. doi: 10.1038/375068a0. [DOI] [PubMed] [Google Scholar]
- Peunova N, Kuzin B, Roberts I, O'Kane C, Enikolopov G. Nitric oxide, cell multiplication, and cell survival. Cold Spring Harbor Symp Quant Biol. 1996;61:417–426. [PubMed] [Google Scholar]
- Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL. Crystal structure of constitutive endothelial nitric oxide synthase: A paradigm for pterin function involving a novel metal center. Cell. 1998;95:939–950. doi: 10.1016/s0092-8674(00)81718-3. [DOI] [PubMed] [Google Scholar]
- Raub TJ, Aldrich HC. Sporangia, spherules, and microcysts. In: Aldrich HC, Daniel JW, editors. Cell biology of Physarum and Didymium. New York: Academic Press; 1982. pp. 21–75. [Google Scholar]
- Saeij JP, Stet RJ, Groeneveld A, Verburg-van Kemenade LB, van Muiswinkel WB, Wiegertjes GF. Molecular and functional characterization of a fish inducible-type nitric oxide synthase. Immunogenetics. 2000;51:339–346. doi: 10.1007/s002510050628. [DOI] [PubMed] [Google Scholar]
- Salerno, J.C., Harris, D.E., Irizarry, K., Patel, B., Morales, A.J., Smith, S.M.E., Martasek, P., Roman, L.J., Masters, B.S.S., Jones, C.L., et al. An autoinhibitory control element defines calcium-regulated isoforms of nitric oxide synthase. J. Biol. Chem. 272: 29769–29777. [DOI] [PubMed]
- Sari MA, Moali C, Boucher JL, Jaouen M, Mansuy D. Detection of a nitric oxide synthase possibly involved in the regulation of the Rhodococcus sp R312 nitrile hydratase. Biochem Biophys Res Commun. 1998;250:364–368. doi: 10.1006/bbrc.1998.9320. [DOI] [PubMed] [Google Scholar]
- Starostzik C, Marwan W. A photoreceptor with characteristics of phytochrome triggers sporulation in the true slime mold Physarum polycephalum. FEBS Lett. 1995;370:146–148. doi: 10.1016/0014-5793(95)00820-y. [DOI] [PubMed] [Google Scholar]
- Tao YP, Misko TP, Howlett AC, Klein C. Nitric oxide, an endogenous regulator of Dictyostelium discoideum differentiation. Development. 1997;124:3587–3595. doi: 10.1242/dev.124.18.3587. [DOI] [PubMed] [Google Scholar]
- Vodovotz Y, Russell D, Xie QW, Bogdan C, Nathan C. Vesicle membrane association of nitric oxide synthase in primary mouse macrophages. J Immunol. 1995;154:2914–2925. [PubMed] [Google Scholar]
- Werner ER, Wachter H, Werner-Felmayer G. Determination of tetrahydrobiopterin biosynthetic activities by high-performance liquid chromatography with fluorescence detection. Meth Enzymol. 1997;281:53–61. doi: 10.1016/s0076-6879(97)81008-7. [DOI] [PubMed] [Google Scholar]
- Werner ER, Werner-Felmayer G, Mayer B. Tetrahydrobiopterin, cytokines, and nitric oxide synthesis. Proc Soc Exp Biol Med. 1998;219:171–182. doi: 10.3181/00379727-219-44331. [DOI] [PubMed] [Google Scholar]
- Werner-Felmayer G, Golderer G, Werner ER, Gröbner P, Wachter H. Pteridine biosynthesis and nitric oxide synthase in Physarum polycephalum. Biochem J. 1994;304:105–111. doi: 10.1042/bj3040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RJ, Sauer HW. Developmental biology of slime molds: An overview. In: Aldrich HC, Daniel JW, editors. Cell biology of Physarum and Didymium. New York: Academic Press; 1982. pp. 3–20. [Google Scholar]
- Yamamoto H, Uchiyama S, Nugroho FA, Sekiguchi J. Cloning and sequencing of a 35.7 kb in the 70Ε-73Ε region of the Bacillus subtilis genome reveal genes for a new two-component system, three spore germination proteins, an iron uptake system and a general stress response protein. Gene. 1997;194:191–199. doi: 10.1016/s0378-1119(97)00130-3. [DOI] [PubMed] [Google Scholar]